

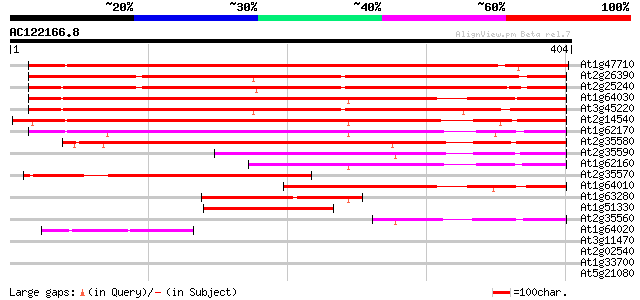
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.8 + phase: 0
(404 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g47710 serpin, putative 394 e-110
At2g26390 putative serpin 334 6e-92
At2g25240 putative serpin 331 5e-91
At1g64030 hypothetical protein 330 6e-91
At3g45220 serpin-like protein 328 4e-90
At2g14540 putative serpin 313 1e-85
At1g62170 hypothetical protein 302 2e-82
At2g35580 putative serpin 278 3e-75
At2g35590 putative serpin 206 2e-53
At1g62160 hypothetical protein 182 2e-46
At2g35570 putative serpin 172 3e-43
At1g64010 hypothetical protein 172 4e-43
At1g63280 hypothetical protein 132 3e-31
At1g51330 hypothetical protein 95 6e-20
At2g35560 putative serpin 82 5e-16
At1g64020 hypothetical protein 67 2e-11
At3g11470 unknown protein 34 0.16
At2g02540 unknown protein 31 1.1
At1g33700 unknown protein 30 3.1
At5g21080 putative protein 28 6.9
>At1g47710 serpin, putative
Length = 391
Score = 394 bits (1013), Expect = e-110
Identities = 206/393 (52%), Positives = 274/393 (69%), Gaps = 9/393 (2%)
Query: 14 IGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGPTQNQLLSFL 73
+ +S++ VS+N+ KH+++ N NV+FSP S+N VL +I AGS G T++Q+LSFL
Sbjct: 3 VRESISLQNQVSMNLAKHVITTVSQNS-NVIFSPASINVVLSIIAAGSAGATKDQILSFL 61
Query: 74 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKAT 133
+ ST L S S++VS+VL+DG+ GGP LS NG W+++SL +PSFKQL+ +KA
Sbjct: 62 KFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLSFKPSFKQLLEDSYKAA 121
Query: 134 LAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVWKRPF 193
DF +KA EVI EVN WA KET G I ++LP GS DS+T LIFANALYFKG W F
Sbjct: 122 SNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFKGTWNEKF 181
Query: 194 DTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGNDKRAFSIYFFLP 253
D S T++ +F LL+G V PFMTSK Q++S++DGFKVLGLPY QG DKR FS+YF+LP
Sbjct: 182 DESLTQEGEFHLLDGNKVTAPFMTSKKKQYVSAYDGFKVLGLPYLQGQDKRQFSMYFYLP 241
Query: 254 DEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTLPF 313
D +GLS L+DK+ S FL+ +PR QVKV +F IP+F SF +AS +L LGLT PF
Sbjct: 242 DANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVLKGLGLTSPF 301
Query: 314 S-KGGLTKMVDSP-ISQELSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPFR--LPPP 369
S + GLT+MV+SP + + L V++IF K+ IE+NEEGT AAA ++ G R L
Sbjct: 302 SGEEGLTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAA----ASAGVIKLRGLLMEE 357
Query: 370 PPIDFVADHPFLFLIREEFSGTILFVGKVVNPL 402
IDFVADHPFL ++ E +G +LF+G+VV+PL
Sbjct: 358 DEIDFVADHPFLLVVTENITGVVLFIGQVVDPL 390
>At2g26390 putative serpin
Length = 389
Score = 334 bits (856), Expect = 6e-92
Identities = 180/395 (45%), Positives = 256/395 (64%), Gaps = 18/395 (4%)
Query: 14 IGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGPTQNQLLSFL 73
