

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122165.6 + phase: 0
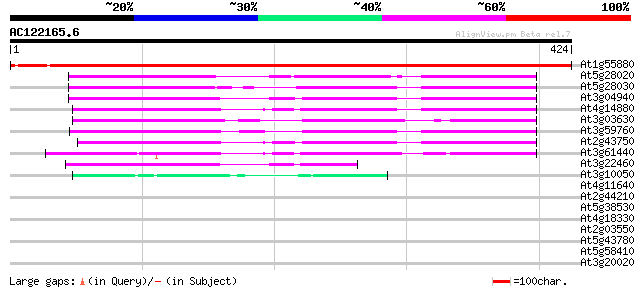
(424 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g55880 unknown protein 638 0.0
At5g28020 cysteine synthase 151 8e-37
At5g28030 cysteine synthase - like 150 1e-36
At3g04940 cysteine synthase (AtcysD1) 145 5e-35
At4g14880 cytosolic O-acetylserine(thiol)lyase (EC 4.2.99.8) 142 3e-34
At3g03630 O-acetylserine (thiol) lyase 139 3e-33
At3g59760 cysteine synthase 137 9e-33
At2g43750 cysteine synthase (cpACS1) 135 3e-32
At3g61440 cysteine synthase (AtcysC1) 131 8e-31
At3g22460 unknown protein 99 4e-21
At3g10050 threonine dehydratase/deaminase (OMR1) 49 7e-06
At4g11640 unknown protein 33 0.23
At2g44210 unknown protein 33 0.30
At5g38530 tryptophan synthase beta chain 32 0.66
At4g18330 translation initiation factor eIF-2 gamma chain-like p... 32 0.87
At2g03550 putative esterase 31 1.1
At5g43780 ATP sulfurylase precursor (gb|AAD26634.1) 30 1.9
At5g58410 unknown protein 28 7.3
At3g20020 arginine methyltransferase like protein 28 9.6
>At1g55880 unknown protein
Length = 421
Score = 638 bits (1646), Expect = 0.0
Identities = 321/425 (75%), Positives = 364/425 (85%), Gaps = 5/425 (1%)
Query: 1 MAPVAARNAGAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRI 60
MAPV GAV A+ + ++FF RL K+K+K +NG++DAIGNTPLIRI
Sbjct: 1 MAPV--NMTGAVVAAAALLMLTSYSFFF--RLSEKKKRKEKLTMRNGLVDAIGNTPLIRI 56
Query: 61 NSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGSAGSTAISI 120
NSLS+ATGCEILGKCEFLNPGGSVKDRVAV+II+EALESG+L GGIVTEGSAGSTAIS+
Sbjct: 57 NSLSEATGCEILGKCEFLNPGGSVKDRVAVKIIQEALESGKLFPGGIVTEGSAGSTAISL 116
Query: 121 ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVNIARRRASEAN 180
ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGA+VERVRPVSITHKDH+VNIARRRA EAN
Sbjct: 117 ATVAPAYGCKCHVVIPDDAAIEKSQIIEALGASVERVRPVSITHKDHYVNIARRRADEAN 176
Query: 181 EFAFKHRK-SQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFENLANFRAHYEGT 239
E A K R S+ G ++ NGC + K +LF + GGFFADQFENLAN+RAHYEGT
Sbjct: 177 ELASKRRLGSETNGIHQEKTNGCTVEEVKEPSLFSDSVTGGFFADQFENLANYRAHYEGT 236
Query: 240 GPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVM 299
GPEIW QT GN+DAFVAAAGTGGT+AGVS+FLQ+KN +KC+LIDPPGSGL+NKV RGVM
Sbjct: 237 GPEIWHQTQGNIDAFVAAAGTGGTLAGVSRFLQDKNERVKCFLIDPPGSGLYNKVTRGVM 296
Query: 300 YTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGL 359
YT+EEAEGRRLKNPFDTITEGIGINR+TKNF AKLDG FRGTD EAVEM+RFL+KNDGL
Sbjct: 297 YTREEAEGRRLKNPFDTITEGIGINRLTKNFLMAKLDGGFRGTDKEAVEMSRFLLKNDGL 356
Query: 360 FLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDSGMRHLSKFYNAEYLSQLGLTPKATGLE 419
F+GSSSAMNCVGAVR+AQ++GPGHTIVTILCDSGMRHLSKF++ +YL+ GL+P A GLE
