

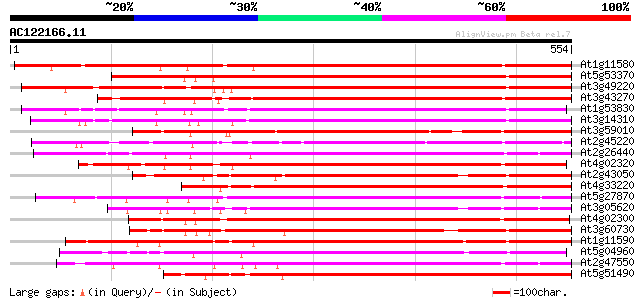
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.11 + phase: 0
(554 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g11580 unknown protein 593 e-170
At5g53370 pectinesterase 434 e-122
At3g49220 pectinesterase - like protein 427 e-120
At3g43270 pectinesterase like protein 424 e-119
At1g53830 unknown protein 421 e-118
At3g14310 pectin methylesterase like protein 421 e-118
At3g59010 pectinesterase precursor-like protein 401 e-112
At2g45220 pectinesterase like protein 394 e-110
At2g26440 putative pectinesterase 384 e-107
At4g02320 pectinesterase - like protein 383 e-106
At2g43050 putative pectinesterase 382 e-106
At4g33220 pectinesterase like protein 376 e-104
At5g27870 pectin methyl-esterase - like protein 375 e-104
At3g05620 putative pectinesterase 374 e-104
At4g02300 putative pectinesterase 373 e-103
At3g60730 pectinesterase - like protein 372 e-103
At1g11590 putative pectin methylesterase 371 e-103
At5g04960 pectinesterase 369 e-102
At2g47550 unknown protein 369 e-102
At5g51490 pectinesterase 366 e-101
>At1g11580 unknown protein
Length = 557
Score = 593 bits (1530), Expect = e-170
Identities = 320/560 (57%), Positives = 397/560 (70%), Gaps = 18/560 (3%)
Query: 5 QSLLDKPRKSIPKTFWLVLSLVAIISSSALIISHL---NKPISFFNLSSAPNLCEHALDT 61
Q LL KP+ K LVLS VAI+ S A + L N + +L + +C A D
Sbjct: 6 QPLLSKPKSLKHKNLCLVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTTSQICHGAHDQ 65
Query: 62 KSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEIA 121
SC +SE +TLS +K ++L L L S + M + + R N R++
Sbjct: 66 DSCQALLSEF---TTLSLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGVRDKAG 122
Query: 122 LNDCEELMDLSMDRVWDSVLTLTKNN--IDSQHDAHTWLSSVLTNHATCLDGLEG---SS 176
DCEE+MD+S DR+ S+ L N ++S + HTWLSSVLTN+ TCL+ + +S
Sbjct: 123 FADCEEMMDVSKDRMMSSMEELRGGNYNLESYSNVHTWLSSVLTNYMTCLESISDVSVNS 182
Query: 177 RVVMESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVG 236
+ +++ L DL+SRAR +LA+ VSVLP + + I ++ FPSW+T+ DR+LLESS
Sbjct: 183 KQIVKPQLEDLVSRARVALAIFVSVLPARDDLKMI---ISNRFPSWLTALDRKLLESSPK 239
Query: 237 DIK--ANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVML 294
+K ANVVVA+DG+GKFKTV +AVA+AP+N TRYVIYVKKG YKE I+IGKKK N+ML
Sbjct: 240 TLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKETIDIGKKKKNLML 299
Query: 295 VGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGA 354
VGDG DATIITGSLN IDG+TTF+SATVAA GDGF+AQDI FQNTAGP KHQAVALRV A
Sbjct: 300 VGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAGPAKHQAVALRVSA 359
Query: 355 DQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQ 414
DQ+VINRC+IDA+QDTLY H+ RQFYRDSYITGTVDFIFGN+AVVFQ + AR P A Q
Sbjct: 360 DQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQNCDIVARNPGAGQ 419
Query: 415 KNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVD 474
KNM+TAQGRED NQNTA SIQ+C + SSDL PV+GS+KT+LGRPWK YSRTV++QS +D
Sbjct: 420 KNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWKLYSRTVIMQSFID 479
Query: 475 GHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVT 534
HIDPAGW WD + L TLYYGEY N+G GA T KRV W G+ +IK++ EA +FTV
Sbjct: 480 NHIDPAGWFPWD--GEFALSTLYYGEYANTGPGADTSKRVNWKGFKVIKDSKEAEQFTVA 537
Query: 535 QLIQGNVWLKNTGVAFIEGL 554
+LIQG +WLK TGV F E L
Sbjct: 538 KLIQGGLWLKPTGVTFQEWL 557
>At5g53370 pectinesterase
Length = 587
Score = 434 bits (1116), Expect = e-122
Identities = 225/468 (48%), Positives = 301/468 (64%), Gaps = 16/468 (3%)
Query: 101 KAMDTANVIKRRVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQH-DAHTWLS 159
KA+ T++ I PR A + C EL+D S+D + ++ ++ + D H D TWLS
Sbjct: 120 KALYTSSTITYTQMPPRVRSAYDSCLELLDDSVDALTRALSSVVVVSGDESHSDVMTWLS 179
Query: 160 SVLTNHATCLDG---LEGSSRVVMES------DLHDLISRARSSLAVLV---SVLPPKAN 207
S +TNH TC DG +EG V + DL +++S + A V S +P N
Sbjct: 180 SAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVKDLSEMVSNCLAIFAGKVKDLSGVPVVNN 239
Query: 208 DGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGK 267
+ + + P+W+ +DR LL + I+A++ V++DGSG FKT+A+A+ AP++
Sbjct: 240 RKLLGTEETEELPNWLKREDRELLGTPTSAIQADITVSKDGSGTFKTIAEAIKKAPEHSS 299
Query: 268 TRYVIYVKKGTYKE-NIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVG 326
R+VIYVK G Y+E N+++G+KKTN+M +GDG T+ITG + D TTF +AT AA G
Sbjct: 300 RRFVIYVKAGRYEEENLKVGRKKTNLMFIGDGKGKTVITGGKSIADDLTTFHTATFAATG 359
Query: 327 DGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYIT 386
GFI +D+ F+N AGP KHQAVALRVG D +V+ RC I +QD LY HSNRQF+R+ I
Sbjct: 360 AGFIVRDMTFENYAGPAKHQAVALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIY 419
Query: 387 GTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLK 446
GTVDFIFGNAAV+ Q + ARKPMA QK +TAQ R+DPNQNT SI C ++ + DL+
Sbjct: 420 GTVDFIFGNAAVILQSCNIYARKPMAQQKITITAQNRKDPNQNTGISIHACKLLATPDLE 479
Query: 447 PVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGA 506
+GS TYLGRPWK YSR V + S + HIDP GW EW+ L +LYYGEYMN G
Sbjct: 480 ASKGSYPTYLGRPWKLYSRVVYMMSDMGDHIDPRGWLEWNGPFA--LDSLYYGEYMNKGL 537
Query: 507 GAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
G+G G+RV WPGYH+I + EASKFTV Q I G+ WL +TGV+F GL