+GKS+ N NV + K ++ N NVVFSP+S+N +L +I AGS T+ ++LSFL
Sbjct: 3 LGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILSFL 62
Query: 74 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKAT 133
S ST L ++ +++ G CLS +GVW+++S L+PSFK+L+ +KA+
Sbjct: 63 MSPSTDHLNAVLAKIADG----GTERSDLCLSTAHGVWIDKSSYLKPSFKELLENSYKAS 118
Query: 134 LAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSL-----TSLIFANALYFKGV 188
+ VDF K EVI EVN+WA+ T G I +L R D++ ++LI ANA+YFK
Sbjct: 119 CSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAVYFKAA 178
Query: 189 WKRPFDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGNDKRAFSI 248
W R FD TKD DF LL+G +VKVPFM S +Q++ +DGF+VL LPY + DKR FS+
Sbjct: 179 WSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKDQYLRGYDGFQVLRLPYVE--DKRHFSM 236
Query: 249 YFFLPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKLG 308
Y +LP++KDGL+AL++K++++ FL+ +P ++ V IP+ N SFE +ASE+L +G
Sbjct: 237 YIYLPNDKDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKASEVLKDMG 296
Query: 309 LTLPF-SKGGLTKMVDSPIS-QELSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPFRL 366
LT PF SKG LT+MVDSP + +L V+SI K+ IE++EEGT AAA + R
Sbjct: 297 LTSPFTSKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIMMPQCLMRN 356
Query: 367 PPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
P DFVADHPFLF +RE+ SG ILF+G+V++P
Sbjct: 357 P-----DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>At2g25240 putative serpin
Length = 385
Score = 331 bits (848), Expect = 5e-91
Identities = 182/392 (46%), Positives = 256/392 (64%), Gaps = 16/392 (4%)
Query: 14 IGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGPTQNQLLSFL 73
+GKS+ N +V + +TKH+++ N N+VFSP+S+N +L +I AGS T+ Q+LSFL
Sbjct: 3 LGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILSFL 61
Query: 74 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKAT 133
ST L + +Q++ G LS NGVW+++ L+ SFK L+ +KAT
Sbjct: 62 MLPSTDHLNLVLAQIIDG----GTEKSDLRLSIANGVWIDKFFSLKLSFKDLLENSYKAT 117
Query: 134 LAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTS--LIFANALYFKGVWKR 191
+ VDF +K EVI EVN WA T G I +L R S+D++ S L+ ANA+YFKG W
Sbjct: 118 CSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFKGAWSS 177
Query: 192 PFDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGNDKRAFSIYFF 251
FD + TK DF LL+G SVKVPFMT+ +Q++ S+DGFKVL LPY + D+R FS+Y +
Sbjct: 178 KFDANMTKKNDFHLLDGTSVKVPFMTNYEDQYLRSYDGFKVLRLPYIE--DQRQFSMYIY 235
Query: 252 LPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTL 311
LP++K+GL+ L++K+ S+ F + +P + + VG F IP+F SFE ASE+L +GLT
Sbjct: 236 LPNDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMGLTS 295
Query: 312 PFSK-GGLTKMVDSPIS-QELSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPFRLPPP 369
PF+ GGLT+MVDSP + +L V+SI K+ IE++EEGT AAA + G FR P