Sbjct: 357 FVGSSSAMNCVGAVRVAQTLGPGHTIVTILCDSGMRHLSKFHDPKYLNLYGLSPTAIGLE 416
Query: 420 FLGIK 424
FLGIK
Sbjct: 417 FLGIK 421
>At5g28020 cysteine synthase
Length = 323
Score = 151 bits (381), Expect = 8e-37
Identities = 110/356 (30%), Positives = 161/356 (44%), Gaps = 59/356 (16%)
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
KN I + IGNTP++ +N++ D I K E + P SVKDR+A +I++A + G +
Sbjct: 8 KNDITELIGNTPMVYLNNVVDGCVARIAAKLEMMEPCSSVKDRIAYSMIKDAEDKGLITP 67
Query: 105 G-GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
G + E +AG+T I +A + A G K +V+P ++E+ I+ ALGA +
Sbjct: 68 GKSTLIEPTAGNTGIGLACMGAARGYKVILVMPSTMSLERRIILRALGAELHL------- 120
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
S Q G K K + + GG+
Sbjct: 121 --------------------------------SDQRIGLKGMLEKTEAIL-SKTPGGYIP 147
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
QFEN AN HY TGPEIW + G +D VA GTGGT GV KFL+E+N +IK ++
Sbjct: 148 QQFENPANPEIHYRTTGPEIWRDSAGKVDILVAGVGTGGTATGVGKFLKEQNKDIKVCVV 207
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTD 343
+P S V+ G P + +GIG V N +D +
Sbjct: 208 EPVES----PVLSG-------------GQPGPHLIQGIGSGIVPFNLDLTIVDEIIQVAG 250
Query: 344 MEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
EA+E A+ L +GL +G SS A+++A + G IV + G R+LS
Sbjct: 251 EEAIETAKLLALKEGLLVGISSGAAAAAALKVAKRPENAGKLIVVVFPSGGERYLS 306
>At5g28030 cysteine synthase - like
Length = 323
Score = 150 bits (379), Expect = 1e-36
Identities = 104/356 (29%), Positives = 161/356 (45%), Gaps = 59/356 (16%)
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
KN + + IGNTP++ +N + D I K E + P S+KDR+A +I++A + G +
Sbjct: 8 KNDVTELIGNTPMVYLNKIVDGCVARIAAKLEMMEPCSSIKDRIAYSMIKDAEDKGLITP 67
Query: 105 G-GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
G + E + G+T I +A++ + G K +++P ++E+ I+ ALGA V + +SI
Sbjct: 68 GKSTLIEATGGNTGIGLASIGASRGYKVILLMPSTMSLERRIILRALGAEVH-LTDISIG 126
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
K +A E K GG+
Sbjct: 127 IKGQL--------EKAKEILSK-------------------------------TPGGYIP 147
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
QF N N HY TGPEIW + G +D VA GTGGTV G KFL+EKN +IK ++
Sbjct: 148 HQFINPENPEIHYRTTGPEIWRDSAGKVDILVAGVGTGGTVTGTGKFLKEKNKDIKVCVV 207
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTD 343
+P S + + P + +GIG + N + +D + T
Sbjct: 208 EPSESAVLSG-----------------GKPGPHLIQGIGSGEIPANLDLSIVDEIIQVTG 250
Query: 344 MEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
EA+E + L +GL +G SS + A+++A + G IV I G R+LS
Sbjct: 251 EEAIETTKLLAIKEGLLVGISSGASAAAALKVAKRPENVGKLIVVIFPSGGERYLS 306
>At3g04940 cysteine synthase (AtcysD1)
Length = 324
Score = 145 bits (365), Expect = 5e-35
Identities = 100/356 (28%), Positives = 160/356 (44%), Gaps = 59/356 (16%)
