

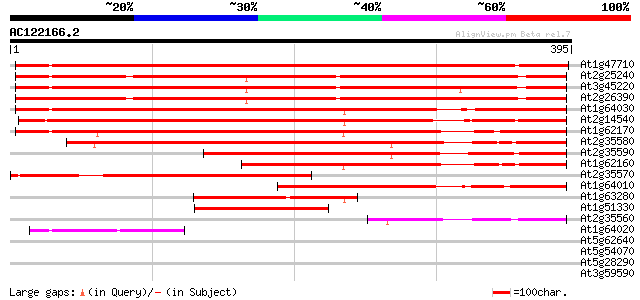
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.2 + phase: 0
(395 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g47710 serpin, putative 395 e-110
At2g25240 putative serpin 338 2e-93
At3g45220 serpin-like protein 335 3e-92
At2g26390 putative serpin 332 2e-91
At1g64030 hypothetical protein 330 6e-91
At2g14540 putative serpin 319 2e-87
At1g62170 hypothetical protein 313 1e-85
At2g35580 putative serpin 290 1e-78
At2g35590 putative serpin 213 1e-55
At1g62160 hypothetical protein 194 9e-50
At2g35570 putative serpin 182 3e-46
At1g64010 hypothetical protein 168 5e-42
At1g63280 hypothetical protein 129 4e-30
At1g51330 hypothetical protein 100 1e-21
At2g35560 putative serpin 93 2e-19
At1g64020 hypothetical protein 70 2e-12
At5g62640 unknown protein 30 2.3
At5g54070 putative protein 30 3.0
At5g28290 serine (threonine) protein kinase - like 30 3.0
At3g59590 putative protein 30 3.0
>At1g47710 serpin, putative
Length = 391
Score = 395 bits (1015), Expect = e-110
Identities = 202/391 (51%), Positives = 278/391 (70%), Gaps = 5/391 (1%)
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFL 64
+ +S + +SMN+ KH+++ + NV+FSP S+N VLS+IA GS G T+ Q+LSFL
Sbjct: 3 VRESISLQNQVSMNLAKHVITTVS-QNSNVIFSPASINVVLSIIAAGSAGATKDQILSFL 61
Query: 65 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAA 124
+ ST L S S++VS+VL+DG+ GGP LS NG W+++++ +PSFKQL+ +KAA
Sbjct: 62 KFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLSFKPSFKQLLEDSYKAA 121
Query: 125 FAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQF 184
DF +KA EV EVN WAEKET GLI +LP GS +S+T+LIFANALYFKG W ++F
Sbjct: 122 SNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFKGTWNEKF 181
Query: 185 DTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDERAFSIYFFLP 244
D + T++ +F LL+G V PFMTSK Q++S+ DGFKVLGLPY QG+D+R FS+YF+LP
Sbjct: 182 DESLTQEGEFHLLDGNKVTAPFMTSKKKQYVSAYDGFKVLGLPYLQGQDKRQFSMYFYLP 241
Query: 245 DKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLALPF 304
D +GLS+L+DK+ S FL+ ++PRR+V+V +F+IP+F SF +AS +LK LGL PF
Sbjct: 242 DANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVLKGLGLTSPF 301
Query: 305 T-LGGLTKMVDSP-ISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPP 362
+ GLT+MV+SP + + L VS IF K+ IEVNEEGT+AAA + I R
Sbjct: 302 SGEEGLTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLRGLLM--EEDE 359
Query: 363 IDFVADHPFLFLIREEFSGTILFVGKVVNPL 393
IDFVADHPFL ++ E +G +LF+G+VV+PL
Sbjct: 360 IDFVADHPFLLVVTENITGVVLFIGQVVDPL 390
>At2g25240 putative serpin
Length = 385
Score = 338 bits (868), Expect = 2e-93
Identities = 184/392 (46%), Positives = 259/392 (65%), Gaps = 16/392 (4%)
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFL 64
+GKS N ++ + +TKH+++ N+VFSP+S+N +LS+IA GS T++Q+LSFL
Sbjct: 3 LGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILSFL 61
Query: 65 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAA 124
ST L + +Q++ G LS NGVW+++ L+ SFK L+ +KA
Sbjct: 62 MLPSTDHLNLVLAQIIDG----GTEKSDLRLSIANGVWIDKFFSLKLSFKDLLENSYKAT 117
Query: 125 FAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSL--TRLIFANALYFKGVWKQ 182