Sbjct: 538 GSGIGQRVKWPGYHVITSTVEASKFTVAQFISGSSWLPSTGVSFFSGL 585
>At3g49220 pectinesterase - like protein
Length = 598
Score = 427 bits (1098), Expect = e-120
Identities = 239/566 (42%), Positives = 342/566 (60%), Gaps = 35/566 (6%)
Query: 12 RKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPN-----LCEHALDTKSCLT 66
+K + + L +SL+ + A + S L S L+ P+ CE + C+
Sbjct: 43 KKLVVSSIVLAISLILAAAIFAGVRSRLKLNQSVPGLARKPSQAISKACELTRFPELCVD 102
Query: 67 HVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAH-IRKAMDTANVIKRRVNSPREEIALNDC 125
+ + S++KD L+ + T H A+ ++ + PR A + C
Sbjct: 103 SLMDFPGSLAASSSKD-----LIHVTVNMTLHHFSHALYSSASLSFVDMPPRARSAYDSC 157
Query: 126 EELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSRVVMESDLH 185
EL+D S+D + ++ ++ ++ Q D TWLS+ LTNH TC +G +G V + +
Sbjct: 158 VELLDDSVDALSRALSSVVSSSAKPQ-DVTTWLSAALTNHDTCTEGFDG----VDDGGVK 212
Query: 186 DLISRARSSLAVLVS---VLPPKANDG-------FIDEKLNG------DFPSWVTSKDRR 229
D ++ A +L+ LVS + ++DG + +L G FP W+ K+R
Sbjct: 213 DHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEEREEKFPRWMRPKERE 272
Query: 230 LLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKEN-IEIGKK 288
+LE V I+A+++V++DG+G KT+++A+ AP N R +IYVK G Y+EN +++G+K
Sbjct: 273 ILEMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKAGRYEENNLKVGRK 332
Query: 289 KTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAV 348
K N+M VGDG T+I+G + D TTF +A+ AA G GFIA+DI F+N AGP KHQAV
Sbjct: 333 KINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDITFENWAGPAKHQAV 392
Query: 349 ALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAAR 408
ALR+GAD +VI RC I +QDTLY HSNRQF+R+ I GTVDFIFGNAAVV Q + AR
Sbjct: 393 ALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNAAVVLQNCSIYAR 452
Query: 409 KPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVV 468
KPM QKN +TAQ R+DPNQNT SI V+ +SDL+ GS +TYLGRPWK +SRTV
Sbjct: 453 KPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYLGRPWKLFSRTVY 512
Query: 469 LQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEA 528
+ S + GH+ GW EW+ L TLYYGEY+NSG G+G G+RV+WPGY +I + AEA
Sbjct: 513 MMSYIGGHVHTRGWLEWNTTFA--LDTLYYGEYLNSGPGSGLGQRVSWPGYRVINSTAEA 570
Query: 529 SKFTVTQLIQGNVWLKNTGVAFIEGL 554
++FTV + I G+ WL +TGV+F+ GL
Sbjct: 571 NRFTVAEFIYGSSWLPSTGVSFLAGL 596
>At3g43270 pectinesterase like protein
Length = 527
Score = 424 bits (1089), Expect = e-119
Identities = 229/491 (46%), Positives = 312/491 (62%), Gaps = 41/491 (8%)
Query: 87 TLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEIALNDCEELMDLSMDRV-WDSVLTLTK 145
T+V +TK+ A + K ++ R A+ DC +L+D + + + W + +
Sbjct: 53 TVVDAITKAVAIVSK-------FDKKAGKSRVSNAIVDCVDLLDSAAEELSWIISASQSP 105
Query: 146 NNIDSQ-----HDAHTWLSSVLTNHATCLDGLEGSSRVVME---SDLHDLISRARSSLAV 197
N D+ D TW+S+ L+N TCLDG EG++ ++ + L + + R+ L +
Sbjct: 106 NGKDNSTGDVGSDLRTWISAALSNQDTCLDGFEGTNGIIKKIVAGGLSKVGTTVRNLLTM 165
Query: 198 LVSVLPP--------------KANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKANVV 243
+ S PP KA+ GF FPSWV DR+LL++ + A+ V
Sbjct: 166 VHS--PPSKPKPKPIKAQTMTKAHSGF------SKFPSWVKPGDRKLLQTDNITV-ADAV 216
Query: 244 VAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATI 303
VA DG+G F T++ AV +APD RYVI+VK+G Y EN+EI KKK N+M+VGDG+DAT+
Sbjct: 217 VAADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGVYVENVEIKKKKWNIMMVGDGIDATV 276
Query: 304 ITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCK 363
ITG+ +FIDG TTF+SAT A G GFIA+DI FQNTAGP+KHQAVA+R D V RC
Sbjct: 277 ITGNRSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCA 336
Query: 364 IDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGR 423
+ +QDTLYAHS RQF+R+ ITGTVDFIFG+A VFQ ++ A++ + NQKN +TAQGR
Sbjct: 337 MRGYQDTLYAHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGR 396
Query: 424 EDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWA 483
+DPN+ T +IQ ++ +DL + TYLGRPWK YSRTV +Q+ + I+P GW
Sbjct: 397 KDPNEPTGFTIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWL 456
Query: 484 EWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWL 543
EW+ L TLYYGEYMNSG GA +RV WPGYH++ +AEA+ FTV+QLIQGN+WL
Sbjct: 457 EWN--GNFALDTLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWL 514
Query: 544 KNTGVAFIEGL 554
+TG+ FI GL
Sbjct: 515 PSTGITFIAGL 525
>At1g53830 unknown protein
Length = 587
Score = 421 bits (1082), Expect = e-118
Identities = 254/576 (44%), Positives = 336/576 (58%), Gaps = 47/576 (8%)
Query: 12 RKSIPKTFWLVLSLVAIISSSALIISHLNKPISFFNLSSAPN-----LCEHALDTKSCLT 66
+K I + + L L+A I A ++ NK LSS + +C L + C +
Sbjct: 18 KKLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELCFS 77
Query: 67 HVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEIALNDCE 126
V+ G L++ K+ + ++L TK+ H A+ + KR+ +PRE AL+DC
Sbjct: 78 AVA-ATGGKELTSQKE-VIEASLNLTTKAVKHNYFAVKKL-IAKRKGLTPREVTALHDCL 134
Query: 127 ELMDLSMDRVWDSVLTL-----TKNNIDSQHDAHTWLSSVLTNHATCLDG---------- 171
E +D ++D + +V L K+ D T +SS +TN TCLDG
Sbjct: 135 ETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKV 194
Query: 172 ----LEGSSRV-------------VMESDLHDLISRARSSLAVLVSVLPPKANDGFIDEK 214
L+G V + E+D+ + R +SS + K G +D