Query: 45 KNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRR 104
K+ IGNTP++ +N++ D I K E + P SVK+R+A +I++A + G +
Sbjct: 9 KDDATQLIGNTPMVYLNNIVDGCVARIAAKLEMMEPCSSVKERIAYGMIKDAEDKGLITP 68
Query: 105 G-GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSIT 163
G + E ++G+T I +A + A G K + +P ++E+ I+ ALGA V P
Sbjct: 69 GKSTLIEATSGNTGIGLAFIGAAKGYKVVLTMPSSMSLERKIILLALGAEVHLTDP---- 124
Query: 164 HKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFA 223
S+ + G + + P+
Sbjct: 125 --------------------------------SKGVQGIIDKAEEICSKNPDS----IML 148
Query: 224 DQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLI 283
+QF+N +N + HY TGPEIW + G +D VA GTGGT++G +FL+EKN + K Y +
Sbjct: 149 EQFKNPSNPQTHYRTTGPEIWRDSAGEVDILVAGVGTGGTLSGSGRFLKEKNKDFKVYGV 208
Query: 284 DPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTD 343
+P S + + P + +GIG + N LD + T
Sbjct: 209 EPTESAVISG-----------------GKPGTHLIQGIGAGLIPDNLDFNVLDEVIQVTS 251
Query: 344 MEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
+EA+E A+ L +GL +G SS A+++A + G IV I G R+LS
Sbjct: 252 VEAIETAKLLALKEGLLVGISSGAAAAAAIKVAKRPENAGKLIVVIFPSGGERYLS 307
>At4g14880 cytosolic O-acetylserine(thiol)lyase (EC 4.2.99.8)
Length = 322
Score = 142 bits (359), Expect = 3e-34
Identities = 99/353 (28%), Positives = 159/353 (44%), Gaps = 59/353 (16%)
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG-G 106
+ + IGNTPL+ +N++++ + K E + P SVKDR+ +I +A + G ++ G
Sbjct: 9 VTELIGNTPLVYLNNVAEGCVGRVAAKLEMMEPCSSVKDRIGFSMISDAEKKGLIKPGES 68
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
++ E ++G+T + +A A A G K + +P + E+ I+ A G + P
Sbjct: 69 VLIEPTSGNTGVGLAFTAAAKGYKLIITMPASMSTERRIILLAFGVELVLTDPA------ 122
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
KG + G + + PN G+ QF
Sbjct: 123 -------------------------KG-----MKGAIAKAEEILAKTPN----GYMLQQF 148
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPP 286
EN AN + HYE TGPEIW+ T G +D FV+ GTGGT+ G K+L+E+N N+K Y ++P
Sbjct: 149 ENPANPKIHYETTGPEIWKGTGGKIDGFVSGIGTGGTITGAGKYLKEQNANVKLYGVEPV 208
Query: 287 GSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEA 346
S + + P +GIG + +D + + E+
Sbjct: 209 ESAILSG-----------------GKPGPHKIQGIGAGFIPSVLNVDLIDEVVQVSSDES 251
Query: 347 VEMARFLVKNDGLFLGSSSAMNCVGAVRIAQ-SIGPGHTIVTILCDSGMRHLS 398
++MAR L +GL +G SS A+++AQ G V I G R+LS
Sbjct: 252 IDMARQLALKEGLLVGISSGAAAAAAIKLAQRPENAGKLFVAIFPSFGERYLS 304
>At3g03630 O-acetylserine (thiol) lyase
Length = 404
Score = 139 bits (350), Expect = 3e-33
Identities = 105/353 (29%), Positives = 158/353 (44%), Gaps = 69/353 (19%)
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLR-RGG 106
+ IG+TP++ +N ++D +I K E + P SVKDR+ + +I EA SG + R
Sbjct: 103 VTQLIGSTPMVYLNRVTDGCLADIAAKLESMEPCRSVKDRIGLSMINEAENSGAITPRKT 162
