

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.7 - phase: 0
(317 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
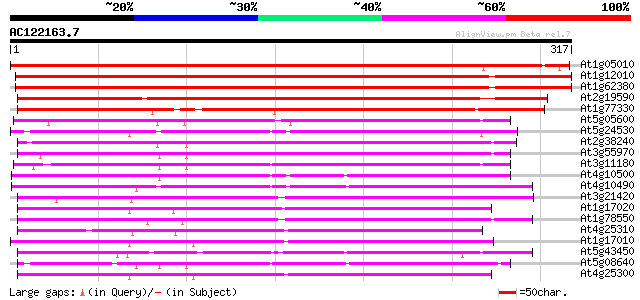
Score E
Sequences producing significant alignments: (bits) Value
At1g05010 1-aminocyclopropane-1-carboxylate oxidase 501 e-142
At1g12010 putative amino-cyclopropane-carboxylic acid oxidase (A... 473 e-134
At1g62380 Unknown protein (F24O1.10) 470 e-133
At2g19590 1-aminocyclopropane-1-carboxylate oxidase 305 2e-83
At1g77330 unknown protein 301 2e-82
At5g05600 leucoanthocyanidin dioxygenase-like protein 195 2e-50
At5g24530 flavanone 3-hydroxylase-like protein 195 3e-50
At2g38240 putative anthocyanidin synthase 193 9e-50
At3g55970 leucoanthocyanidin dioxygenase -like protein 189 2e-48
At3g11180 putative leucoanthocyanidin dioxygenase 186 1e-47
At4g10500 putative Fe(II)/ascorbate oxidase 177 5e-45
At4g10490 putative flavanone 3-beta-hydroxylase 176 2e-44
At3g21420 unknown protein 173 1e-43
At1g17020 SRG1-like protein 167 9e-42
At1g78550 unknown protein 165 3e-41
At4g25310 SRG1-like protein 164 6e-41
At1g17010 SRG1-like protein 164 6e-41
At5g43450 1-aminocyclopropane-1-carboxylate oxidase 160 1e-39
At5g08640 flavonol synthase (FLS) (sp|Q96330) 160 1e-39
At4g25300 SRG1-like protein 159 2e-39
>At1g05010 1-aminocyclopropane-1-carboxylate oxidase
Length = 323
Score = 501 bits (1289), Expect = e-142
Identities = 240/323 (74%), Positives = 281/323 (86%), Gaps = 8/323 (2%)
Query: 1 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 60
ME+FP++++ KLN EER TME IKDACENWGFFECVNH IS+EL+DKVEK+TKEHYKKC
Sbjct: 1 MESFPIINLEKLNGEERAITMEKIKDACENWGFFECVNHGISLELLDKVEKMTKEHYKKC 60
Query: 61 MEQRFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEK 120
ME+RFKE + ++GL+ ++SE+ND+DWESTF+L+HLP SNIS++PDLD+DYR MK+FA K
Sbjct: 61 MEERFKESIKNRGLDSLRSEVNDVDWESTFYLKHLPVSNISDVPDLDDDYRTLMKDFAGK 120
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDA 180
+EKL+EELLDLLCENLGLEKGYLKKVFYGSK P FGTKVSNYPPCP PDL+KGLRAHTDA
Sbjct: 121 IEKLSEELLDLLCENLGLEKGYLKKVFYGSKRPTFGTKVSNYPPCPNPDLVKGLRAHTDA 180
Query: 181 GGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQT 240
GGIILLFQDDKVSGLQLLKD +W+DVPP+ HSIV+NLGDQLEVITNGKYKSV HRV++QT
Sbjct: 181 GGIILLFQDDKVSGLQLLKDGEWVDVPPVKHSIVVNLGDQLEVITNGKYKSVEHRVLSQT 240
Query: 241 DG-ARMSIASFYNPGNDAVISPASTLL---KEDETSEIYPKFIFDDYMKLYMGLKFQAKE 296
DG RMSIASFYNPG+D+VI PA L+ E E E YP+F+F+DYMKLY +KFQAKE
Sbjct: 241 DGEGRMSIASFYNPGSDSVIFPAPELIGKEAEKEKKENYPRFVFEDYMKLYSAVKFQAKE 300
Query: 297 PRFEAMMKAMSSV---EVGPVVT 316
PRFEA MKAM + VGP+ T
Sbjct: 301 PRFEA-MKAMETTVANNVGPLAT 322
>At1g12010 putative amino-cyclopropane-carboxylic acid oxidase (ACC
oxidase)
Length = 320
Score = 473 bits (1216), Expect = e-134
Identities = 219/314 (69%), Positives = 265/314 (83%), Gaps = 3/314 (0%)