Sbjct: 195 RKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKLKEVTGDLD-- 252
Query: 215 LNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYV 274
+ +P W++ DRRLL+ S IKA+ VA DGSG F TVA AVA+AP+ R+VI++
Sbjct: 253 -SDGWPKWLSVGDRRLLQGST--IKADATVADDGSGDFTTVAAAVAAAPEKSNKRFVIHI 309
Query: 275 KKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDI 334
K G Y+EN+E+ KKKTN+M +GDG TIITGS N +DG+TTF SATVAAVG+ F+A+DI
Sbjct: 310 KAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGERFLARDI 369
Query: 335 RFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFG 394
FQNTAGP KHQAVALRVG+D S +C + A+QDTLY HSNRQF+ +ITGTVDFIFG
Sbjct: 370 TFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTVDFIFG 429
Query: 395 NAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKT 454
NAA V Q + AR+P + QKNMVTAQGR DPNQNT IQ C + +SDL V+G+ T
Sbjct: 430 NAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVKGTFPT 489
Query: 455 YLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRV 514
YLGRPWK+YSRTV++QS + I P GW EW + L TL Y EY+N G GAGT RV
Sbjct: 490 YLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFA--LDTLTYREYLNRGGGAGTANRV 547
Query: 515 TWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAF 550
W GY +I + EA FT Q I G WL +TG F
Sbjct: 548 KWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPF 583
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 421 bits (1081), Expect = e-118
Identities = 251/580 (43%), Positives = 339/580 (58%), Gaps = 53/580 (9%)
Query: 21 LVLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALDTKSCLTH------VSEVVQ- 73
LVL A+ ++ ++ S N + HA+ SC + +S VV
Sbjct: 20 LVLLSAAVALLFVAAVAGISAGASKANEKRTLSPSSHAVLRSSCSSTRYPELCISAVVTA 79
Query: 74 -GSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVN-SPREEIALNDCEELMDL 131
G L++ KD + V+L + H +IK+R +PRE+ AL+DC E +D
Sbjct: 80 GGVELTSQKD-VIEASVNLTITAVEH--NYFTVKKLIKKRKGLTPREKTALHDCLETIDE 136
Query: 132 SMDRVWDSVLTL----TKNNI-DSQHDAHTWLSSVLTNHATCLDGLEGSS------RVVM 180
++D + ++V L TK + + D T +SS +TN TCLDG + ++
Sbjct: 137 TLDELHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSHDDADKQVRKALL 196
Query: 181 ESDLH------DLISRARSSLAVLVSVLPPKANDGFIDEKLNGD---------------- 218
+ +H + ++ ++ ++ KA + KL +
Sbjct: 197 KGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAGEL 256
Query: 219 ----FPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYV 274
+P+W+++ DRRLL+ S +KA+ VA DGSG FKTVA AVA+AP+N RYVI++
Sbjct: 257 DSEGWPTWLSAGDRRLLQGS--GVKADATVAADGSGTFKTVAAAVAAAPENSNKRYVIHI 314
Query: 275 KKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDI 334
K G Y+EN+E+ KKK N+M +GDG TIITGS N +DG+TTF SATVAAVG+ F+A+DI
Sbjct: 315 KAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGERFLARDI 374
Query: 335 RFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFG 394
FQNTAGP KHQAVALRVG+D S C + A+QDTLY HSNRQF+ I GTVDFIFG
Sbjct: 375 TFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFIFG 434
Query: 395 NAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKT 454
NAAVV Q + AR+P + QKNMVTAQGR DPNQNT IQ+C + +SDL+ V+GS T
Sbjct: 435 NAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSFPT 494
Query: 455 YLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRV 514
YLGRPWK+YS+TV++QS + I P GW+EW L TL Y EY N+GAGAGT RV
Sbjct: 495 YLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFA--LNTLTYREYSNTGAGAGTANRV 552
Query: 515 TWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
W G+ +I AAEA K+T Q I G WL +TG F GL
Sbjct: 553 KWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLGL 592
>At3g59010 pectinesterase precursor-like protein
Length = 529
Score = 401 bits (1031), Expect = e-112
Identities = 221/442 (50%), Positives = 290/442 (65%), Gaps = 24/442 (5%)
Query: 122 LNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGLEGSSR---- 177
+NDC EL+D ++D ++ V+ K++++ D HTWLS+ LTN TC L S
Sbjct: 102 VNDCLELLDDTLDMLYRIVVIKRKDHVND--DVHTWLSAALTNQETCKQSLSEKSSFNKE 159
Query: 178 -VVMESDLHDLISRARSSLAVLVSVLPPKANDGFID--EKL--NGDFPSWVTSKDRRLLE 232
+ ++S +L +SL + VS ++ + KL + DFP+WV+S DR+LLE
Sbjct: 160 GIAIDSFARNLTGLLTNSLDMFVSDKQKSSSSSNLTGGRKLLSDHDFPTWVSSSDRKLLE 219
Query: 233 SSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNV 292
+SV +++ + VVA DGSG +VA+A+AS + G R VI++ GTYKEN+ I K+ NV
Sbjct: 220 ASVEELRPHAVVAADGSGTHMSVAEALASL-EKGSGRSVIHLTAGTYKENLNIPSKQKNV 278
Query: 293 MLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRV 352
MLVGDG T+I GS + G T++SATVAA+GDGFIA+DI F N+AGP QAVALRV
Sbjct: 279 MLVGDGKGKTVIVGSRSNRGGWNTYQSATVAAMGDGFIARDITFVNSAGPNSEQAVALRV 338
Query: 353 GADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMA 412
G+D+SV+ RC ID +QD+LY S RQFYR++ ITGTVDFIFGN+AVVFQ L +RK +
Sbjct: 339 GSDRSVVYRCSIDGYQDSLYTLSKRQFYRETDITGTVDFIFGNSAVVFQSCNLVSRKGSS 398
Query: 413 NQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSV 472
+Q N VTAQGR DPNQNT SI C + GS KTYLGRPWK+YSRTVV+QS
Sbjct: 399 DQ-NYVTAQGRSDPNQNTGISIHNC---------RITGSTKTYLGRPWKQYSRTVVMQSF 448
Query: 473 VDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFT 532
+DG I P+GW+ W +S L+TLYYGE+ NSG G+ RV+W GYH EA FT