Query: 107 IVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKD 166
++ E + G+T + IA VA A G K V +P IE+ ++ ALGA + P
Sbjct: 163 VLVEPTTGNTGLGIAFVAAAKGYKLIVTMPASINIERRMLLRALGAEIVLTNPEKGL--- 219
Query: 167 HFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQF 226
+ +A E K + + + QF
Sbjct: 220 ------KGAVDKAKEIVLKTKNA-------------------------------YMFQQF 242
Query: 227 ENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPP 286
+N AN + H+E TGPEIWE T GN+D FVA GTGGTV G FL+ N +IK ++P
Sbjct: 243 DNTANTKIHFETTGPEIWEDTMGNVDIFVAGIGTGGTVTGTGGFLKMMNKDIKVVGVEPS 302
Query: 287 GSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEA 346
+ + G + GI V LD F+ ++ EA
Sbjct: 303 ERSVISGDNPGYL---------------------PGILDV------KLLDEVFKVSNGEA 335
Query: 347 VEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSIGPGHTIVTILCDS-GMRHLS 398
+EMAR L +GL +G SS V AV +A+ ++T+L S G R+++
Sbjct: 336 IEMARRLALEEGLLVGISSGAAAVAAVSLAKRAENAGKLITVLFPSHGERYIT 388
>At3g59760 cysteine synthase
Length = 430
Score = 137 bits (346), Expect = 9e-33
Identities = 94/355 (26%), Positives = 160/355 (44%), Gaps = 59/355 (16%)
Query: 46 NGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG 105
+ + IG TP++ +NS++ I K E + P SVKDR+ ++ +A + G + G
Sbjct: 115 DNVSQLIGKTPMVYLNSIAKGCVANIAAKLEIMEPCCSVKDRIGYSMVTDAEQKGFISPG 174
Query: 106 -GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITH 164
++ E ++G+T I +A +A + G + + +P ++E+ +++A GA + P
Sbjct: 175 KSVLVEPTSGNTGIGLAFIAASRGYRLILTMPASMSMERRVLLKAFGAELVLTDPA---- 230
Query: 165 KDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFAD 224
+ + A + A + K+ P +
Sbjct: 231 ---------KGMTGAVQKAEEILKNTPDA---------------------------YMLQ 254
Query: 225 QFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLID 284
QF+N AN + HYE TGPEIW+ T G +D FVA GTGGT+ GV +F++EKNP + ++
Sbjct: 255 QFDNPANPKIHYETTGPEIWDDTKGKVDIFVAGIGTGGTITGVGRFIKEKNPKTQVIGVE 314
Query: 285 PPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDM 344
P S + + P +GIG + KN + +D +
Sbjct: 315 PTESDILSG-----------------GKPGPHKIQGIGAGFIPKNLDQKIMDEVIAISSE 357
Query: 345 EAVEMARFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
EA+E A+ L +GL +G SS A+++A + G I + G R+LS
Sbjct: 358 EAIETAKQLALKEGLMVGISSGAAAAAAIKVAKRPENAGKLIAVVFPSFGERYLS 412
>At2g43750 cysteine synthase (cpACS1)
Length = 392
Score = 135 bits (341), Expect = 3e-32
Identities = 96/349 (27%), Positives = 153/349 (43%), Gaps = 59/349 (16%)
Query: 52 IGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRG-GIVTE 110
IG TP++ +N++ + K E + P SVKDR+ +I +A E G + G ++ E
Sbjct: 83 IGKTPMVYLNNVVKGCVASVAAKLEIMEPCCSVKDRIGYSMITDAEEKGLITPGKSVLVE 142
Query: 111 GSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDHFVN 170