Query: 4 FPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
FPV+D+ KLN EER TM +I DAC+NWGFFE VNH + +LMD +E++TKEHYKK MEQ
Sbjct: 7 FPVIDLSKLNGEERDQTMALIDDACQNWGFFELVNHGLPYDLMDNIERMTKEHYKKHMEQ 66
Query: 64 RFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEKLEK 123
+FKEM+ SKGL+ +++E+ D+DWESTF+L HLP SN+ +IPD+ +YR MK+F ++LE
Sbjct: 67 KFKEMLRSKGLDTLETEVEDVDWESTFYLHHLPQSNLYDIPDMSNEYRLAMKDFGKRLEI 126
Query: 124 LAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDAGGI 183
LAEELLDLLCENLGLEKGYLKKVF+G+ GP F TK+SNYPPCPKP++IKGLRAHTDAGG+
Sbjct: 127 LAEELLDLLCENLGLEKGYLKKVFHGTTGPTFATKLSNYPPCPKPEMIKGLRAHTDAGGL 186
Query: 184 ILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQTDGA 243
ILLFQDDKVSGLQLLKD W+DVPP+ HSIVINLGDQLEVITNGKYKSVMHRV+ Q +G
Sbjct: 187 ILLFQDDKVSGLQLLKDGDWVDVPPLKHSIVINLGDQLEVITNGKYKSVMHRVMTQKEGN 246
Query: 244 RMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAKEPRFEAMM 303
RMSIASFYNPG+DA ISPA++L+ +D YP F+FDDYMKLY GLKFQAKEPRFEAM
Sbjct: 247 RMSIASFYNPGSDAEISPATSLVDKDSK---YPSFVFDDYMKLYAGLKFQAKEPRFEAMK 303
Query: 304 KAMSSVEVGPVVTI 317
A ++ ++ PV +
Sbjct: 304 NAEAAADLNPVAVV 317
>At1g62380 Unknown protein (F24O1.10)
Length = 320
Score = 470 bits (1210), Expect = e-133
Identities = 216/314 (68%), Positives = 265/314 (83%), Gaps = 3/314 (0%)
Query: 4 FPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
FPVVD+ KLN EER TM +I +ACENWGFFE VNH + +LMDK+EK+TK+HYK C EQ
Sbjct: 7 FPVVDLSKLNGEERDQTMALINEACENWGFFEIVNHGLPHDLMDKIEKMTKDHYKTCQEQ 66
Query: 64 RFKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEKLEK 123
+F +M+ SKGL+ +++E+ D+DWESTF++RHLP SN+++I D+ ++YR MK+F ++LE
Sbjct: 67 KFNDMLKSKGLDNLETEVEDVDWESTFYVRHLPQSNLNDISDVSDEYRTAMKDFGKRLEN 126
Query: 124 LAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDAGGI 183
LAE+LLDLLCENLGLEKGYLKKVF+G+KGP FGTKVSNYPPCPKP++IKGLRAHTDAGGI
Sbjct: 127 LAEDLLDLLCENLGLEKGYLKKVFHGTKGPTFGTKVSNYPPCPKPEMIKGLRAHTDAGGI 186
Query: 184 ILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVIAQTDGA 243
ILLFQDDKVSGLQLLKD WIDVPP+ HSIVINLGDQLEVITNGKYKSV+HRV+ Q +G
Sbjct: 187 ILLFQDDKVSGLQLLKDGDWIDVPPLNHSIVINLGDQLEVITNGKYKSVLHRVVTQQEGN 246
Query: 244 RMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAKEPRFEAMM 303
RMS+ASFYNPG+DA ISPA++L+++D YP F+FDDYMKLY G+KFQ KEPRF AM
Sbjct: 247 RMSVASFYNPGSDAEISPATSLVEKDSE---YPSFVFDDYMKLYAGVKFQPKEPRFAAMK 303
Query: 304 KAMSSVEVGPVVTI 317
A + E+ P +
Sbjct: 304 NASAVTELNPTAAV 317
>At2g19590 1-aminocyclopropane-1-carboxylate oxidase
Length = 310
Score = 305 bits (782), Expect = 2e-83
Identities = 148/302 (49%), Positives = 206/302 (68%), Gaps = 12/302 (3%)
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D +L+ E+R TM ++ AC+ WGFF NH I ELM+KV+K+ HY++ ++++
Sbjct: 12 PVIDFAELDGEKRSKTMSLLDHACDKWGFFMVDNHGIDKELMEKVKKMINSHYEEHLKEK 71
Query: 65 FKEMVASKGLECVQSEINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAEKLEKL 124
F + K L + + +D DWES+FF+ H P+SNI +IP++ E+ KTM E+ +L K
Sbjct: 72 FYQSEMVKALS--EGKTSDADWESSFFISHKPTSNICQIPNISEELSKTMDEYVCQLHKF 129
Query: 125 AEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAHTDAGGII 184
AE L L+CENLGL++ + F G KGP FGTKV+ YP CP+P+L++GLR HTDAGGII
Sbjct: 130 AERLSKLMCENLGLDQEDIMNAFSGPKGPAFGTKVAKYPECPRPELMRGLREHTDAGGII 189
Query: 185 LLFQDDKVSGLQLLKDDQWIDVPPMP-HSIVINLGDQLEVITNGKYKSVMHRVIAQTDGA 243
LL QDD+V GL+ KD +W+ +PP ++I +N GDQLE+++NG+YKSV+HRV+ G+
Sbjct: 190 LLLQDDQVPGLEFFKDGKWVPIPPSKNNTIFVNTGDQLEILSNGRYKSVVHRVMTVKHGS 249
Query: 244 RMSIASFYNPGNDAVISPASTLLKEDETSEIYPK-FIFDDYMKLYMGLKFQAKEPRFEAM 302
R+SIA+FYNP DA+ISPA LL YP + F DY+KLY KF K PR E M
Sbjct: 250 RLSIATFYNPAGDAIISPAPKLL--------YPSGYRFQDYLKLYSTTKFGDKGPRLETM 301
Query: 303 MK 304
K
Sbjct: 302 KK 303
>At1g77330 unknown protein
Length = 307
Score = 301 bits (772), Expect = 2e-82
Identities = 156/308 (50%), Positives = 213/308 (68%), Gaps = 17/308 (5%)
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+D KLN EER+ T+ I ACE WGFF+ VNH I +EL++KV+KL+ + YK E+
Sbjct: 4 PVIDFSKLNGEEREKTLSEIARACEEWGFFQLVNHGIPLELLNKVKKLSSDCYKTEREEA 63
Query: 65 FKEMVASKGL-ECVQS----EINDLDWESTFFLRHLPSSNISEIPDLDEDYRKTMKEFAE 119
FK K L E VQ ++ ++DWE F L N +E P + ++TM E+ E
Sbjct: 64 FKTSNPVKLLNELVQKNSGEKLENVDWEDVFTLL---DHNQNEWPS---NIKETMGEYRE 117
Query: 120 KLEKLAEELLDLLCENLGLEKGYLKKVFY-----GSKGPNFGTKVSNYPPCPKPDLIKGL 174
++ KLA ++++++ ENLGL KGY+KK F G + FGTKVS+YPPCP P+L+ GL
Sbjct: 118 EVRKLASKMMEVMDENLGLPKGYIKKAFNEGMEDGEETAFFGTKVSHYPPCPHPELVNGL 177
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMH 234
RAHTDAGG++LLFQDD+ GLQ+LKD +WIDV P+P++IVIN GDQ+EV++NG+YKS H
Sbjct: 178 RAHTDAGGVVLLFQDDEYDGLQVLKDGEWIDVQPLPNAIVINTGDQIEVLSNGRYKSAWH 237
Query: 235 RVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQA 294
RV+A+ +G R SIASFYNP A I PA+ + +E+ + + YPKF+F DYM +Y KF
Sbjct: 238 RVLAREEGNRRSIASFYNPSYKAAIGPAA-VAEEEGSEKKYPKFVFGDYMDVYANQKFMP 296
Query: 295 KEPRFEAM 302
KEPRF A+
Sbjct: 297 KEPRFLAV 304
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 195 bits (496), Expect = 2e-50
Identities = 106/293 (36%), Positives = 162/293 (55%), Gaps = 16/293 (5%)
Query: 3 NFPVVDMGKLNTEERKAT---MEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK 59
N P++D+ L +EE + M I +AC WGFF+ VNH + ELMD + +E +
Sbjct: 61 NIPIIDLEGLFSEEGLSDDVIMARISEACRGWGFFQVVNHGVKPELMDAARENWREFFHM 120
Query: 60 CMEQRFKEMVASKGLECVQSEIN-----DLDWESTFFLRHLPS--SNISEIPDLDEDYRK 112
+ + + + E S + LDW +FL LP + ++ P R+
Sbjct: 121 PVNAKETYSNSPRTYEGYGSRLGVEKGASLDWSDYYFLHLLPHHLKDFNKWPSFPPTIRE 180
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGT--KVSNYPPCPKPDL 170