+ VDF +K +EV +EVN WAE T GLIK +L S++++ + L+ ANA+YFKG W
Sbjct: 118 CSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFKGAWSS 177
Query: 183 QFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDERAFSIYFF 242
+FD TK DF LL+G SVKVPFMT+ DQ++ S DGFKVL LPY + D+R FS+Y +
Sbjct: 178 KFDANMTKKNDFHLLDGTSVKVPFMTNYEDQYLRSYDGFKVLRLPYIE--DQRQFSMYIY 235
Query: 243 LPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLAL 302
LP+ K+GL+ L++K+ S+ F + ++P + VG FRIP+F SFE ASE+LK +GL
Sbjct: 236 LPNDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMGLTS 295
Query: 303 PFTL-GGLTKMVDSPIS-QELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPP 360
PF GGLT+MVDSP + +LYVS I K+ IEV+EEGT+AAAV+V +S S P
Sbjct: 296 PFNNGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEAAAVSVGVVSCTSFRRNP-- 353
Query: 361 PPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
DFVAD PFLF +RE+ SG ILF+G+V++P
Sbjct: 354 ---DFVADRPFLFTVREDKSGVILFMGQVLDP 382
>At3g45220 serpin-like protein
Length = 393
Score = 335 bits (858), Expect = 3e-92
Identities = 175/397 (44%), Positives = 261/397 (65%), Gaps = 18/397 (4%)
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFL 64
+GKS N T++ + + KH++ N+VFSP+S+N +L +IA GS T++Q+LSF+
Sbjct: 3 LGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILSFI 61
Query: 65 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAA 124
S+ L ++ ++ VS L+DG LS GVW+++++ +PSFK L+ + A
Sbjct: 62 MLPSSDYLNAVLAKTVSVALNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLENSYNAT 121
Query: 125 FAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSL--TRLIFANALYFKGVWKQ 182
VDF K EV EVN WAE T GLIK +L S+ ++ + LI ANA+YFKG W +
Sbjct: 122 CNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFKGAWSK 181
Query: 183 QFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDERAFSIYFF 242
+FD TK YDF LL+G VKVPFMT+ Q++ DGFKVL LPY + D+R F++Y +
Sbjct: 182 KFDAKLTKSYDFHLLDGTMVKVPFMTNYKKQYLEYYDGFKVLRLPYVE--DQRQFAMYIY 239
Query: 243 LPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLAL 302
LP+ +DGL L+++++S FL+ ++PR+++ F+IP+F SFE +AS++LK++GL L
Sbjct: 240 LPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVLKEMGLTL 299
Query: 303 PFTLGGLTKMVDSP-------ISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRY 355
PFT G LT+MV+SP +++ L+VS +F K+ IEV+EEGT+AAAV+V+ ++
Sbjct: 300 PFTHGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSVASMTKDMLL 359
Query: 356 SPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
DFVADHPFLF +REE SG ILF+G+V++P
Sbjct: 360 MG------DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>At2g26390 putative serpin
Length = 389
Score = 332 bits (851), Expect = 2e-91
Identities = 177/395 (44%), Positives = 258/395 (64%), Gaps = 18/395 (4%)
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQLLSFL 64
+GKS N N+ + K ++ NVVFSP+S+N +LS+IA GS T++++LSFL
Sbjct: 3 LGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILSFL 62
Query: 65 QSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAA 124
S ST L ++ +++ G CLS +GVW++++ L+PSFK+L+ +KA+