Sbjct: 296 PFNNGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVV-SCTSFRRNP- 353
Query: 370 PPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
DFVAD PFLF +RE+ SG ILF+G+V++P
Sbjct: 354 ---DFVADRPFLFTVREDKSGVILFMGQVLDP 382
>At1g64030 hypothetical protein
Length = 385
Score = 330 bits (847), Expect = 6e-91
Identities = 178/392 (45%), Positives = 253/392 (64%), Gaps = 27/392 (6%)
Query: 14 IGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGP-TQNQLLSF 72
+ +++ N T+V++ ++ H+LS+ + NV+FSP S+N+ + M AG G Q+LSF
Sbjct: 3 VREAMKNQTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQILSF 61
Query: 73 LQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKA 132
L+S S +LK++ +L S V +D + GGP ++ NG+W+++SLP P FK L FKA
Sbjct: 62 LRSSSIDELKTVFRELASVVYADRSATGGPKITAANGLWIDKSLPTDPKFKDLFENFFKA 121
Query: 133 TLAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVWKRP 192
VDF ++A+EV KEVN W T I DLLP GSV SLT+ I+ANAL FKG WKRP
Sbjct: 122 VYVPVDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWKRP 181
Query: 193 FDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGND--KRAFSIYF 250
F+ T+D DF L+NG SV VPFM+S NQ++ ++DGFKVL LPY++G+D R FS+YF
Sbjct: 182 FEKYYTRDNDFYLVNGTSVSVPFMSSYENQYVRAYDGFKVLRLPYQRGSDDTNRKFSMYF 241
Query: 251 FLPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKLGLT 310
+LPD+KDGL L++K+AS FL+ +P + ++ KF IP+F I F + +L++LG
Sbjct: 242 YLPDKKDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRLG-- 299
Query: 311 LPFSKGGLTKMVDSPISQELSVTSIFQKSFIELNEEGT-IAAATARGSTGGAAPFRLPPP 369
L S++ K+ +E++EEG AAATA G G + F + PP
Sbjct: 300 -------------------LRSMSMYHKACVEIDEEGAEAAAATADGDCGCSLDF-VEPP 339
Query: 370 PPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
IDFVADHPFLFLIREE +GT+LFVG++ +P
Sbjct: 340 KKIDFVADHPFLFLIREEKTGTVLFVGQIFDP 371
>At3g45220 serpin-like protein
Length = 393
Score = 328 bits (840), Expect = 4e-90
Identities = 174/397 (43%), Positives = 255/397 (63%), Gaps = 18/397 (4%)
Query: 14 IGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGPTQNQLLSFL 73
+GKS+ N T+V + + KH++ N N+VFSP+S+N +L +I AGS T+ Q+LSF+
Sbjct: 3 LGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILSFI 61
Query: 74 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKAT 133
S+ L ++ ++ VS L+DG LS GVW+++SL +PSFK L+ + AT
Sbjct: 62 MLPSSDYLNAVLAKTVSVALNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLENSYNAT 121
Query: 134 LAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSL--TSLIFANALYFKGVWKR 191
VDF K EVI EVN WA T G I ++L S+ ++ + LI ANA+YFKG W +
Sbjct: 122 CNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFKGAWSK 181
Query: 192 PFDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGNDKRAFSIYFF 251
FD TK YDF LL+G VKVPFMT+ Q++ +DGFKVL LPY + D+R F++Y +
Sbjct: 182 KFDAKLTKSYDFHLLDGTMVKVPFMTNYKKQYLEYYDGFKVLRLPYVE--DQRQFAMYIY 239