+ E+ E+L KL+ ++ +L NLGL++ ++ F G N G +V+ YP CP+P+L
Sbjct: 181 VIDEYGEELVKLSGRIMRVLSTNLGLKEDKFQEAFGGE---NIGACLRVNYYPKCPRPEL 237
Query: 171 IKGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYK 230
GL H+D GG+ +L DD+V GLQ+ KDD WI V P PH+ ++N+GDQ+++++N YK
Sbjct: 238 ALGLSPHSDPGGMTILLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTYK 297
Query: 231 SVMHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDY 283
SV HRVI +D R+S+A FYNP +D I P L+ +YP FD Y
Sbjct: 298 SVEHRVIVNSDKERVSLAFFYNPKSDIPIQPLQELV-STHNPPLYPPMTFDQY 349
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 195 bits (495), Expect = 3e-50
Identities = 103/298 (34%), Positives = 174/298 (57%), Gaps = 19/298 (6%)
Query: 1 MENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKC 60
+E+FP++D L++ +R ++ I AC +GFF+ +NH ++ +++D++ + +E +
Sbjct: 35 LEDFPLID---LSSTDRSFLIQQIHQACARFGFFQVINHGVNKQIIDEMVSVAREFFSMS 91
Query: 61 MEQRFK--------EMVASKGLECVQSEINDLDWESTFFLRHLPSSN-ISEIPDLDEDYR 111
ME++ K S + E+N+ W L P ++E P ++
Sbjct: 92 MEEKMKLYSDDPTKTTRLSTSFNVKKEEVNN--WRDYLRLHCYPIHKYVNEWPSNPPSFK 149
Query: 112 KTMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLI 171
+ + +++ ++ ++ ++ +L+ E+LGLEK Y+KKV G +G + V+ YPPCP+P+L
Sbjct: 150 EIVSKYSREVREVGFKIEELISESLGLEKDYMKKVL-GEQGQHMA--VNYYPPCPEPELT 206
Query: 172 KGLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKS 231
GL AHTD + +L QD V GLQ+L D QW V P P + VIN+GDQL+ ++NG YKS
Sbjct: 207 YGLPAHTDPNALTILLQDTTVCGLQILIDGQWFAVNPHPDAFVINIGDQLQALSNGVYKS 266
Query: 232 VMHRVIAQTDGARMSIASFYNPGNDAVISPASTL--LKEDETSEIYPKFIFDDYMKLY 287
V HR + T+ R+S+ASF P + AV+SPA L ++DET +Y F + +Y K +
Sbjct: 267 VWHRAVTNTENPRLSVASFLCPADCAVMSPAKPLWEAEDDETKPVYKDFTYAEYYKKF 324
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 193 bits (491), Expect = 9e-50
Identities = 100/289 (34%), Positives = 167/289 (57%), Gaps = 10/289 (3%)
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PV+DM + + + +++ ACE WGFF+ VNH ++ LM++V +E ++ +E++
Sbjct: 49 PVLDMN--DVWGKPEGLRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEK 106
Query: 65 FKEMVASKGLECVQSEIN-----DLDWESTFFLRHLPSS--NISEIPDLDEDYRKTMKEF 117
K + E S + LDW FFL +LPSS N S+ P R+ ++++
Sbjct: 107 RKYANSPDTYEGYGSRLGVVKDAKLDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKY 166
Query: 118 AEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRAH 177
E++ KL E L + L E+LGL+ L + G + + YP CP+P L GL +H
Sbjct: 167 GEEVRKLCERLTETLSESLGLKPNKLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSH 226
Query: 178 TDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRVI 237
+D GGI +L D+KV+GLQ+ + D W+ + +P+++++N+GDQL++++NG YKSV H+VI
Sbjct: 227 SDPGGITILLPDEKVAGLQVRRGDGWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVI 286
Query: 238 AQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKL 286