++G+T I +A +A + G K + +P ++E+ ++ A GA + P
Sbjct: 143 STSGNTGIGLAFIAASKGYKLILTMPASMSLERRVLLRAFGAELVLTEPA---------- 192
Query: 171 IARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFENLA 230
KG + G + PN + QF+N A
Sbjct: 193 ---------------------KG-----MTGAIQKAEEILKKTPNS----YMLQQFDNPA 222
Query: 231 NFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDPPGSGL 290
N + HYE TGPEIWE T G +D VA GTGGT+ GV +F++E+ P +K ++P S +
Sbjct: 223 NPKIHYETTGPEIWEDTRGKIDILVAGIGTGGTITGVGRFIKERKPELKVIGVEPTESAI 282
Query: 291 FNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINRVTKNFAEAKLDGAFRGTDMEAVEMA 350
+ P +GIG V KN A +D + EA+E +
Sbjct: 283 LSG-----------------GKPGPHKIQGIGAGFVPKNLDLAIVDEYIAISSEEAIETS 325
Query: 351 RFLVKNDGLFLGSSSAMNCVGAVRIA-QSIGPGHTIVTILCDSGMRHLS 398
+ L +GL +G SS A+++A + G I + G R+LS
Sbjct: 326 KQLALQEGLLVGISSGAAAAAAIQVAKRPENAGKLIAVVFPSFGERYLS 374
>At3g61440 cysteine synthase (AtcysC1)
Length = 368
Score = 131 bits (329), Expect = 8e-31
Identities = 106/374 (28%), Positives = 163/374 (43%), Gaps = 61/374 (16%)
Query: 28 FCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDR 87
F RL + K +K IG TPL+ +N +++ + K E P S+KDR
Sbjct: 33 FAQRLRDLPKDFPSTNAKRDASLLIGKTPLVFLNKVTEGCEAYVAAKQEHFQPTCSIKDR 92
Query: 88 VAVQIIEEALESGQLRRGGIVT--EGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQ 145
A+ +I +A E +L G T E ++G+ IS+A +A G + + +P ++E+
Sbjct: 93 PAIAMIADA-EKKKLIIPGKTTLIEPTSGNMGISLAFMAAMKGYRIIMTMPSYTSLERRV 151
Query: 146 IIEALGATVERVRPVSITHKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSD 205
+ + GA + P KG + G
Sbjct: 152 TMRSFGAELVLTDPA-------------------------------KG-----MGGTVKK 175
Query: 206 GHKHSTLFPNDCQGGFFADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVA 265
+ P+ F QF N AN + H++ TGPEIWE T GN+D FV G+GGTV+
Sbjct: 176 AYDLLDSTPD----AFMCQQFANPANTQIHFDTTGPEIWEDTLGNVDIFVMGIGSGGTVS 231
Query: 266 GVSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIGINR 325
GV ++L+ KNPN+K Y ++P S + N G P G+G
Sbjct: 232 GVGRYLKSKNPNVKIYGVEPAESNILNGGKPG---------------PHAITGNGVGFKP 276
Query: 326 VTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQ-SIGPGHT 384
+ ++ + +A++MAR L +GL +G SS N V A+R+A+ G
Sbjct: 277 EILDM--DVMESVLEVSSEDAIKMARELALKEGLMVGISSGANTVAAIRLAKMPENKGKL 334
Query: 385 IVTILCDSGMRHLS 398
IVTI G R+LS
Sbjct: 335 IVTIHASFGERYLS 348
>At3g22460 unknown protein
Length = 188
Score = 99.0 bits (245), Expect = 4e-21
Identities = 64/222 (28%), Positives = 101/222 (44%), Gaps = 41/222 (18%)
Query: 43 KSKNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQL 102
K + + IGNTPL+ +N ++ + K E + P SVKDR+ +I +A G +
Sbjct: 7 KIAKDVTELIGNTPLVYLNKVAKDCVGHVAAKLEMMEPCSSVKDRIGYSMIADAEAKGLI 66