Query: 252 LPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTL 311
LP+++DGL L+++++S FL+ +PR ++ F IP+F SFE +AS++L ++GLTL
Sbjct: 240 LPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVLKEMGLTL 299
Query: 312 PFSKGGLTKMVDSP-------ISQELSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPF 364
PF+ G LT+MV+SP +++ L V+++F K+ IE++EEGT AAA + A
Sbjct: 300 PFTHGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVS------VASM 353
Query: 365 RLPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
DFVADHPFLF +REE SG ILF+G+V++P
Sbjct: 354 TKDMLLMGDFVADHPFLFTVREEKSGVILFMGQVLDP 390
>At2g14540 putative serpin
Length = 407
Score = 313 bits (801), Expect = 1e-85
Identities = 179/406 (44%), Positives = 247/406 (60%), Gaps = 33/406 (8%)
Query: 3 PQKRRTSEQNLIG--KSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAG 60
P +T+++ I +++ N VSL + ++S N N VFSP S+N VL + A
Sbjct: 18 PSLSKTNKKQKIDMQEAMKNQNEVSLLLVGKVISAVAKNS-NCVFSPASINAVLTVTAAN 76
Query: 61 SEGPT-QNQLLSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQ 119
++ T ++ +LSFL+S ST + ++ +L S V DG+ GGP ++ VNGVW+EQSL
Sbjct: 77 TDNKTLRSFILSFLKSSSTEETNAIFHELASVVFKDGSETGGPKIAAVNGVWMEQSLSCN 136
Query: 120 PSFKQLMTTDFKATLAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIF 179
P ++ L FKA+ A VDF +KA+EV +VN WA++ T I ++LPRGSV SLT+ I+
Sbjct: 137 PDWEDLFLNFFKASFAKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIY 196
Query: 180 ANALYFKGVWKRPFDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQ 239
NALYFKG W++ FD S T+D F LLNGKSV VPFM S QFI ++DGFKVL LPY+Q
Sbjct: 197 GNALYFKGAWEKAFDKSMTRDKPFHLLNGKSVSVPFMRSYEKQFIEAYDGFKVLRLPYRQ 256
Query: 240 GND--KRAFSIYFFLPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFE 297
G D R FS+Y +LPD+K L L++++ S+ FL+ +P +V VG F IP+F I F
Sbjct: 257 GRDDTNREFSMYLYLPDKKGELDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFG 316
Query: 298 IEASELLNKLGLTLPFSKGGLTKMVDSPISQELSVTSIFQKSFIELNEEGTIAAA--TAR 355
EAS + N L + S+ QK+ IE++EEGT AAA T
Sbjct: 317 FEASSVFNDFELNV----------------------SLHQKALIEIDEEGTEAAAATTVV 354
Query: 356 GSTGGAAPFRLPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
TG P IDFVADHPFLFLIRE+ +GT+LF G++ +P
Sbjct: 355 VVTGSCL---WEPKKKIDFVADHPFLFLIREDKTGTLLFAGQIFDP 397
>At1g62170 hypothetical protein
Length = 433
Score = 302 bits (773), Expect = 2e-82
Identities = 171/396 (43%), Positives = 238/396 (59%), Gaps = 36/396 (9%)
Query: 14 IGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGPTQNQL---- 69
+G+++ +V++ +T ++S+ N N VFSP S+N L M+ A S G +L
Sbjct: 64 VGEAMKKQNDVAIFLTGIVISSVAKNS-NFVFSPASINAALTMVAASSGGEQGEELRSFI 122
Query: 70 LSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTD 129
LSFL+S ST +L ++ ++ S VL DG+ GGP ++ VNG+W++QSL + P K L
Sbjct: 123 LSFLKSSSTDELNAIFREIASVVLVDGSKKGGPKIAVVNGMWMDQSLSVNPLSKDLFKNF 182