Sbjct: 63 MSPSTDHLNAVLAKIADG----GTERSDLCLSTAHGVWIDKSSYLKPSFKELLENSYKAS 118
Query: 125 FAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSL-----TRLIFANALYFKGV 179
+ VDF K EV +EVN WA+ T GLIK +L +++ + LI ANA+YFK
Sbjct: 119 CSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAVYFKAA 178
Query: 180 WKQQFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDERAFSI 239
W ++FD TKD DF LL+G +VKVPFM S DQ++ DGF+VL LPY + D+R FS+
Sbjct: 179 WSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKDQYLRGYDGFQVLRLPYVE--DKRHFSM 236
Query: 240 YFFLPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLG 299
Y +LP+ KDGL+ L++K++++ FL+ ++P + V RIP+ N SFE +ASE+LK +G
Sbjct: 237 YIYLPNDKDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKASEVLKDMG 296
Query: 300 LALPFT-LGGLTKMVDSPIS-QELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSP 357
L PFT G LT+MVDSP + +L+VS I K+ IEV+EEGT+AAAV+V+ + +
Sbjct: 297 LTSPFTSKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIMMPQCLMRN 356
Query: 358 PPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
P DFVADHPFLF +RE+ SG ILF+G+V++P
Sbjct: 357 P-----DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>At1g64030 hypothetical protein
Length = 385
Score = 330 bits (847), Expect = 6e-91
Identities = 176/391 (45%), Positives = 252/391 (64%), Gaps = 25/391 (6%)
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGP-TQKQLLSF 63
+ ++ N T+++M ++ H+LS+ K+ NV+FSP S+N+ ++M A G G Q+LSF
Sbjct: 3 VREAMKNQTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQILSF 61
Query: 64 LQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKA 123
L+S S +LK++ +L S V +D + GGP ++ NG+W+++++P P FK L FKA
Sbjct: 62 LRSSSIDELKTVFRELASVVYADRSATGGPKITAANGLWIDKSLPTDPKFKDLFENFFKA 121
Query: 124 AFAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQ 183
+ VDF ++A EV +EVN W E T LIK+LLP GSV SLT I+ANAL FKG WK+
Sbjct: 122 VYVPVDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWKRP 181
Query: 184 FDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDE--RAFSIYF 241
F+ T+D DF L+NG SV VPFM+S +Q++ + DGFKVL LPY++G D+ R FS+YF
Sbjct: 182 FEKYYTRDNDFYLVNGTSVSVPFMSSYENQYVRAYDGFKVLRLPYQRGSDDTNRKFSMYF 241
Query: 242 FLPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLA 301
+LPDKKDGL +L++K+AS FL+ ++P + E+ KFRIP+F I F + +L +LGL
Sbjct: 242 YLPDKKDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRLGLR 301
Query: 302 LPFTLGGLTKMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPP 361
S +Y K+ +E++EEG +AAA T S PP
Sbjct: 302 ----------------SMSMY-----HKACVEIDEEGAEAAAATADGDCGCSLDFVEPPK 340
Query: 362 PIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
IDFVADHPFLFLIREE +GT+LFVG++ +P
Sbjct: 341 KIDFVADHPFLFLIREEKTGTVLFVGQIFDP 371
>At2g14540 putative serpin
Length = 407
Score = 319 bits (817), Expect = 2e-87
Identities = 176/389 (45%), Positives = 244/389 (62%), Gaps = 27/389 (6%)
Query: 7 KSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQ-LLSFLQ 65
++ N +S+ + ++S K N VFSP S+N VL++ A ++ T + +LSFL+
Sbjct: 33 EAMKNQNEVSLLLVGKVIS-AVAKNSNCVFSPASINAVLTVTAANTDNKTLRSFILSFLK 91
Query: 66 SESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKAAF 125