Sbjct: 449 IDGSIHPSGWSPW--SSNFALKTLYYGEFGNSGPGSSVSGRVSWAGYHPALTLTEAQGFT 506
Query: 533 VTQLIQGNVWLKNTGVAFIEGL 554
V+ I GN WL +TGV F GL
Sbjct: 507 VSGFIDGNSWLPSTGVVFDSGL 528
>At2g45220 pectinesterase like protein
Length = 511
Score = 394 bits (1013), Expect = e-110
Identities = 243/543 (44%), Positives = 327/543 (59%), Gaps = 42/543 (7%)
Query: 22 VLSLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALDTKSC---LTHVS--EVVQGST 76
+++ A I + ++ + +S +N C + K C LTH S E ++ +
Sbjct: 1 MMAFRAYIINFVILCILVASTVSGYNQKDVKAWCSQTPNPKPCEYFLTHNSNNEPIKSES 60
Query: 77 LSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEIALNDCEELMDLSMDRV 136
KL ++L K+ A + + RE+ A DC +L DL++ ++
Sbjct: 61 EFLKISMKLVLDRAILAKTHAF---------TLGPKCRDTREKAAWEDCIKLYDLTVSKI 111
Query: 137 WDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDG-LE-GSSRVV---MESDLHDLISRA 191
+ T+ N S+ DA TWLS+ LTN TC G LE G + +V M +++ +L+
Sbjct: 112 NE---TMDPNVKCSKLDAQTWLSTALTNLDTCRAGFLELGVTDIVLPLMSNNVSNLLCNT 168
Query: 192 RSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGK 251
+ V + PP+ DGF PSWV DR+LL+SS K N VVA+DGSG
Sbjct: 169 LAINKVPFNYTPPE-KDGF---------PSWVKPGDRKLLQSSTP--KDNAVVAKDGSGN 216
Query: 252 FKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFI 311
FKT+ +A+ +A +G R+VIYVK+G Y EN+EI KK NVML GDG+ TIITGS +
Sbjct: 217 FKTIKEAIDAASGSG--RFVIYVKQGVYSENLEIRKK--NVMLRGDGIGKTIITGSKSVG 272
Query: 312 DGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTL 371
GTTTF SATVAAVGDGFIA+ I F+NTAG QAVALR G+D SV +C +A+QDTL
Sbjct: 273 GGTTTFNSATVAAVGDGFIARGITFRNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTL 332
Query: 372 YAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTA 431
Y HSNRQFYRD + GTVDFIFGNAA V Q + AR+P ++ N +TAQGR DPNQNT
Sbjct: 333 YVHSNRQFYRDCDVYGTVDFIFGNAAAVLQNCNIFARRP-RSKTNTITAQGRSDPNQNTG 391
Query: 432 TSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKD 491
I V +SDL+PV GS KTYLGRPW++YSRTV +++ +D IDP GW EWD
Sbjct: 392 IIIHNSRVTAASDLRPVLGSTKTYLGRPWRQYSRTVFMKTSLDSLIDPRGWLEWD--GNF 449
Query: 492 FLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFI 551
L+TL+Y E+ N+G GA T RVTWPG+ ++ +A+EASKFTV + G W+ ++ V F
Sbjct: 450 ALKTLFYAEFQNTGPGASTSGRVTWPGFRVLGSASEASKFTVGTFLAGGSWIPSS-VPFT 508
Query: 552 EGL 554
GL
Sbjct: 509 SGL 511
>At2g26440 putative pectinesterase
Length = 547
Score = 384 bits (986), Expect = e-107
Identities = 217/544 (39%), Positives = 320/544 (57%), Gaps = 19/544 (3%)
Query: 24 SLVAIISSSALIISHLNKPISFFNLSSAPNLCEHALDTKSCLTHVSEVVQGSTLSNTKDH 83
SL+ ++ + + S+ +P + +SA + C++ +C T + + + N
Sbjct: 10 SLLFLLFFTPSVFSYSYQPSLNPHETSATSFCKNTPYPDACFTSLKLSISINISPNILSF 69
Query: 84 KLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSPREEIALNDCEELMDLSMDRVWDSVLTL 143
L TL + L+++ + + A V V R +L DC++L ++ + S+ +
Sbjct: 70 LLQTLQTALSEA-GKLTDLLSGAGVSNNLVEGQRG--SLQDCKDLHHITSSFLKRSISKI 126
Query: 144 TKNNIDSQH--DAHTWLSSVLTNHATCLDGLEGSSR------VVMESDLHDLISRARSSL 195
DS+ DA +LS+ LTN TCL+GLE +S V + + IS + S+L
Sbjct: 127 QDGVNDSRKLADARAYLSAALTNKITCLEGLESASGPLKPKLVTSFTTTYKHISNSLSAL 186
Query: 196 AVLVSVLPPKANDGFIDEKLNGDFPSWVTSKDRRLLESSVG-----DIKANVVVAQDGSG 250
PK + +L G FP WV KD R LE S D ++VVA DG+G
Sbjct: 187 PKQRRTTNPKTGGNTKNRRLLGLFPDWVYKKDHRFLEDSSDGYDEYDPSESLVVAADGTG 246
Query: 251 KFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNF 310
F T+ +A++ AP+ R +IYVK+G Y ENI+I KTN++L+GDG D T ITG+ +
Sbjct: 247 NFSTINEAISFAPNMSNDRVLIYVKEGVYDENIDIPIYKTNIVLIGDGSDVTFITGNRSV 306
Query: 311 IDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDT 370
DG TTF+SAT+A G+GF+A+DI NTAGP+KHQAVALRV AD + RC ID +QDT
Sbjct: 307 GDGWTTFRSATLAVSGEGFLARDIMITNTAGPEKHQAVALRVNADFVALYRCVIDGYQDT 366
Query: 371 LYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNT 430
LY HS RQFYR+ I GT+D+IFGNAAVVFQ + ++ PM Q ++TAQ R+ +++T
Sbjct: 367 LYTHSFRQFYRECDIYGTIDYIFGNAAVVFQGCNIVSKLPMPGQFTVITAQSRDTQDEDT 426
Query: 431 ATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASK 490
S+Q C ++ S DL +K+YLGRPW+++SRTVV++S +D ID +GW++W+
Sbjct: 427 GISMQNCSILASEDLFNSSNKVKSYLGRPWREFSRTVVMESYIDEFIDGSGWSKWNGG-- 484
Query: 491 DFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAF 550
+ L TLYYGEY N+G G+ T KRV WPG+HI+ +A FT T+ I G+ WL +T +
Sbjct: 485 EALDTLYYGEYNNNGPGSETVKRVNWPGFHIM-GYEDAFNFTATEFITGDGWLGSTSFPY 543
Query: 551 IEGL 554
G+
Sbjct: 544 DNGI 547
>At4g02320 pectinesterase - like protein
Length = 518
Score = 383 bits (984), Expect = e-106
Identities = 213/505 (42%), Positives = 304/505 (60%), Gaps = 38/505 (7%)
Query: 69 SEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVNSP---REEIALNDC 125
S V Q T S D +V+ L ++ +++ + + + +R+ S R+ A +DC
Sbjct: 25 SHVTQRFTSS---DDITELVVTALNQTISNVNLSSSNFSDLLQRLGSNLSHRDLCAFDDC 81
Query: 126 EELMDLSMDRVWDSVLTLTKNNIDSQ--HDAHTWLSSVLTNHATCLDGLEGSSR------ 177
EL+D D V+D ++K S H+ LS+ +TN TCLDG S
Sbjct: 82 LELLD---DTVFDLTTAISKLRSHSPELHNVKMLLSAAMTNTRTCLDGFASSDNDENLNN 138
Query: 178 ---------VVMESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGD--FPSWVTSK 226
++ L ++ S SLA+L ++ G I K+ D FP WV+