Query: 130 FKATLAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVW 189
F A A VDF +KA+EV EVN WA+ T G I DLLPRGSV SLT ++ +ALYFKG W
Sbjct: 183 FSAAFAQVDFRSKAEEVRTEVNAWASSHTNGLIKDLLPRGSVTSLTDRVYGSALYFKGTW 242
Query: 190 KRPFDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGND--KRAFS 247
+ + S TK F LLNG SV VPFM+S Q+I+++DGFKVL LPY+QG D R F+
Sbjct: 243 EEKYSKSMTKCKPFYLLNGTSVSVPFMSSFEKQYIAAYDGFKVLRLPYRQGRDNTNRNFA 302
Query: 248 IYFFLPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKL 307
+Y +LPD+K L L++++ S FL+ P +VKVGKF IP+F I F EAS +
Sbjct: 303 MYIYLPDKKGELDDLLERMTSTPGFLDSHNPERRVKVGKFRIPKFKIEFGFEASSAFSDF 362
Query: 308 GLTLPFSKGGLTKMVDSPISQELSVTSIFQKSFIELNEEGT--IAAATARGSTGGAAPFR 365
L + F +QK+ IE++E+GT + R + G A +
Sbjct: 363 ELDVSF----------------------YQKTLIEIDEKGTEAVTFTAFRSAYLGCALVK 400
Query: 366 LPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
PIDFVADHPFLFLIREE +GT+LF G++ +P
Sbjct: 401 -----PIDFVADHPFLFLIREEQTGTVLFAGQIFDP 431
>At2g35580 putative serpin
Length = 374
Score = 278 bits (712), Expect = 3e-75
Identities = 158/372 (42%), Positives = 228/372 (60%), Gaps = 32/372 (8%)
Query: 39 NEKNVVF---SPLSLNTVLIMITAGSEGPTQ--NQLLSFLQSESTGDLKSLCSQLVSSVL 93
N+ ++V +PL +N +L +I A S G T ++++S LQ+ ST L ++ S++V++VL
Sbjct: 23 NQNDIVLRLTAPL-INVILSIIAASSPGDTDTADKIVSLLQASSTDKLHAVSSEIVTTVL 81
Query: 94 SDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKATLAAVDFINKADEVIKEVNLW 153
+D +GGP +S NG+W+E++L ++PSFK L+ +KA VDF KADEV +EVN W
Sbjct: 82 ADSTASGGPTISAANGLWIEKTLNVEPSFKDLLLNSYKAAFNRVDFRTKADEVNREVNSW 141
Query: 154 ANKETKGFINDLLPRGSVDS-LTSLIFANALYFKGVWKRPFDTSKTKDYDFDLLNGKSVK 212
K+T G I +LLP + LT IFANAL+F G W FD S TKD DF LL+G V+
Sbjct: 142 VEKQTNGLITNLLPSNPKSAPLTDHIFANALFFNGRWDSQFDPSLTKDSDFHLLDGTKVR 201
Query: 213 VPFMTSKNNQFISSFDGFKVLGLPYKQG-NDKRAFSIYFFLPDEKDGLSALIDKVASDSE 271
VPFMT + ++ ++GFKV+ L Y++G D R+FS+ +LPDEKDGL ++++++AS
Sbjct: 202 VPFMTGASCRYTHVYEGFKVINLQYRRGREDSRSFSMQIYLPDEKDGLPSMLERLASTRG 261
Query: 272 FLE--QKLPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTLPFSKGGLTKMVDSPISQE 329
FL+ + LP + + + IPRF F EASE L GL +P S
Sbjct: 262 FLKDNEVLPSHSAVIKELKIPRFKFDFAFEASEALKGFGLVVPLS--------------- 306
Query: 330 LSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPFRLPPPPPIDFVADHPFLFLIREEFS 389
I KS IE++E G+ AAA A G R PPP DFVADHPFLF+++E S
Sbjct: 307 ----MIMHKSCIEVDEVGSKAAAAAAFRGIGC---RRPPPEKHDFVADHPFLFIVKEYRS 359
Query: 390 GTILFVGKVVNP 401
G +LF+G+V++P
Sbjct: 360 GLVLFLGQVMDP 371
>At2g35590 putative serpin
Length = 331
Score = 206 bits (524), Expect = 2e-53
Identities = 114/257 (44%), Positives = 153/257 (59%), Gaps = 25/257 (9%)
Query: 148 KEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVWKRPFDTSKTKDYDFDLLN 207
KEVN W ++T G I DLLP S LT LIFANAL+F G W F+ S TK+ DF LL+