S ST + ++ +L S V DG+ GGP ++ VNGVW+EQ++ P ++ L FKA+F
Sbjct: 92 SSSTEETNAIFHELASVVFKDGSETGGPKIAAVNGVWMEQSLSCNPDWEDLFLNFFKASF 151
Query: 126 AAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFD 185
A VDF +KA EV +VN WA + T LIK +LP GSV SLT I+ NALYFKG W++ FD
Sbjct: 152 AKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIYGNALYFKGAWEKAFD 211
Query: 186 TTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDE--RAFSIYFFL 243
+ T+D F LLNGKSV VPFM S QFI + DGFKVL LPY+QG+D+ R FS+Y +L
Sbjct: 212 KSMTRDKPFHLLNGKSVSVPFMRSYEKQFIEAYDGFKVLRLPYRQGRDDTNREFSMYLYL 271
Query: 244 PDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLALP 303
PDKK L NL++++ S+ FL+ ++P +V+VG FRIP+F I F EAS +
Sbjct: 272 PDKKGELDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSVFNDF----- 326
Query: 304 FTLGGLTKMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPI 363
EL VS + QK+ IE++EEGT+AAA T + + S P I
Sbjct: 327 ----------------ELNVS-LHQKALIEIDEEGTEAAAATTVVVVTGSCLW-EPKKKI 368
Query: 364 DFVADHPFLFLIREEFSGTILFVGKVVNP 392
DFVADHPFLFLIRE+ +GT+LF G++ +P
Sbjct: 369 DFVADHPFLFLIREDKTGTLLFAGQIFDP 397
>At1g62170 hypothetical protein
Length = 433
Score = 313 bits (801), Expect = 1e-85
Identities = 174/395 (44%), Positives = 248/395 (62%), Gaps = 34/395 (8%)
Query: 5 IGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQL---- 60
+G++ ++++ +T ++S+ K N VFSP S+N L+M+A S G ++L
Sbjct: 64 VGEAMKKQNDVAIFLTGIVISSVA-KNSNFVFSPASINAALTMVAASSGGEQGEELRSFI 122
Query: 61 LSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTD 120
LSFL+S ST +L ++ ++ S VL DG+ GGP ++ VNG+W++Q++ + P K L
Sbjct: 123 LSFLKSSSTDELNAIFREIASVVLVDGSKKGGPKIAVVNGMWMDQSLSVNPLSKDLFKNF 182
Query: 121 FKAAFAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVW 180
F AAFA VDF +KA EV EVN WA T GLIK+LLP GSV SLT ++ +ALYFKG W
Sbjct: 183 FSAAFAQVDFRSKAEEVRTEVNAWASSHTNGLIKDLLPRGSVTSLTDRVYGSALYFKGTW 242
Query: 181 KQQFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKD--ERAFS 238
++++ + TK F LLNG SV VPFM+S Q+I++ DGFKVL LPY+QG+D R F+
Sbjct: 243 EEKYSKSMTKCKPFYLLNGTSVSVPFMSSFEKQYIAAYDGFKVLRLPYRQGRDNTNRNFA 302
Query: 239 IYFFLPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKL 298
+Y +LPDKK L +L++++ S FL+ + P R+V+VGKFRIP+F I F EAS
Sbjct: 303 MYIYLPDKKGELDDLLERMTSTPGFLDSHNPERRVKVGKFRIPKFKIEFGFEASSAFSDF 362
Query: 299 GLALPFTLGGLTKMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRY-SP 357
L + F +QK+ IE++E+GT+A V+F + RS Y
Sbjct: 363 ELDVSF----------------------YQKTLIEIDEKGTEA----VTFTAFRSAYLGC 396
Query: 358 PPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
PIDFVADHPFLFLIREE +GT+LF G++ +P
Sbjct: 397 ALVKPIDFVADHPFLFLIREEQTGTVLFAGQIFDP 431
>At2g35580 putative serpin
Length = 374
Score = 290 bits (741), Expect = 1e-78
Identities = 160/358 (44%), Positives = 228/358 (62%), Gaps = 28/358 (7%)
Query: 41 LNTVLSMIATGSEGPTQK--QLLSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYV 98
+N +LS+IA S G T +++S LQ+ ST L ++ S++V++VL+D +GGP +S
Sbjct: 36 INVILSIIAASSPGDTDTADKIVSLLQASSTDKLHAVSSEIVTTVLADSTASGGPTISAA 95