Sbjct: 139 NDNKTYGVAESLKESLFNISSHVSDSLAMLENI------PGHIPGKVKEDVGFPMWVSGS 192
Query: 227 DRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIG 286
DR LL+ V + K N+VVAQ+G+G + T+ +A+++AP++ +TR+VIY+K G Y ENIEI
Sbjct: 193 DRNLLQDPVDETKVNLVVAQNGTGNYTTIGEAISAAPNSSETRFVIYIKCGEYFENIEIP 252
Query: 287 KKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQ 346
++KT +M +GDG+ T+I + ++ DG T F SATV G GFIA+D+ F N AGP+KHQ
Sbjct: 253 REKTMIMFIGDGIGRTVIKANRSYADGWTAFHSATVGVRGSGFIAKDLSFVNYAGPEKHQ 312
Query: 347 AVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLA 406
AVALR +D S RC +++QDT+Y HS++QFYR+ I GTVDFIFG+A+VVFQ L
Sbjct: 313 AVALRSSSDLSAYYRCSFESYQDTIYVHSHKQFYRECDIYGTVDFIFGDASVVFQNCSLY 372
Query: 407 ARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRT 466
AR+P NQK + TAQGRE+ + T SI ++ + DL PVQ + K YLGRPW+ YSRT
Sbjct: 373 ARRPNPNQKIIYTAQGRENSREPTGISIISSRILAAPDLIPVQANFKAYLGRPWQLYSRT 432
Query: 467 VVLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNA 525
V+++S +D +DPAGW +W DF L+TLYYGEYMN G G+ RV WPG+ I+
Sbjct: 433 VIMKSFIDDLVDPAGWLKW---KDDFALETLYYGEYMNEGPGSNMTNRVQWPGFKRIETV 489
Query: 526 AEASKFTVTQLIQGNVWLKNTGVAF 550
EAS+F+V I GN WL +T + F
Sbjct: 490 EEASQFSVGPFIDGNKWLNSTRIPF 514
>At2g43050 putative pectinesterase
Length = 518
Score = 382 bits (980), Expect = e-106
Identities = 214/443 (48%), Positives = 283/443 (63%), Gaps = 34/443 (7%)
Query: 122 LNDCEELMDLSMDRVWDSVLTLTKNNIDS-QHDAHTWLSSVLTNHATCLDGLEGSSRVVM 180
++DC EL+D ++D L++ + D+ + D HTWLS+ LTN TC L+ S
Sbjct: 98 IHDCLELLDDTLDM-------LSRIHADNDEEDVHTWLSAALTNQDTCEQSLQEKSESYK 150
Query: 181 ESDLHDLISR-----ARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWV-TSKDRRLLESS 234
D ++R SSL + VSV K+ + K FP++V +S+ RRLLE+
Sbjct: 151 HGLAMDFVARNLTGLLTSSLDLFVSV---KSKHRKLLSKQEY-FPTFVPSSEQRRLLEAP 206
Query: 235 VGDIKANVVVAQDGSGKFKTVAQAVAS---APDNGKTRYVIYVKKGTYKENIEIGKKKTN 291
V ++ + VVA DGSG KT+ +A+ S A G+T+ IY+K GTY ENI I K+ N
Sbjct: 207 VEELNVDAVVAPDGSGTHKTIGEALLSTSLASSGGRTK--IYLKAGTYHENINIPTKQKN 264
Query: 292 VMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALR 351
VMLVGDG T+I GS + G TT+K+ATVAA+G+GFIA+D+ F N AGP+ QAVALR
Sbjct: 265 VMLVGDGKGKTVIVGSRSNRGGWTTYKTATVAAMGEGFIARDMTFVNNAGPKSEQAVALR 324
Query: 352 VGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPM 411
VGAD+SV++RC ++ +QD+LY HS RQFYR++ ITGTVDFIFGN+AVVFQ +AARKP+
Sbjct: 325 VGADKSVVHRCSVEGYQDSLYTHSKRQFYRETDITGTVDFIFGNSAVVFQSCNIAARKPL 384
Query: 412 ANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQS 471
Q+N VTAQGR +P QNT +IQ C + S TYLGRPWK+YSRTVV+QS
Sbjct: 385 PGQRNFVTAQGRSNPGQNTGIAIQNCRITAES---------MTYLGRPWKEYSRTVVMQS 435
Query: 472 VVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKF 531
+ G I P+GW+ W L++L+YGEY NSG G+ RV W G H EA KF
Sbjct: 436 FIGGSIHPSGWSPWSGGFG--LKSLFYGEYGNSGPGSSVSGRVKWSGCHPSLTVTEAEKF 493
Query: 532 TVTQLIQGNVWLKNTGVAFIEGL 554
TV I GN+WL +TGV+F GL
Sbjct: 494 TVASFIDGNIWLPSTGVSFDPGL 516
>At4g33220 pectinesterase like protein
Length = 404
Score = 376 bits (965), Expect = e-104
Identities = 197/405 (48%), Positives = 261/405 (63%), Gaps = 27/405 (6%)
Query: 170 DGLEGSSRVVMESDLHDLISRARSSLAVLVSVLPPKA-------------------NDGF 210
DG G + ++ L L S R L ++ PKA D
Sbjct: 5 DGTSGLVKSLVAGSLDQLYSMLRELLPLVQPEQKPKAVSKPGPIAKGPKAPPGRKLRDTD 64
Query: 211 IDEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRY 270
DE L FP WV DR+LLES+ +V VA DG+G F + A+ APD TR+
Sbjct: 65 EDESLQ--FPDWVRPDDRKLLESNGRTY--DVSVALDGTGNFTKIMDAIKKAPDYSSTRF 120
Query: 271 VIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFI 330
VIY+KKG Y EN+EI KKK N++++GDG+D T+I+G+ +FIDG TTF+SAT A G GF+
Sbjct: 121 VIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNRSFIDGWTTFRSATFAVSGRGFL 180
Query: 331 AQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVD 390
A+DI FQNTAGP+KHQAVALR +D SV RC + +QDTLY H+ RQFYR+ ITGTVD
Sbjct: 181 ARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECTITGTVD 240
Query: 391 FIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQG 450
FIFG+ VVFQ ++ A++ + NQKN +TAQGR+D NQ + SIQ ++ +DL P
Sbjct: 241 FIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADADLVPYLN 300
Query: 451 SIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAG 509
+ +TYLGRPWK YSRTV +++ + + P GW EW+A DF L TL+YGE+MN G G+G
Sbjct: 301 TTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNA---DFALDTLFYGEFMNYGPGSG 357
Query: 510 TGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
RV WPGYH+ N+ +A+ FTV+Q I+GN+WL +TGV F +GL
Sbjct: 358 LSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGL 402
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 375 bits (962), Expect = e-104
Identities = 208/553 (37%), Positives = 320/553 (57%), Gaps = 31/553 (5%)
Query: 26 VAIISSSALIISHLNKPISFFNLSSAPNLCEHALDTK-------SCLTHVSEVVQGSTLS 78
V I SS L+IS + +++ + N + + T T E + +
Sbjct: 17 VIISISSVLLISMVVAVTIGVSVNKSDNAGDEEITTSVKAIKDVCAPTDYKETCEDTLRK 76
Query: 79 NTKDHKLSTLVSLLTKSTAHIRKAMDTANVIKRRVN---SPREEIALNDCEELMDLSMDR 135
+ KD L + T A +++ D A + + PR ++AL+ C+ELMD ++
Sbjct: 77 DAKDTS-DPLELVKTAFNATMKQISDVAKKSQTMIELQKDPRAKMALDQCKELMDYAIGE 135