Sbjct: 94 KEVNSWVEEQTNGLITDLLPPNSASPLTDLIFANALFFNGRWDSQFNPSLTKESDFHLLD 153
Query: 208 GKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQG-NDKRAFSIYFFLPDEKDGLSALIDKV 266
G V+VPFMT + + ++ FKVL LPY++G D R FS+ +LPDEKDGL ++++ +
Sbjct: 154 GTKVRVPFMTGAHEDSLDVYEAFKVLNLPYREGREDSRGFSMQIYLPDEKDGLPSMLESL 213
Query: 267 ASDSEFLEQK--LPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTLPFSKGGLTKMVDS 324
AS FL+ LP + V + IPRF +F+ EAS+ L LGL +P S
Sbjct: 214 ASTRGFLKDNKVLPSQKAGVKELKIPRFKFAFDFEASKALKGLGLKVPLS---------- 263
Query: 325 PISQELSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPFRLPPPPPIDFVADHPFLFLI 384
+I KS IE++E G+ AAA A + G F PP DFVADHPFLF++
Sbjct: 264 ---------TIIHKSCIEVDEVGSKAAAAAALRSCGGCYF---PPKKYDFVADHPFLFIV 311
Query: 385 REEFSGTILFVGKVVNP 401
+E SG +LF+G V++P
Sbjct: 312 KEYISGLVLFLGHVMDP 328
>At1g62160 hypothetical protein
Length = 265
Score = 182 bits (463), Expect = 2e-46
Identities = 103/232 (44%), Positives = 139/232 (59%), Gaps = 29/232 (12%)
Query: 173 SLTSLIFANALYFKGVWKRPFDTSKTKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKV 232
+LT I+ NALYFKG W++ F S TK F L+NG SV VPFM+S +Q+I ++DGFKV
Sbjct: 58 NLTKRIYGNALYFKGTWEKKFSKSMTKRKPFYLVNGISVSVPFMSSSKDQYIEAYDGFKV 117
Query: 233 LGLPYKQGND--KRAFSIYFFLPDEKDGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIP 290
L LPY+QG D R FS+YF+LPD+K L L+ ++ S FL+ PR +V+V +F IP
Sbjct: 118 LRLPYRQGRDNTNRNFSMYFYLPDKKGELDDLLKRMTSTPGFLDSHTPRERVEVDEFRIP 177
Query: 291 RFNISFEIEASELLNKLGLTLPFSKGGLTKMVDSPISQELSVTSIFQKSFIELNEEGT-I 349
+F I F EAS + + + + F +QK+ IE++EEGT
Sbjct: 178 KFKIEFGFEASSVFSDFEIDVSF----------------------YQKALIEIDEEGTEA 215
Query: 350 AAATARGSTGGAAPFRLPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 401
AAATA F +DFVADHPFLFLIREE +GT+LF G++ +P
Sbjct: 216 AAATAFVDNEDGCGF----VETLDFVADHPFLFLIREEQTGTVLFAGQIFDP 263
Score = 36.2 bits (82), Expect = 0.033
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 5/51 (9%)
Query: 57 ITAGSEGPTQNQ-----LLSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGP 102
+ A S G Q+ +LSFL++ ST +L ++ ++ SSV DG+ GGP
Sbjct: 1 MVAASSGDEQDDELRFFILSFLKASSTDELNAVLRKIASSVFVDGSKKGGP 51
>At2g35570 putative serpin
Length = 213
Score = 172 bits (436), Expect = 3e-43
Identities = 90/207 (43%), Positives = 129/207 (61%), Gaps = 17/207 (8%)
Query: 11 QNLIGKSLTNLTNVSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAGSEGPTQNQLL 70
+NL G S+ NL ++ L +T+H++++ N+VFSP L+ +L
Sbjct: 15 ENLKG-SVGNLNDIILRLTQHVIASTAGKTSNLVFSPALLSVIL---------------- 57
Query: 71 SFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDF 130
SF+ + S +L ++ S +V+++L+D P+GGP +S NGVW+E++L ++PSFK L+ +
Sbjct: 58 SFVAASSPDELNAVSSGIVTTILADSTPSGGPTISAANGVWIEKTLYVEPSFKDLLLNSY 117
Query: 131 KATLAAVDFINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVWK 190