+ R+S+A FYNP +D + P L+ + + +Y FD+Y L
Sbjct: 287 VNSGMERVSLAFFYNPRSDIPVGPIEELVTANRPA-LYKPIRFDEYRSL 334
>At3g55970 leucoanthocyanidin dioxygenase -like protein
Length = 363
Score = 189 bits (479), Expect = 2e-48
Identities = 99/290 (34%), Positives = 168/290 (57%), Gaps = 12/290 (4%)
Query: 5 PVVDMGKLNTEE---RKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
P++D+G+L T++ + T++ I AC GFF+ VNH +S +LMD+ + +E + M
Sbjct: 53 PIIDLGRLYTDDLTLQAKTLDEISKACRELGFFQVVNHGMSPQLMDQAKATWREFFNLPM 112
Query: 62 EQRFKEMVASKGLECVQSEIND-----LDWESTFFLRHLPSS--NISEIPDLDEDYRKTM 114
E + + K E S + LDW ++L + PSS + ++ P L R+ +
Sbjct: 113 ELKNMHANSPKTYEGYGSRLGVEKGAILDWSDYYYLHYQPSSLKDYTKWPSLPLHCREIL 172
Query: 115 KEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGL 174
+++ +++ KL E L+ +L +NLGL++ L+ F G + +V+ YP CP+P+L G+
Sbjct: 173 EDYCKEMVKLCENLMKILSKNLGLQEDRLQNAFGGKEESGGCLRVNYYPKCPQPELTLGI 232
Query: 175 RAHTDAGGIILLFQDDKVSGLQLL-KDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVM 233
H+D GG+ +L D++V+ LQ+ DD WI V P PH+ ++N+GDQ+++++N YKSV
Sbjct: 233 SPHSDPGGLTILLPDEQVASLQVRGSDDAWITVEPAPHAFIVNMGDQIQMLSNSIYKSVE 292
Query: 234 HRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDY 283
HRVI + R+S+A FYNP + I P L+ D + +Y +D Y
Sbjct: 293 HRVIVNPENERLSLAFFYNPKGNVPIEPLKELVTVDSPA-LYSSTTYDRY 341
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 186 bits (473), Expect = 1e-47
Identities = 101/291 (34%), Positives = 160/291 (54%), Gaps = 16/291 (5%)
Query: 3 NFPVVDMGKL---NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK 59
N P++D+ L N +++K I +AC WGFF+ +NH + ELMD + K +
Sbjct: 94 NIPIIDLDSLFSGNEDDKKR----ISEACREWGFFQVINHGVKPELMDAARETWKSFFNL 149
Query: 60 CMEQRFKEMVASKGLECVQSEIND-----LDWESTFFLRHLPSS--NISEIPDLDEDYRK 112
+E + + + E S + LDW ++L LP + + ++ P L + R+
Sbjct: 150 PVEAKEVYSNSPRTYEGYGSRLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIRE 209
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 172
E+ ++L KL L+ +L NLGL L++ F G + +V+ YP CP+P+L
Sbjct: 210 MNDEYGKELVKLGGRLMTILSSNLGLRAEQLQEAF-GGEDVGACLRVNYYPKCPQPELAL 268
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSV 232
GL H+D GG+ +L DD+V GLQ+ D WI V P+ H+ ++N+GDQ+++++N KYKSV
Sbjct: 269 GLSPHSDPGGMTILLPDDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSV 328
Query: 233 MHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDY 283
HRVI ++ R+S+A FYNP +D I P L+ +YP FD Y
Sbjct: 329 EHRVIVNSEKERVSLAFFYNPKSDIPIQPMQQLV-TSTMPPLYPPMTFDQY 378
>At4g10500 putative Fe(II)/ascorbate oxidase
Length = 349
Score = 177 bits (450), Expect = 5e-45
Identities = 96/289 (33%), Positives = 158/289 (54%), Gaps = 11/289 (3%)
Query: 2 ENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
++ P++D+ L+ R ++ + AC +GFF+ NH + ++K++ + +E + +
Sbjct: 42 DSIPLIDLRDLHGPNRAVIVQQLASACSTYGFFQIKNHGVPDTTVNKMQTVAREFFHQPE 101