Query: 103 RRG-GIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVS 161
+ G ++ E ++G+T + +A A A G K + +P +IE+ I+ A GA + P
Sbjct: 67 KPGESVLIEPTSGNTGVGLAFTAAAKGYKLVITMPASMSIERRIILLAFGAELILTDP-- 124
Query: 162 ITHKDHFVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGF 221
++ + G + + PN G+
Sbjct: 125 ----------------------------------AKGMKGAVAKAEEILAKTPN----GY 146
Query: 222 FADQFENLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGT 263
QFEN AN + HYE TGPEIW+ + G +D FV+ GTGGT
Sbjct: 147 MLQQFENPANPKIHYETTGPEIWKGSGGKVDGFVSGIGTGGT 188
>At3g10050 threonine dehydratase/deaminase (OMR1)
Length = 592
Score = 48.5 bits (114), Expect = 7e-06
Identities = 61/238 (25%), Positives = 91/238 (37%), Gaps = 47/238 (19%)
Query: 48 IIDAIGNTPLIRINSLSDATGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGI 107
+ D +PL LS G + K E L P S K R A ++ + L + QL +G I
Sbjct: 104 VYDIAIESPLQLAKKLSKRLGVRMYLKREDLQPVFSFKLRGAYNMMVK-LPADQLAKGVI 162
Query: 108 VTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIEKSQIIEALGATVERVRPVSITHKDH 167
+ SAG+ A +A A GC +V+P K Q +E LGATV + H
Sbjct: 163 CS--SAGNHAQGVALSASKLGCTAVIVMPVTTPEIKWQAVENLGATVVLFGDSYDQAQAH 220
Query: 168 FVNIARRRASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPNDCQGGFFADQFE 227
A+ RA E +G F F+
Sbjct: 221 ----AKIRAEE---------------------------------------EGLTFIPPFD 237
Query: 228 NLANFRAHYEGTGPEIWEQTNGNLDAFVAAAGTGGTVAGVSKFLQEKNPNIKCYLIDP 285
+ + A G EI Q G L A G GG +AG++ +++ +P +K ++P
Sbjct: 238 H-PDVIAGQGTVGMEITRQAKGPLHAIFVPVGGGGLIAGIAAYVKRVSPEVKIIGVEP 294
>At4g11640 unknown protein
Length = 346
Score = 33.5 bits (75), Expect = 0.23
Identities = 29/113 (25%), Positives = 51/113 (44%), Gaps = 18/113 (15%)
Query: 42 KKSKNGIIDAIGNTPLIRINSLSDATGCEILGKCEFLNPG---------------GSVKD 86
K++ + I I TP++ SL+ +G + KCE L G G+ K
Sbjct: 16 KEAHDRIKPYIHRTPVLTSESLNSISGRSLFFKCECLQKGIECRSFYFSVYCGKSGAFKF 75
Query: 87 RVAVQIIEEALESGQLRRGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDA 139
R A + +L++ Q +G V S+G+ A +++ A G ++V+P A
Sbjct: 76 RGACNAV-LSLDAEQAAKG--VVTHSSGNHAAALSLAAKIQGIPAYIVVPKGA 125
>At2g44210 unknown protein
Length = 415
Score = 33.1 bits (74), Expect = 0.30
Identities = 14/42 (33%), Positives = 23/42 (54%)
Query: 175 RASEANEFAFKHRKSQPKGTDSQQINGCKSDGHKHSTLFPND 216
RAS + K++KS PK S+ N +GH+H+ ++ D
Sbjct: 134 RASSVENYGMKNQKSIPKPKSSEPPNVLTQNGHQHAIMYVED 175
>At5g38530 tryptophan synthase beta chain
Length = 506
Score = 32.0 bits (71), Expect = 0.66
Identities = 27/81 (33%), Positives = 35/81 (42%), Gaps = 5/81 (6%)
Query: 55 TPLIRINSLSDA--TGCEILGKCEFLNPGGSVKDRVAVQIIEEALESGQLRRGGIVTEGS 112
TPLIR L T I K E +P GS K AV +A + + +VTE
Sbjct: 134 TPLIRAKRLEKLLQTPARIYFKYEGGSPAGSHKPNTAVP---QAYYNAKEGVKNVVTETG 190
Query: 113 AGSTAISIATVAPAYGCKCHV 133