KA VDF KADEV KEVN W ++T G I DLLP S S T LIFANAL+F G W
Sbjct: 118 KAAFNQVDFRTKADEVNKEVNSWVEEQTNGLITDLLPPNSASSWTDLIFANALFFNGRWD 177
Query: 191 RPFDTSKTKDYDFDLLNGKSVKVPFMT 217
F+ S TK+ DF LL+G V+VPFMT
Sbjct: 178 SQFNPSLTKESDFHLLDGTKVRVPFMT 204
>At1g64010 hypothetical protein
Length = 185
Score = 172 bits (435), Expect = 4e-43
Identities = 96/207 (46%), Positives = 130/207 (62%), Gaps = 30/207 (14%)
Query: 198 TKDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGND-KRAFSIYFFLPDEK 256
TKD DF L+NG SV V M+S +Q+I ++DGFKVL LP++QGND R FS++F+LPDEK
Sbjct: 2 TKDRDFHLINGTSVSVSLMSSYKDQYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYLPDEK 61
Query: 257 DGLSALIDKVASDSEFLEQKLPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTLPFSKG 316
DGL L++K+AS FL+ +P +VKVG+F IP+F I F AS N+LG
Sbjct: 62 DGLDNLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLG-------- 113
Query: 317 GLTKMVDSPISQELSVTSIFQKSFIELNEEG--TIAAATARGSTGGAAPFRLPPPPPIDF 374
L +++QK+ +E++EEG IAA G G A R IDF
Sbjct: 114 -------------LDEMALYQKACVEIDEEGAEAIAATAVVGGFGCAFVKR------IDF 154
Query: 375 VADHPFLFLIREEFSGTILFVGKVVNP 401
VADHPFLF+IRE+ +GT+LFVG++ +P
Sbjct: 155 VADHPFLFMIREDKTGTVLFVGQIFDP 181
>At1g63280 hypothetical protein
Length = 120
Score = 132 bits (333), Expect = 3e-31
Identities = 69/118 (58%), Positives = 83/118 (69%), Gaps = 4/118 (3%)
Query: 139 FINKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVWKRPFDTSKT 198
F+ KA +V +E+N WA+ T G I DLLPRGSV S T ++ NALYFKG W+ FD S T
Sbjct: 5 FVLKAVQVRQELNKWASDHTNGLIIDLLPRGSVKSETVQVYGNALYFKGAWENKFDKSST 64
Query: 199 KDYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGND--KRAFSIYFFLPD 254
KD +F GK V VPFM S +Q+I + DGFKVLGLPY+QG D KR FSIYF+LPD
Sbjct: 65 KDNEFH--QGKEVHVPFMRSYESQYIMACDGFKVLGLPYQQGLDDTKRKFSIYFYLPD 120
>At1g51330 hypothetical protein
Length = 193
Score = 95.1 bits (235), Expect = 6e-20
Identities = 47/94 (50%), Positives = 61/94 (64%)
Query: 140 INKADEVIKEVNLWANKETKGFINDLLPRGSVDSLTSLIFANALYFKGVWKRPFDTSKTK 199
+N A+EV EVN WA + T G I +LLP GSV + T I+ NALYFKG W+ F S T
Sbjct: 29 VNGAEEVRMEVNSWALRHTNGLIKNLLPPGSVTNQTIKIYGNALYFKGAWENKFGKSMTI 88
Query: 200 DYDFDLLNGKSVKVPFMTSKNNQFISSFDGFKVL 233
F L+NGK V VPFM S +++ +++GFKVL
Sbjct: 89 HKPFHLVNGKQVLVPFMKSYERKYMKAYNGFKVL 122
>At2g35560 putative serpin
Length = 121
Score = 82.0 bits (201), Expect = 5e-16
Identities = 55/142 (38%), Positives = 75/142 (52%), Gaps = 26/142 (18%)
Query: 262 LIDKVASDSEFLEQK--LPRNQVKVGKFMIPRFNISFEIEASELLNKLGLTLPFSKGGLT 319
+++++AS FL K +P + VG+ IPRF F EASE L LGL +P
Sbjct: 1 MLERLASTRGFLNGKEDIPSHWADVGELKIPRFKFDFGFEASEALKGLGLEVP------- 53
Query: 320 KMVDSPISQELSVTSIFQKSFIELNEEGTIAAATARGSTGGAAPFRLPPPPPIDFVADHP 379
++I KS IE++E G+ AAA A G P DFVADHP
Sbjct: 54 -------------STIIHKSCIEVDEVGSKAAAAAVIFVGCCGPAE----KRYDFVADHP 96