Query: 99 NGVWVEQTIPLQPSFKQLMNTDFKAAFAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLP 158
NG+W+E+T+ ++PSFK L+ +KAAF VDF KA+EV EVN W EK+T GLI NLLP
Sbjct: 96 NGLWIEKTLNVEPSFKDLLLNSYKAAFNRVDFRTKADEVNREVNSWVEKQTNGLITNLLP 155
Query: 159 PGSVNS-LTRLIFANALYFKGVWKQQFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISS 217
++ LT IFANAL+F G W QFD + TKD DF LL+G V+VPFMT + ++
Sbjct: 156 SNPKSAPLTDHIFANALFFNGRWDSQFDPSLTKDSDFHLLDGTKVRVPFMTGASCRYTHV 215
Query: 218 LDGFKVLGLPYKQGK-DERAFSIYFFLPDKKDGLSNLIDKVASDSEFLERN--LPRRKVE 274
+GFKV+ L Y++G+ D R+FS+ +LPD+KDGL ++++++AS FL+ N LP
Sbjct: 216 YEGFKVINLQYRRGREDSRSFSMQIYLPDEKDGLPSMLERLASTRGFLKDNEVLPSHSAV 275
Query: 275 VGKFRIPRFNISFEIEASELLKKLGLALPFTLGGLTKMVDSPISQELYVSGIFQKSFIEV 334
+ + +IPRF F EASE LK GL +P ++ I KS IEV
Sbjct: 276 IKELKIPRFKFDFAFEASEALKGFGLVVPLSM-------------------IMHKSCIEV 316
Query: 335 NEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
+E G+KAAA +F R PPP DFVADHPFLF+++E SG +LF+G+V++P
Sbjct: 317 DEVGSKAAA-AAAFRGIGCR--RPPPEKHDFVADHPFLFIVKEYRSGLVLFLGQVMDP 371
>At2g35590 putative serpin
Length = 331
Score = 213 bits (543), Expect = 1e-55
Identities = 116/259 (44%), Positives = 162/259 (61%), Gaps = 25/259 (9%)
Query: 137 VGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDTTKTKDYDFDL 196
+ +EVN W E++T GLI +LLPP S + LT LIFANAL+F G W QF+ + TK+ DF L
Sbjct: 92 LNKEVNSWVEEQTNGLITDLLPPNSASPLTDLIFANALFFNGRWDSQFNPSLTKESDFHL 151
Query: 197 LNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGK-DERAFSIYFFLPDKKDGLSNLID 255
L+G V+VPFMT ++ + + FKVL LPY++G+ D R FS+ +LPD+KDGL ++++
Sbjct: 152 LDGTKVRVPFMTGAHEDSLDVYEAFKVLNLPYREGREDSRGFSMQIYLPDEKDGLPSMLE 211
Query: 256 KVASDSEFLERN--LPRRKVEVGKFRIPRFNISFEIEASELLKKLGLALPFTLGGLTKMV 313
+AS FL+ N LP +K V + +IPRF +F+ EAS+ LK LGL +P
Sbjct: 212 SLASTRGFLKDNKVLPSQKAGVKELKIPRFKFAFDFEASKALKGLGLKVP---------- 261
Query: 314 DSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHPFLF 373
+S I KS IEV+E G+KAAA + PP DFVADHPFLF
Sbjct: 262 ---------LSTIIHKSCIEVDEVGSKAAAAAALRSCGGCYF---PPKKYDFVADHPFLF 309
Query: 374 LIREEFSGTILFVGKVVNP 392
+++E SG +LF+G V++P
Sbjct: 310 IVKEYISGLVLFLGHVMDP 328
>At1g62160 hypothetical protein
Length = 265
Score = 194 bits (492), Expect = 9e-50
Identities = 105/231 (45%), Positives = 146/231 (62%), Gaps = 27/231 (11%)
Query: 164 SLTRLIFANALYFKGVWKQQFDTTKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKV 223
+LT+ I+ NALYFKG W+++F + TK F L+NG SV VPFM+S DQ+I + DGFKV
Sbjct: 58 NLTKRIYGNALYFKGTWEKKFSKSMTKRKPFYLVNGISVSVPFMSSSKDQYIEAYDGFKV 117
Query: 224 LGLPYKQGKD--ERAFSIYFFLPDKKDGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIP 281
L LPY+QG+D R FS+YF+LPDKK L +L+ ++ S FL+ + PR +VEV +FRIP
Sbjct: 118 LRLPYRQGRDNTNRNFSMYFYLPDKKGELDDLLKRMTSTPGFLDSHTPRERVEVDEFRIP 177
Query: 282 RFNISFEIEASELLKKLGLALPFTLGGLTKMVDSPISQELYVSGIFQKSFIEVNEEGTKA 341
+F I F EAS + + + F +QK+ IE++EEGT+A
Sbjct: 178 KFKIEFGFEASSVFSDFEIDVSF----------------------YQKALIEIDEEGTEA 215
Query: 342 AAVTVSFISSRSRYSPPPPPPIDFVADHPFLFLIREEFSGTILFVGKVVNP 392
AA T +F+ + +DFVADHPFLFLIREE +GT+LF G++ +P
Sbjct: 216 AAAT-AFVDNEDGCG--FVETLDFVADHPFLFLIREEQTGTVLFAGQIFDP 263
