

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122165.12 + phase: 1 /pseudo
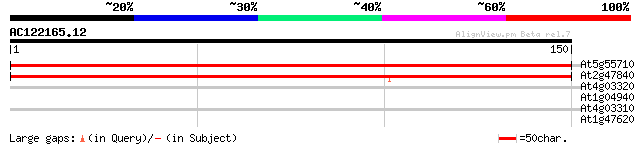
(150 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g55710 unknown protein 259 5e-70
At2g47840 unknown protein 139 6e-34
At4g03320 putative chloroplast protein import component 37 0.005
At1g04940 unknown protein 29 1.1
At4g03310 hypothetical protein 27 4.1
At1g47620 hypothetical protein 26 9.2
>At5g55710 unknown protein
Length = 209
Score = 259 bits (661), Expect = 5e-70
Identities = 122/150 (81%), Positives = 137/150 (91%)
Query: 1 ASDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTL 60
ASDR+ISA+CYFYPFFDGIQYGKF+ITQ+ P Q ++QPL PAIR FKSFPFNGFL+F+TL
Sbjct: 59 ASDRIISAVCYFYPFFDGIQYGKFIITQYQPFQILIQPLFPAIRAFKSFPFNGFLIFITL 118
Query: 61 YFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNPRGGFGLDLLMSLDSTVFLFL 120
YF VVRNPNFS+YVRFNTMQAIVLDVLLIFPDLLER FNPR GFGLD++MSLDSTVFLFL
Sbjct: 119 YFVVVRNPNFSRYVRFNTMQAIVLDVLLIFPDLLERSFNPRDGFGLDVVMSLDSTVFLFL 178
Query: 121 LVCLIYGSSSCILGQLPRLPIVADAADRQV 150
LV LIYG S+C+ G PRLP+VA+AADRQV
Sbjct: 179 LVSLIYGFSACLFGLTPRLPLVAEAADRQV 208
>At2g47840 unknown protein
Length = 208
Score = 139 bits (350), Expect = 6e-34
Identities = 68/152 (44%), Positives = 100/152 (65%), Gaps = 2/152 (1%)
Query: 1 ASDRLISALCYFYPFFDGIQYGKFVITQFYPIQAIVQPLIPAIRVFKSFPFNGFLVFLTL 60
A++R+IS Y PFF+ +QYG+F+ Q+ + + +P+ P + +++S P+ F+ F L
Sbjct: 57 ATERVISIASYALPFFNSLQYGRFLFAQYPRLGLLFEPIFPILNLYRSVPYASFVAFFGL 116
Query: 61 YFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLERGFNP--RGGFGLDLLMSLDSTVFL 118
Y VVRN +FS+YVRFN MQA+ LDVLL P LL R +P GGFG+ +M + VF+
Sbjct: 117 YLGVVRNTSFSRYVRFNAMQAVTLDVLLAVPVLLTRILDPGQGGGFGMKAMMWGHTGVFV 176
Query: 119 FLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
F +C +YG S +LG+ P +P VADAA RQ+
Sbjct: 177 FSFMCFVYGVVSSLLGKTPYIPFVADAAGRQL 208
>At4g03320 putative chloroplast protein import component
Length = 284
Score = 36.6 bits (83), Expect = 0.005
Identities = 28/102 (27%), Positives = 48/102 (46%), Gaps = 18/102 (17%)
Query: 54 FLVFLTL-YFFVVRNPNFSKYVRFNTMQAIVLDVLL--------IFPDLLERGFNPRGGF 104
F+V+ L Y +VV+N Y+RF+ M ++L+ L FP + + +G F
Sbjct: 176 FMVYCYLGYMWVVKNKELPHYLRFHMMMGMLLETALQVIWCTSNFFPLI-----HFKGRF 230
Query: 105 GLDLLMSLDSTVFLFLLVCLIYGSSSCILGQLPRLPIVADAA 146
G+ M++ T LL C+ + G ++P + DAA
Sbjct: 231 GMYYWMAIGFTYICLLLECI----RCALAGVYAQIPFMTDAA 268
>At1g04940 unknown protein
Length = 274
Score = 28.9 bits (63), Expect = 1.1
Identities = 26/115 (22%), Positives = 48/115 (41%), Gaps = 11/115 (9%)
Query: 46 FKSFPFNG--------FLV--FLTLYFFVVRNPNFSKYVRFNTMQAIVLDVLLIFPDLLE 95
F ++PF G FL+ F Y +VR + + RF+ + ++L++ L +
Sbjct: 158 FLTYPFLGAIGRLPSWFLMAYFFVAYLGIVRRKEWPHFFRFHVVMGMLLEIALQVIGTVS 217
Query: 96 RGFNPRGGFGLDLLMSLDSTVFLFLLVCLIYGSSSCILGQLPRLPIVADAADRQV 150
+ + P G + M + V L ++ + G +P V DAA Q+
Sbjct: 218 K-WMPLGVYWGKFGMHFWTAVAFAYLFTVLESIRCALAGMYADIPFVCDAAYIQI 271
>At4g03310 hypothetical protein
Length = 562
Score = 26.9 bits (58), Expect = 4.1
Identities = 11/32 (34%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query: 116 VFLFLLVCLIYGSSSCIL-GQLPRLPIVADAA 146
V F+ +C++ C + G P++PI++DAA
Sbjct: 506 VIWFIFLCVLLECLRCAMAGSYPQIPIISDAA 537
>At1g47620 hypothetical protein
Length = 520
Score = 25.8 bits (55), Expect = 9.2
Identities = 18/48 (37%), Positives = 28/48 (57%), Gaps = 3/48 (6%)
Query: 51 FNGFLVFLTLYFFVVRNPNFSKYVRFNTMQAIVLD--VLLIFPDLLER 96
F GFL FL ++F+V+ P FS + T+Q+ + VL + P +L R
Sbjct: 10 FIGFLCFLISFYFLVKKP-FSYLLIKKTLQSYPWNWPVLGMLPGVLLR 56
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.335 0.153 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,337,664
Number of Sequences: 26719
Number of extensions: 135771
Number of successful extensions: 430
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 425
Number of HSP's gapped (non-prelim): 6
length of query: 150
length of database: 11,318,596
effective HSP length: 90
effective length of query: 60
effective length of database: 8,913,886
effective search space: 534833160
effective search space used: 534833160
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC122165.12