Query: 136 VWDSVLTLTKNNIDSQHDA----HTWLSSVLTNHATCLDGLEGS---SRVVMESDLHDLI 188
+ S L K +A WLS+ +++ TCLDG +G+ + ++ L +
Sbjct: 136 LSKSFEELGKFEFHKVDEALVKLRIWLSATISHEQTCLDGFQGTQGNAGETIKKALKTAV 195
Query: 189 SRARSSLAVLVSVLP-------PKANDGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKAN 241
+ LA++ + P+ N + L+ +FPSW+ ++ RRLL + + ++K +
Sbjct: 196 QLTHNGLAMVTEMSNYLGQMQIPEMNSRRL---LSQEFPSWMDARARRLLNAPMSEVKPD 252
Query: 242 VVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDA 301
+VVAQDGSG++KT+ +A+ P T +V+++K+G YKE +++ + T+++ +GDG D
Sbjct: 253 IVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEGIYKEYVQVNRSMTHLVFIGDGPDK 312
Query: 302 TIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINR 361
T+I+GS ++ DG TT+K+ATVA VGD FIA++I F+NTAG KHQAVA+RV AD+S+
Sbjct: 313 TVISGSKSYKDGITTYKTATVAIVGDHFIAKNIAFENTAGAIKHQAVAIRVLADESIFYN 372
Query: 362 CKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQ 421
CK D +QDTLYAHS+RQFYRD I+GT+DF+FG+AA VFQ L RKP+ NQ +TA
Sbjct: 373 CKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDAAAVFQNCTLLVRKPLLNQACPITAH 432
Query: 422 GREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAG 481
GR+DP ++T +Q C ++ D V+ KTYLGRPWK+YSRT+++ + + + P G
Sbjct: 433 GRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTYLGRPWKEYSRTIIMNTFIPDFVPPEG 492
Query: 482 WAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNV 541
W W + L TL+Y E N+G GA KRVTWPG + + E KFT Q IQG+
Sbjct: 493 WQPW--LGEFGLNTLFYSEVQNTGPGAAITKRVTWPGIKKLSD-EEILKFTPAQYIQGDA 549
Query: 542 WLKNTGVAFIEGL 554
W+ GV +I GL
Sbjct: 550 WIPGKGVPYILGL 562
>At3g05620 putative pectinesterase
Length = 543
Score = 374 bits (961), Expect = e-104
Identities = 221/490 (45%), Positives = 285/490 (58%), Gaps = 53/490 (10%)
Query: 97 AHIRKAMDTANVIKRRVNS-------PREEIALNDCEELMDLSMDRVWDSVLTLTKNN-- 147
A +++A D A + R+ + RE++A+ DC+EL+ S+ + S+L + K +
Sbjct: 73 AAVKEAHDKAKLAMERIPTVMMLSIRSREQVAIEDCKELVGFSVTELAWSMLEMNKLHGG 132
Query: 148 -----IDSQHDA-------HTWLSSVLTNHATCLDGLEGSSRVVME------SDLHDLIS 189
D HDA TWLS+ ++N TCL+G EG+ R E + L+S
Sbjct: 133 GGIDLDDGSHDAAAAGGNLKTWLSAAMSNQDTCLEGFEGTERKYEELIKGSLRQVTQLVS 192
Query: 190 RARSSLAVLVSVLPPKA--NDGFIDEKLNGDFPSWVTSKDRRLL---ESSVGDIKANVVV 244
L + LP KA N+ I P W+T D L+ + SV + N VV
Sbjct: 193 NVLDMYTQL-NALPFKASRNESVIAS------PEWLTETDESLMMRHDPSV--MHPNTVV 243
Query: 245 AQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATII 304
A DG GK++T+ +A+ AP++ RYVIYVKKG YKENI++ KKKTN+MLVGDG+ TII
Sbjct: 244 AIDGKGKYRTINEAINEAPNHSTKRYVIYVKKGVYKENIDLKKKKTNIMLVGDGIGQTII 303
Query: 305 TGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKI 364
TG NF+ G TTF++ATVA G GFIA+DI F+NTAGPQ QAVALRV +DQS RC +
Sbjct: 304 TGDRNFMQGLTTFRTATVAVSGRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAFYRCSV 363
Query: 365 DAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGRE 424
+ +QDTLYAHS RQFYRD I GT+DFIFGN A V Q K+ R P+ QK +TAQGR+
Sbjct: 364 EGYQDTLYAHSLRQFYRDCEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRK 423
Query: 425 DPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAE 484
PNQNT IQ V+ + TYLGRPWK YSRTV + + + + P GW E
Sbjct: 424 SPNQNTGFVIQNSYVLATQ---------PTYLGRPWKLYSRTVYMNTYMSQLVQPRGWLE 474
Query: 485 WDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLK 544
W L TL+YGEY N G G + RV WPGYHI+ + A FTV I G WL
Sbjct: 475 W--FGNFALDTLWYGEYNNIGPGWRSSGRVKWPGYHIM-DKRTALSFTVGSFIDGRRWLP 531
Query: 545 NTGVAFIEGL 554
TGV F GL
Sbjct: 532 ATGVTFTAGL 541
>At4g02300 putative pectinesterase
Length = 532
Score = 373 bits (957), Expect = e-103
Identities = 202/452 (44%), Positives = 283/452 (61%), Gaps = 26/452 (5%)
Query: 118 EEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGL----- 172
E A DC L+D ++ + +V L ++++ +D L++V+T TCLDG
Sbjct: 90 ERCAFEDCLGLLDDTISDLETAVSDLRSSSLEF-NDISMLLTNVMTYQDTCLDGFSTSDN 148
Query: 173 EGSSRVVMES---------DLHDLISRARSSLAVLVSVLP-PKANDGFIDEKLNGDFPSW 222
E ++ + E D+ + +S + L V+ P PK+++ ++ ++PSW
Sbjct: 149 ENNNDMTYELPENLKEIILDISNNLSNSLHMLQVISRKKPSPKSSE------VDVEYPSW 202
Query: 223 VTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKEN 282
++ D+RLLE+ V + N+ VA DG+G F T+ AV +AP+ +TR++IY+K G Y EN
Sbjct: 203 LSENDQRLLEAPVQETNYNLSVAIDGTGNFTTINDAVFAAPNMSETRFIIYIKGGEYFEN 262
Query: 283 IEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGP 342
+E+ KKKT +M +GDG+ T+I + + IDG +TF++ TV G G+IA+DI F N+AGP
Sbjct: 263 VELPKKKTMIMFIGDGIGKTVIKANRSRIDGWSTFQTPTVGVKGKGYIAKDISFVNSAGP 322
Query: 343 QKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQK 402
K QAVA R G+D S RC+ D +QDTLY HS +QFYR+ I GT+DFIFGNAAVVFQ
Sbjct: 323 AKAQAVAFRSGSDHSAFYRCEFDGYQDTLYVHSAKQFYRECDIYGTIDFIFGNAAVVFQN 382
Query: 403 SKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKK 462
S L ARKP K TAQ R +Q T SI C ++ + DL PV+ + K YLGRPW+K
Sbjct: 383 SSLYARKPNPGHKIAFTAQSRNQSDQPTGISILNCRILAAPDLIPVKENFKAYLGRPWRK 442
Query: 463 YSRTVVLQSVVDGHIDPAGWAEWDAASKDF-LQTLYYGEYMNSGAGAGTGKRVTWPGYHI 521
YSRTV+++S +D I PAGW E KDF L+TLYYGEYMN G GA KRVTWPG+
Sbjct: 443 YSRTVIIKSFIDDLIHPAGWLE---GKKDFALETLYYGEYMNEGPGANMAKRVTWPGFRR 499