Query: 62 EQRFKEMVASKGLECVQSEIND------LDWESTFFLRHLPSSN-ISEIPDLDEDYRKTM 114
+R K A S + L+W L P + I E P +R+
Sbjct: 102 SERVKHYSADPTKTTRLSTSFNVGADKVLNWRDFLRLHCFPIEDFIEEWPSSPISFREVT 161
Query: 115 KEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGL 174
E+A + L LL+ + E+LGLE ++ + G + + YPPCP+P+L GL
Sbjct: 162 AEYATSVRALVLRLLEAISESLGLESDHISNIL-GKHAQHMA--FNYYPPCPEPELTYGL 218
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMH 234
H D I +L QD +VSGLQ+ KDD+W+ V P+P++ ++N+GDQ++VI+N KYKSV+H
Sbjct: 219 PGHKDPTVITVLLQD-QVSGLQVFKDDKWVAVSPIPNTFIVNIGDQMQVISNDKYKSVLH 277
Query: 235 RVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDY 283
R + T+ R+SI +FY P DAVI PA L+ E ++ IY + F +Y
Sbjct: 278 RAVVNTENERLSIPTFYFPSTDAVIGPAHELVNEQDSLAIYRTYPFVEY 326
>At4g10490 putative flavanone 3-beta-hydroxylase
Length = 348
Score = 176 bits (445), Expect = 2e-44
Identities = 102/304 (33%), Positives = 163/304 (53%), Gaps = 16/304 (5%)
Query: 2 ENFPVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
++ P++D+ L+ R + AC + GFF+ NH + E + K+ +E +++
Sbjct: 40 DSIPLIDLHDLHGPNRADIINQFAHACSSCGFFQIKNHGVPEETIKKMMNAAREFFRQSE 99
Query: 62 EQRFKEMVA--------SKGLECVQSEINDLDWESTFFLRHLPSSN-ISEIPDLDEDYRK 112
+R K A S + ++++ W L P + I+E P +R+
Sbjct: 100 SERVKHYSADTKKTTRLSTSFNVSKEKVSN--WRDFLRLHCYPIEDFINEWPSTPISFRE 157
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 172
E+A + L LL+ + E+LGL K + G G + ++ YP CP+P+L
Sbjct: 158 VTAEYATSVRALVLTLLEAISESLGLAKDRVSNTI-GKHGQHMA--INYYPRCPQPELTY 214
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSV 232
GL H DA I +L QD+ VSGLQ+ KD +WI V P+P++ ++NLGDQ++VI+N KYKSV
Sbjct: 215 GLPGHKDANLITVLLQDE-VSGLQVFKDGKWIAVNPVPNTFIVNLGDQMQVISNEKYKSV 273
Query: 233 MHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETS-EIYPKFIFDDYMKLYMGLK 291
+HR + +D R+SI +FY P DAVISPA L+ E+E S IY F + +Y + +
Sbjct: 274 LHRAVVNSDMERISIPTFYCPSEDAVISPAQELINEEEDSPAIYRNFTYAEYFEKFWDTA 333
Query: 292 FQAK 295
F +
Sbjct: 334 FDTE 337
>At3g21420 unknown protein
Length = 364
Score = 173 bits (438), Expect = 1e-43
Identities = 96/303 (31%), Positives = 171/303 (55%), Gaps = 14/303 (4%)
Query: 5 PVVDMGKLNTEERKATM-EMIK--DACENWGFFECVNHSISIELMDKVEKLTKEHYKKCM 61
PV+D+ KL+ + E++K ACE+WGFF+ +NH I +E+++ +E++ E + +
Sbjct: 56 PVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMPL 115
Query: 62 EQRFKE-----MVASKGLECVQSEINDLDWESTFFLR-HLPSSNISEI-PDLDEDYRKTM 114
E++ K V G + SE LDW + F L H P ++ P + +++
Sbjct: 116 EEKKKYPMEPGTVQGYGQAFIFSEDQKLDWCNMFALGVHPPQIRNPKLWPSKPARFSESL 175
Query: 115 KEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGL 174
+ +++++ +L + LL + +LGL++ +++F + +++ YPPC PDL+ GL
Sbjct: 176 EGYSKEIRELCKRLLKYIAISLGLKEERFEEMFGEAVQ---AVRMNYYPPCSSPDLVLGL 232
Query: 175 RAHTDAGGIILLFQD-DKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVM 233