Score = 35.0 bits (79), Expect = 0.072
Identities = 15/34 (44%), Positives = 24/34 (70%)
Query: 60 LLSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGP 93
+LSFL++ ST +L ++ ++ SSV DG+ GGP
Sbjct: 18 ILSFLKASSTDELNAVLRKIASSVFVDGSKKGGP 51
>At2g35570 putative serpin
Length = 213
Score = 182 bits (462), Expect = 3e-46
Identities = 93/212 (43%), Positives = 139/212 (64%), Gaps = 17/212 (8%)
Query: 1 MQNLIGKSFTNLTNISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATGSEGPTQKQL 60
++NL G S NL +I + +T+H++++ K N+VFSP L+ +LS +A
Sbjct: 14 LENLKG-SVGNLNDIILRLTQHVIASTAGKTSNLVFSPALLSVILSFVA----------- 61
Query: 61 LSFLQSESTGDLKSLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTD 120
+ S +L ++ S +V+++L+D P+GGP +S NGVW+E+T+ ++PSFK L+
Sbjct: 62 -----ASSPDELNAVSSGIVTTILADSTPSGGPTISAANGVWIEKTLYVEPSFKDLLLNS 116
Query: 121 FKAAFAAVDFVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVW 180
+KAAF VDF KA+EV +EVN W E++T GLI +LLPP S +S T LIFANAL+F G W
Sbjct: 117 YKAAFNQVDFRTKADEVNKEVNSWVEEQTNGLITDLLPPNSASSWTDLIFANALFFNGRW 176
Query: 181 KQQFDTTKTKDYDFDLLNGKSVKVPFMTSKND 212
QF+ + TK+ DF LL+G V+VPFMT ++
Sbjct: 177 DSQFNPSLTKESDFHLLDGTKVRVPFMTGAHE 208
>At1g64010 hypothetical protein
Length = 185
Score = 168 bits (425), Expect = 5e-42
Identities = 92/205 (44%), Positives = 129/205 (62%), Gaps = 26/205 (12%)
Query: 189 TKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKD-ERAFSIYFFLPDKK 247
TKD DF L+NG SV V M+S DQ+I + DGFKVL LP++QG D R FS++F+LPD+K
Sbjct: 2 TKDRDFHLINGTSVSVSLMSSYKDQYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYLPDEK 61
Query: 248 DGLSNLIDKVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLALPFTLG 307
DGL NL++K+AS FL+ ++P +KV+VG+F IP+F I F AS +LGL
Sbjct: 62 DGLDNLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRLGL------- 114
Query: 308 GLTKMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVA 367
E+ ++QK+ +E++EEG +A A T + IDFVA
Sbjct: 115 -----------DEM---ALYQKACVEIDEEGAEAIAATAVV----GGFGCAFVKRIDFVA 156
Query: 368 DHPFLFLIREEFSGTILFVGKVVNP 392
DHPFLF+IRE+ +GT+LFVG++ +P
Sbjct: 157 DHPFLFMIREDKTGTVLFVGQIFDP 181
>At1g63280 hypothetical protein
Length = 120
Score = 129 bits (323), Expect = 4e-30
Identities = 67/118 (56%), Positives = 82/118 (68%), Gaps = 4/118 (3%)
Query: 130 FVNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDTTKT 189
FV KA +V +E+N WA T GLI +LLP GSV S T ++ NALYFKG W+ +FD + T
Sbjct: 5 FVLKAVQVRQELNKWASDHTNGLIIDLLPRGSVKSETVQVYGNALYFKGAWENKFDKSST 64
Query: 190 KDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPYKQGKDE--RAFSIYFFLPD 245
KD +F GK V VPFM S Q+I + DGFKVLGLPY+QG D+ R FSIYF+LPD
Sbjct: 65 KDNEFH--QGKEVHVPFMRSYESQYIMACDGFKVLGLPYQQGLDDTKRKFSIYFYLPD 120
>At1g51330 hypothetical protein
Length = 193
Score = 100 bits (250), Expect = 1e-21
Identities = 51/94 (54%), Positives = 63/94 (66%)
Query: 131 VNKANEVGEEVNFWAEKETKGLIKNLLPPGSVNSLTRLIFANALYFKGVWKQQFDTTKTK 190
VN A EV EVN WA + T GLIKNLLPPGSV + T I+ NALYFKG W+ +F + T
Sbjct: 29 VNGAEEVRMEVNSWALRHTNGLIKNLLPPGSVTNQTIKIYGNALYFKGAWENKFGKSMTI 88