AG S+A + +G C V
Sbjct: 191 AGQWGSSLAFASSLFGLDCEV 211
>At4g18330 translation initiation factor eIF-2 gamma chain-like
protein
Length = 471
Score = 31.6 bits (70), Expect = 0.87
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Query: 10 GAVGVGTAIFVSLVVAYFFCDRLCNPSKKKKKKKSKNGIIDAIGNTPLIRINSLSDATGC 69
G +G +FV L V+Y RL K+K+K+ K + + ++ +N S +TG
Sbjct: 358 GEMGTLPDVFVELEVSYQLLTRLIGVRTKEKEKQMK---VSKLTKEEILMVNIGSMSTGA 414
Query: 70 EILG 73
+++G
Sbjct: 415 KVIG 418
>At2g03550 putative esterase
Length = 312
Score = 31.2 bits (69), Expect = 1.1
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query: 222 FADQFENLANFRAHYEGTGPEIWEQTNGNL-DAFVAAAGTGGTVA 265
+ D +++L H GTGPE W +G+ F+A GG ++
Sbjct: 121 YEDSWDSLKWVLTHITGTGPETWINKHGDFGKVFLAGDSAGGNIS 165
>At5g43780 ATP sulfurylase precursor (gb|AAD26634.1)
Length = 469
Score = 30.4 bits (67), Expect = 1.9
Identities = 19/75 (25%), Positives = 36/75 (47%), Gaps = 7/75 (9%)
Query: 263 TVAGVSKFLQEKNPNIKCYLIDPPGSGLFNKVMRGVMYTKEEAEGRRLKNPFDTITEGIG 322
+V + + + + ++K LI+P G L N V+ EE+ R +K+ +T+ I
Sbjct: 34 SVVSLPRQVSRRGLSVKSGLIEPDGGKLMNLVV-------EESRRRVMKHEAETVPARIK 86
Query: 323 INRVTKNFAEAKLDG 337
+NRV + +G
Sbjct: 87 LNRVDLEWVHVLSEG 101
>At5g58410 unknown protein
Length = 1873
Score = 28.5 bits (62), Expect = 7.3
Identities = 30/115 (26%), Positives = 47/115 (40%), Gaps = 16/115 (13%)
Query: 276 PNIKCYLIDPPGSG-----LFNKVMRGVM-YTKEEAEGRRLKNPFDTITEGI-----GIN 324
PN+ + D P SG +F+ ++ G + + K+E EG L N + I
Sbjct: 1066 PNLVSFCKDEPSSGGLLNSIFDILLNGALVHVKDEEEG--LGNMWVDFNNNIVDVVEPFL 1123
Query: 325 RVTKNFAEAKLDGAFRGTDMEAVEMARFLVKNDGLFLGSSSAMNCVGAVRIAQSI 379
R +F G E MA F + D LF+G ++ NC+ + SI
Sbjct: 1124 RALVSFLHILFKEDLWG---EEEAMAAFKMITDKLFIGEETSKNCLRIIPYIMSI 1175
>At3g20020 arginine methyltransferase like protein
Length = 435
Score = 28.1 bits (61), Expect = 9.6
Identities = 19/69 (27%), Positives = 34/69 (48%), Gaps = 2/69 (2%)
Query: 84 VKDRVAVQIIEEALESGQ-LRRGGIVTEGSAGSTAISIATVAPAYGCKCHVVIPDDAAIE 142
+KDR + EA+ Q L G +V + G+ +SI A A + + V D A++
Sbjct: 97 IKDRARTETYREAIMQHQSLIEGKVVVDVGCGTGILSIFC-AQAGAKRVYAVDASDIAVQ 155
Query: 143 KSQIIEALG 151
++++A G
Sbjct: 156 AKEVVKANG 164
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,542,188
Number of Sequences: 26719
Number of extensions: 423041
Number of successful extensions: 1112
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1073
Number of HSP's gapped (non-prelim): 31
length of query: 424
length of database: 11,318,596
effective HSP length: 102
effective length of query: 322
effective length of database: 8,593,258
effective search space: 2767029076
effective search space used: 2767029076
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122165.6