Query: 522 IKNAAEASKFTVTQLIQGNVWLKNTGVAFIEG 553
I+N EA++FTV I G+ WL +TG+ F G
Sbjct: 500 IENQTEATQFTVGPFIDGSTWLNSTGIPFSLG 531
>At3g60730 pectinesterase - like protein
Length = 496
Score = 372 bits (955), Expect = e-103
Identities = 208/452 (46%), Positives = 278/452 (61%), Gaps = 37/452 (8%)
Query: 119 EIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCLDGL----EG 174
++ L++CE+L D S R+ S L + N + D TWLS VL NH TCLDGL +G
Sbjct: 66 KLGLSECEKLYDESEARL--SKLVVDHENFTVE-DVRTWLSGVLANHHTCLDGLIQQRQG 122
Query: 175 SSRVVMESD---LHDLISRARSSLA------VLVSVLPPKANDGFIDEKLNGDFPSWVTS 225
+V + LH+ ++ + S A P + N G + PS
Sbjct: 123 HKPLVHSNVTFVLHEALAFYKKSRARQGHGPTRPKHRPTRPNHG---PGRSHHGPSRPNQ 179
Query: 226 KDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRY---VIYVKKGTYKEN 282
L+ + +A+ VVA+DGS +T+ QA+A+ GK+R +IY+K G Y E
Sbjct: 180 NGGMLVSWNPTSSRADFVVARDGSATHRTINQALAAVSRMGKSRLNRVIIYIKAGVYNEK 239
Query: 283 IEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGP 342
IEI + N+MLVGDGMD TI+T + N DG+TT+ SAT GDGF A+DI F+NTAGP
Sbjct: 240 IEIDRHMKNIMLVGDGMDRTIVTNNRNVPDGSTTYGSATFGVSGDGFWARDITFENTAGP 299
Query: 343 QKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQK 402
KHQAVALRV +D S+ RC +QDTL+ HS RQFYRD +I GT+DFIFG+AA VFQ
Sbjct: 300 HKHQAVALRVSSDLSLFYRCSFKGYQDTLFTHSLRQFYRDCHIYGTIDFIFGDAAAVFQN 359
Query: 403 SKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKK 462
+ R+PM +Q NM+TAQGR+DP+ N S+ + V+G K+YLGRPWKK
Sbjct: 360 CDIFVRRPMDHQGNMITAQGRDDPHTN-------------SEFEAVKGRFKSYLGRPWKK 406
Query: 463 YSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHII 522
YSRTV L++ +D IDP GW EW + L TLYYGE+MN+GAGAGTG+RV WPG+H++
Sbjct: 407 YSRTVFLKTDIDELIDPRGWREWSGSYA--LSTLYYGEFMNTGAGAGTGRRVNWPGFHVL 464
Query: 523 KNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+ EAS FTV++ IQG+ W+ TGV F G+
Sbjct: 465 RGEEEASPFTVSRFIQGDSWIPITGVPFSAGV 496
>At1g11590 putative pectin methylesterase
Length = 524
Score = 371 bits (953), Expect = e-103
Identities = 213/510 (41%), Positives = 312/510 (60%), Gaps = 19/510 (3%)
Query: 56 EHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAM-DTANVIKRRVN 114
E+ D +SC + E+ GS+ S ++ + L+ +L S I M + K+
Sbjct: 23 EYCDDKQSCQNFLLELKAGSS-SLSEIRRRDLLIIVLKNSVRRIDMVMIGVMDDTKQHEE 81
Query: 115 SPREEIALND----CEELMDLSMDRVWDSVLTLTKN---NIDSQHDAHTWLSSVLTNHAT 167
+ + + + EE+M+ + DR+ SV L N S + HTWLSSVLT++ T
Sbjct: 82 MENDLLGVKEDTKLFEEMMESTKDRMIRSVEELLGGEFPNRGSYENVHTWLSSVLTSYIT 141
Query: 168 CLDGL-EGSSRVVMESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTSK 226
C+D + EG+ + +E L DLISRAR +LA+ +S+ P+ N I N PSW+
Sbjct: 142 CIDEIGEGAYKRRVEPKLEDLISRARIALALFISI-SPRDNTELISVIPNS--PSWLFHV 198
Query: 227 DRRLLESSVGDIK--ANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTYKENIE 284
D++ L + +K A+VVVA+DG+GK+ TV A+A+AP + + R+VIY+K G Y E +
Sbjct: 199 DKKDLYLNAEALKKIADVVVAKDGTGKYSTVNAAIAAAPQHSQKRFVIYIKTGIYDEIVV 258
Query: 285 IGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNTAGPQK 344
I K N+ L+GDG D TIIT +L+ + TF +ATVA+ G+GFI D+ F+NTAGP K
Sbjct: 259 IENTKPNLTLIGDGQDLTIITSNLSASNVRRTFNTATVASNGNGFIGVDMCFRNTAGPAK 318
Query: 345 HQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVVFQKSK 404
AVALRV D SVI RC+++ +QD LY HS+RQFYR+ +ITGTVDFI GNA VFQ +
Sbjct: 319 GPAVALRVSGDMSVIYRCRVEGYQDALYPHSDRQFYRECFITGTVDFICGNAVAVFQFCQ 378
Query: 405 LAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRPWKKYS 464
+ AR+P Q N++TAQ R + + +IQ+C++ SSDL ++KTYLGRPW+ +S
Sbjct: 379 IVARQPKMGQSNVITAQSRAFKDIYSGFTIQKCNITASSDLDTT--TVKTYLGRPWRIFS 436
Query: 465 RTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGYHIIKN 524
V+QS + +DPAGW W+ + L TL+Y EY N G GA T +RV W G+ ++K+
Sbjct: 437 TVAVMQSFIGDLVDPAGWTPWEGETG--LSTLHYREYQNRGPGAVTSRRVKWSGFKVMKD 494
Query: 525 AAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+A++FTV +L+ G WLK T + + GL
Sbjct: 495 PKQATEFTVAKLLDGETWLKETRIPYESGL 524
>At5g04960 pectinesterase
Length = 564
Score = 369 bits (948), Expect = e-102
Identities = 214/511 (41%), Positives = 295/511 (56%), Gaps = 27/511 (5%)
Query: 50 SAPNLCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRKAMDTANVI 109
S LC+ L + C + S S + K + V++ + K +D +
Sbjct: 67 SVKALCDVTLHKEKCFETLGSAPNASRSSPEELFKYAVKVTI-----TELSKVLDGFSNG 121
Query: 110 KRRVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKNNIDSQHDAHTWLSSVLTNHATCL 169
+ N+ A+ C EL+ L++D++ + +T + N D D TWLSSV T TC+
Sbjct: 122 EHMDNAT--SAAMGACVELIGLAVDQL-NETMTSSLKNFD---DLRTWLSSVGTYQETCM 175
Query: 170 DGLEGSSRVVM----ESDLHDLISRARSSLAVLVSVLPPKANDGFIDEKLNGDFPSWVTS 225
D L +++ + E+ L + ++LA++ + F +L + V
Sbjct: 176 DALVEANKPSLTTFGENHLKNSTEMTSNALAIITWLGKIADTVKFRRRRLLETGNAKVVV 235
Query: 226 KD------RRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGKTRYVIYVKKGTY 279
D RRLLES KA +VVA+DGSGK++T+ +A+A + + +IYVKKG Y
Sbjct: 236 ADLPMMEGRRLLESGDLKKKATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIYVKKGVY 295
Query: 280 KENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAVGDGFIAQDIRFQNT 339
EN+ + K K NV++VGDG TI++ LNFIDGT TF++AT A G GF+A+D+ F NT