Query: 191 DYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVL 224
F L+NGK V VPFM S +++ + +GFKVL
Sbjct: 89 HKPFHLVNGKQVLVPFMKSYERKYMKAYNGFKVL 122
>At2g35560 putative serpin
Length = 121
Score = 93.2 bits (230), Expect = 2e-19
Identities = 58/142 (40%), Positives = 79/142 (54%), Gaps = 26/142 (18%)
Query: 253 LIDKVASDSEFL--ERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGLALPFTLGGLT 310
+++++AS FL + ++P +VG+ +IPRF F EASE LK LGL +P T
Sbjct: 1 MLERLASTRGFLNGKEDIPSHWADVGELKIPRFKFDFGFEASEALKGLGLEVPST----- 55
Query: 311 KMVDSPISQELYVSGIFQKSFIEVNEEGTKAAAVTVSFISSRSRYSPPPPPPIDFVADHP 370
I KS IEV+E G+KAAA V F+ P DFVADHP
Sbjct: 56 ---------------IIHKSCIEVDEVGSKAAAAAVIFVGCCG----PAEKRYDFVADHP 96
Query: 371 FLFLIREEFSGTILFVGKVVNP 392
FLFL++E SG ILF+G+V++P
Sbjct: 97 FLFLVKEYRSGLILFLGQVMDP 118
>At1g64020 hypothetical protein
Length = 121
Score = 70.1 bits (170), Expect = 2e-12
Identities = 40/110 (36%), Positives = 63/110 (56%), Gaps = 3/110 (2%)
Query: 15 ISMNITKHLLSNQKLKEENVVFSPLSLNTVLSMIATG-SEGPTQKQLLSFLQSESTGDLK 73
++M ++ HL S K N VFSP S+ V +M+A+G Q+LSFL S S +L
Sbjct: 13 VAMILSWHLFSTVA-KHSNNVFSPASITAVFTMMASGPGSSLISDQILSFLGSSSIDELN 71
Query: 74 SLCSQLVSSVLSDGAPAGGPCLSYVNGVWVEQTIPLQPSFKQLMNTDFKA 123
S+ +++++V +DG+ GGP + NG W++Q+ + S K L FKA
Sbjct: 72 SVF-RVITTVFADGSNIGGPTIKVANGAWIDQSFSIDSSSKNLFENFFKA 120
>At5g62640 unknown protein
Length = 520
Score = 30.0 bits (66), Expect = 2.3
Identities = 10/18 (55%), Positives = 13/18 (71%)
Query: 357 PPPPPPIDFVADHPFLFL 374
PPPPPP+D HP +F+
Sbjct: 356 PPPPPPLDMHPPHPGMFV 373
>At5g54070 putative protein
Length = 331
Score = 29.6 bits (65), Expect = 3.0
Identities = 16/45 (35%), Positives = 27/45 (59%), Gaps = 10/45 (22%)
Query: 256 KVASDSEFLERNLPRRKVEVGKFRIPRFNISFEIEASELLKKLGL 300
K+A D F+ER + +RK+++ + E+EA+E +KKL L
Sbjct: 232 KLAKDQRFVERLVKKRKMKIQR----------ELEAAEFVKKLKL 266
>At5g28290 serine (threonine) protein kinase - like
Length = 568
Score = 29.6 bits (65), Expect = 3.0
Identities = 11/15 (73%), Positives = 12/15 (79%)
Query: 350 SSRSRYSPPPPPPID 364
SSR YSPPP PP+D
Sbjct: 444 SSRGSYSPPPEPPLD 458
>At3g59590 putative protein
Length = 454
Score = 29.6 bits (65), Expect = 3.0
Identities = 26/94 (27%), Positives = 37/94 (38%), Gaps = 22/94 (23%)
Query: 192 YDFDLLNGKSVK-------VPFMTSKNDQFISSLDGFKVLGLPYKQGKDERAFSIYFFLP 244
YD+D ++ SV V F + +DQFI S D F Y LP
Sbjct: 127 YDWDFMSSYSVSLNGALYWVAFHMASDDQFIQSFD------------FSTEKFEPYCLLP 174
Query: 245 DKKDGLSNLIDKVASDSE---FLERNLPRRKVEV 275
+KK SN + +LE+N R +E+
Sbjct: 175 NKKCDPSNARSLAVFRGDRFSYLEQNYETRNIEI 208
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,963,373
Number of Sequences: 26719
Number of extensions: 392561
Number of successful extensions: 2396
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 2313
Number of HSP's gapped (non-prelim): 31
length of query: 395
length of database: 11,318,596
effective HSP length: 101
effective length of query: 294
effective length of database: 8,619,977
effective search space: 2534273238
effective search space used: 2534273238
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122166.2