Query: 380 FLFLIREEFSGTILFVGKVVNP 401
FLFL++E SG ILF+G+V++P
Sbjct: 97 FLFLVKEYRSGLILFLGQVMDP 118
>At1g64020 hypothetical protein
Length = 121
Score = 66.6 bits (161), Expect = 2e-11
Identities = 39/110 (35%), Positives = 62/110 (55%), Gaps = 3/110 (2%)
Query: 24 VSLNITKHLLSNQKLNEKNVVFSPLSLNTVLIMITAG-SEGPTQNQLLSFLQSESTGDLK 82
V++ ++ HL S + NV FSP S+ V M+ +G +Q+LSFL S S +L
Sbjct: 13 VAMILSWHLFSTVAKHSNNV-FSPASITAVFTMMASGPGSSLISDQILSFLGSSSIDELN 71
Query: 83 SLCSQLVSSVLSDGAPAGGPCLSHVNGVWVEQSLPLQPSFKQLMTTDFKA 132
S+ +++++V +DG+ GGP + NG W++QS + S K L FKA
Sbjct: 72 SVF-RVITTVFADGSNIGGPTIKVANGAWIDQSFSIDSSSKNLFENFFKA 120
>At3g11470 unknown protein
Length = 300
Score = 33.9 bits (76), Expect = 0.16
Identities = 39/140 (27%), Positives = 61/140 (42%), Gaps = 9/140 (6%)
Query: 241 NDKRAFSIYFFLPDEKDGLSALIDKVASDSEFLEQ-KLPRNQVK-VGK-FMIPRFNISFE 297
+D AF+ F+ DE LS L D EF++ L VK +GK F FN +F
Sbjct: 158 HDILAFAERFYSADEVKFLSTLPDPEVQRKEFIKLWTLKEAYVKALGKGFSAAPFN-TFT 216
Query: 298 IEASELLNKLGLTLPFSKGGLTKMVDSPISQELSVTSIFQKSFIELNEEGTIAAATARGS 357
I++ K+G ++ +T+M S + + ++ S +EL++ A
Sbjct: 217 IQS-----KVGTKGEYNLCKMTEMTASSLEETNKCDGEWKFSLLELDDSHYAAICIEDDE 271
Query: 358 TGGAAPFRLPPPPPIDFVAD 377
G AP R+ I FV D
Sbjct: 272 ASGGAPMRVIVRKTIPFVED 291
>At2g02540 unknown protein
Length = 310
Score = 31.2 bits (69), Expect = 1.1
Identities = 22/65 (33%), Positives = 30/65 (45%), Gaps = 17/65 (26%)
Query: 320 KMVDSPISQEL----SVTSIFQKSFIE--LNEEGTIAAATARGSTGGAAPFRLPPPPPID 373
KM+ SP+ Q++ VT+ S E + EEG GG+ FR PPPPP
Sbjct: 166 KMIKSPLPQQMIMPIGVTTAGSNSESEDLMEEEG-----------GGSLTFRQPPPPPSP 214
Query: 374 FVADH 378
+ H
Sbjct: 215 YSYGH 219
>At1g33700 unknown protein
Length = 947
Score = 29.6 bits (65), Expect = 3.1
Identities = 15/43 (34%), Positives = 27/43 (61%), Gaps = 1/43 (2%)
Query: 134 LAAVDFINKADEVIKEVNLWANKE-TKGFINDLLPRGSVDSLT 175
+A ++I KA E + + N+ ++ T+G +N +LP G VD+ T
Sbjct: 764 IAKEEWIKKALETVYDFNVMRVRDGTRGAVNGMLPDGRVDTST 806
>At5g21080 putative protein
Length = 980
Score = 28.5 bits (62), Expect = 6.9
Identities = 16/37 (43%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Query: 205 LLNGKSVKVPFMTSKNNQFISSFDGFKVLGLPYKQGN 241
LL GK K+ FM++K+N+F SS + + LP GN
Sbjct: 890 LLMGKQEKMSFMSAKSNKF-SSSQTKEAVALPCSGGN 925
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,051,205
Number of Sequences: 26719
Number of extensions: 393026
Number of successful extensions: 1328
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1257
Number of HSP's gapped (non-prelim): 24
length of query: 404
length of database: 11,318,596
effective HSP length: 102
effective length of query: 302
effective length of database: 8,593,258
effective search space: 2595163916
effective search space used: 2595163916
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122166.8