Sbjct: 296 LENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARDMGFINT 355
Query: 340 AGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYITGTVDFIFGNAAVV 399
AGP KHQAVAL V AD SV +C +DAFQDT+YAH+ RQFYRD I GTVDFIFGNAAVV
Sbjct: 356 AGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIFGNAAVV 415
Query: 400 FQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDLKPVQGSIKTYLGRP 459
FQK ++ R+PM Q+N +TAQGR+DPNQNT SI C + P +L +Q T+LGRP
Sbjct: 416 FQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNLTDIQ----TFLGRP 471
Query: 460 WKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSGAGAGTGKRVTWPGY 519
WK +S TV+++S +D I+P GW W + T++Y EY+NSG GA T RV W G
Sbjct: 472 WKDFSTTVIMKSFMDKFINPKGWLPWTGDTAP--DTIFYAEYLNSGPGASTKNRVKWQGL 529
Query: 520 HIIKNAAEASKFTVTQLIQGNVWLKNTGVAF 550
EA+KFTV I GN WL T V F
Sbjct: 530 KTSLTKKEANKFTVKPFIDGNNWLPATKVPF 560
>At2g47550 unknown protein
Length = 560
Score = 369 bits (946), Expect = e-102
Identities = 225/541 (41%), Positives = 300/541 (54%), Gaps = 46/541 (8%)
Query: 47 NLSSAPNLCEHALDTKSCLTHVSEVVQGSTLSNTKDHKLSTLVSLLTKSTAHIRK---AM 103
N +S N+C A D C S L N S L +S + R+ +
Sbjct: 31 NATSPSNICRFAPDPSYCR---------SVLPNQPGDIYSYGRLSLRRSLSRARRFISMI 81
Query: 104 DTANVIKRRVNSPREEIALNDCEELMDLSMDRVWDSVLTLTKN---NIDSQHDAHTWLSS 160
D K +V + AL DC+ L L+MD + S T ++ D HT+LS+
Sbjct: 82 DAELDRKGKVAAKSTVGALEDCKFLASLTMDYLLSSSQTADSTKTLSLSRAEDVHTFLSA 141
Query: 161 VLTNHATCLDGLEGS-SRVVMESDLHDLISRARSSLAVLVS-----------VLPPKAND 208
+TN TCL+GL+ + S + DL + SLA+ + P+A
Sbjct: 142 AITNEQTCLEGLKSTASENGLSGDLFNDTKLYGVSLALFSKGWVPRRQRSRPIWQPQARF 201
Query: 209 GFIDEKLNGDFPSWVTSKDR---------RLLESSVGDIKAN--VVVAQDGSGKFKTVAQ 257
NG P +T + R +LL+S ++ + V V Q+G+G F T+
Sbjct: 202 KKFFGFRNGKLPLKMTERARAVYNTVTRRKLLQSDADAVQVSDIVTVIQNGTGNFTTINA 261
Query: 258 AVASAP---DNGKTRYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGT 314
A+A+AP D ++IYV G Y+E +E+ K K VM++GDG++ T+ITG+ + +DG
Sbjct: 262 AIAAAPNKTDGSNGYFLIYVTAGLYEEYVEVPKNKRYVMMIGDGINQTVITGNRSVVDGW 321
Query: 315 TTFKSATVAAVGDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAH 374
TTF SAT G FI +I +NTAGP K QAVALR G D SV C +A+QDTLY H
Sbjct: 322 TTFNSATFILSGPNFIGVNITIRNTAGPTKGQAVALRSGGDLSVFYSCSFEAYQDTLYTH 381
Query: 375 SNRQFYRDSYITGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSI 434
S RQFYR+ + GTVDFIFGNAAVV Q L R+P Q N VTAQGR DPNQNT T+I
Sbjct: 382 SLRQFYRECDVYGTVDFIFGNAAVVLQNCNLYPRQPRKGQSNEVTAQGRTDPNQNTGTAI 441
Query: 435 QQCDVIPSSDLKPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDF-L 493
C + P+ DL ++KTYLGRPWK+YSRTVV+Q+ +DG ++P+GW W S DF L
Sbjct: 442 HGCTIRPADDLATSNYTVKTYLGRPWKEYSRTVVMQTYIDGFLEPSGWNAW---SGDFAL 498
Query: 494 QTLYYGEYMNSGAGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEG 553
TLYY EY N+G G+ T RVTWPGYH+I NA +AS FTVT + G W+ TGV F+ G
Sbjct: 499 STLYYAEYNNTGPGSDTTNRVTWPGYHVI-NATDASNFTVTNFLVGEGWIGQTGVPFVGG 557
Query: 554 L 554
L
Sbjct: 558 L 558
>At5g51490 pectinesterase
Length = 536
Score = 366 bits (940), Expect = e-101
Identities = 194/409 (47%), Positives = 257/409 (62%), Gaps = 15/409 (3%)
Query: 153 DAHTWLSSVLTNHATCLDGLEGSSRVVMESDLHDLISRAR-----SSLAVLVSVLPPKAN 207
DA TWLS+ LTN TC GSS + + + ++S + S+ + L N
Sbjct: 136 DAQTWLSTALTNTETCR---RGSSDLNVTDFITPIVSNTKISHLISNCLAVNGALLTAGN 192
Query: 208 DGFIDEKLNGDFPSWVTSKDRRLLESSVGDIKANVVVAQDGSGKFKTVAQAVASAPDNGK 267
G G FP+W++ KD+RLL + ++AN+VVA+DGSG F TV A+ A
Sbjct: 193 KGNTTANQKG-FPTWLSRKDKRLLRA----VRANLVVAKDGSGHFNTVQAAIDVAGRRKV 247
Query: 268 T--RYVIYVKKGTYKENIEIGKKKTNVMLVGDGMDATIITGSLNFIDGTTTFKSATVAAV 325
T R+VIYVK+G Y+ENI + ++MLVGDGM +TIITG + G TT+ SAT
Sbjct: 248 TSGRFVIYVKRGIYQENINVRLNNDDIMLVGDGMRSTIITGGRSVQGGYTTYNSATAGIE 307
Query: 326 GDGFIAQDIRFQNTAGPQKHQAVALRVGADQSVINRCKIDAFQDTLYAHSNRQFYRDSYI 385
G FIA+ I F+NTAGP K QAVALR +D S+ +C I+ +QDTL HS RQFYR+ YI
Sbjct: 308 GLHFIAKGITFRNTAGPAKGQAVALRSSSDLSIFYKCSIEGYQDTLMVHSQRQFYRECYI 367
Query: 386 TGTVDFIFGNAAVVFQKSKLAARKPMANQKNMVTAQGREDPNQNTATSIQQCDVIPSSDL 445
GTVDFIFGNAA VFQ + R+P+ Q N++TAQGR DP QNT SI ++P+ DL
Sbjct: 368 YGTVDFIFGNAAAVFQNCLILPRRPLKGQANVITAQGRADPFQNTGISIHNSRILPAPDL 427
Query: 446 KPVQGSIKTYLGRPWKKYSRTVVLQSVVDGHIDPAGWAEWDAASKDFLQTLYYGEYMNSG 505
KPV G++KTY+GRPW K+SRTVVLQ+ +D + P GW+ W S L TL+Y EY N+G
Sbjct: 428 KPVVGTVKTYMGRPWMKFSRTVVLQTYLDNVVSPVGWSPWIEGSVFGLDTLFYAEYKNTG 487
Query: 506 AGAGTGKRVTWPGYHIIKNAAEASKFTVTQLIQGNVWLKNTGVAFIEGL 554
+ T RV+W G+H++ A++AS FTV + I G WL TG+ F GL
Sbjct: 488 PASSTRWRVSWKGFHVLGRASDASAFTVGKFIAGTAWLPRTGIPFTSGL 536
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,626,415
Number of Sequences: 26719
Number of extensions: 478036
Number of successful extensions: 1638
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 80
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 1279
Number of HSP's gapped (non-prelim): 123
length of query: 554
length of database: 11,318,596
effective HSP length: 104
effective length of query: 450
effective length of database: 8,539,820
effective search space: 3842919000
effective search space used: 3842919000
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC122166.11