H+D + +L Q + GLQ+LKD+ W+ V P+P+++VIN+GD +EV++NGKYKSV
Sbjct: 233 SPHSDGSALTVLQQSKNSCVGLQILKDNTWVPVKPLPNALVINIGDTIEVLSNGKYKSVE 292
Query: 234 HRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQ 293
HR + + R++I +FY P + I P S L+ ++ Y + DY Y+ K Q
Sbjct: 293 HRAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCKYRSYNHGDYSYHYVSNKLQ 352
Query: 294 AKE 296
K+
Sbjct: 353 GKK 355
>At1g17020 SRG1-like protein
Length = 358
Score = 167 bits (422), Expect = 9e-42
Identities = 87/276 (31%), Positives = 159/276 (57%), Gaps = 9/276 (3%)
Query: 5 PVVDMGKL-NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
P++DM +L ++ + +E + AC+ WGFF+ VNH I +DKV+ ++ + ME+
Sbjct: 54 PIIDMKRLCSSTTMDSEVEKLDFACKEWGFFQLVNHGIDSSFLDKVKSEIQDFFNLPMEE 113
Query: 64 RFK-----EMVASKGLECVQSEINDLDWESTFF--LRHLPSSNISEIPDLDEDYRKTMKE 116
+ K + + G V SE LDW FF ++ + P L +R T++
Sbjct: 114 KKKFWQRPDEIEGFGQAFVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPLPFRDTLEM 173
Query: 117 FAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRA 176
++ +++ +A+ L+ + L ++ L+K+F +++ YPPCP+PD + GL
Sbjct: 174 YSSEVQSVAKILIAKMARALEIKPEELEKLFDDVDSVQ-SMRMNYYPPCPQPDQVIGLTP 232
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRV 236
H+D+ G+ +L Q + V GLQ+ KD +W+ V P+P++ ++N+GD LE+ITNG Y+S+ HR
Sbjct: 233 HSDSVGLTVLMQVNDVEGLQIKKDGKWVPVKPLPNAFIVNIGDVLEIITNGTYRSIEHRG 292
Query: 237 IAQTDGARMSIASFYNPGNDAVISPASTLLKEDETS 272
+ ++ R+SIA+F+N G + PA +L++ + +
Sbjct: 293 VVNSEKERLSIATFHNVGMYKEVGPAKSLVERQKVA 328
>At1g78550 unknown protein
Length = 356
Score = 165 bits (418), Expect = 3e-41
Identities = 89/299 (29%), Positives = 163/299 (53%), Gaps = 12/299 (4%)
Query: 5 PVVDMGKL-NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
PV+DM +L + + ++ + AC++WGFF+ VNH I ++K+E +E + M++
Sbjct: 54 PVIDMTRLCSVSAMDSELKKLDFACQDWGFFQLVNHGIDSSFLEKLETEVQEFFNLPMKE 113
Query: 64 RFKEMVASKGLEC-----VQSEINDLDWESTFFLRHLP--SSNISEIPDLDEDYRKTMKE 116
+ K S E + SE LDW F L P S L +R+T++
Sbjct: 114 KQKLWQRSGEFEGFGQVNIVSENQKLDWGDMFILTTEPIRSRKSHLFSKLPPPFRETLET 173
Query: 117 FAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRA 176
++ +++ +A+ L + L ++ ++ +F K++ YPPCP+PD + GL
Sbjct: 174 YSSEVKSIAKILFAKMASVLEIKHEEMEDLFDDVWQ---SIKINYYPPCPQPDQVMGLTQ 230
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRV 236
H+DA G+ +L Q ++V GLQ+ KD +W+ V P+ ++V+N+G+ LE+ITNG+Y+S+ HR
Sbjct: 231 HSDAAGLTILLQVNQVEGLQIKKDGKWVVVKPLRDALVVNVGEILEIITNGRYRSIEHRA 290
Query: 237 IAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKFQAK 295
+ ++ R+S+A F++PG + +I PA +L+ + ++ +Y + K K
Sbjct: 291 VVNSEKERLSVAMFHSPGKETIIRPAKSLVDRQKQC-LFKSMSTQEYFDAFFTQKLNGK 348
>At4g25310 SRG1-like protein
Length = 353
Score = 164 bits (415), Expect = 6e-41
Identities = 90/271 (33%), Positives = 154/271 (56%), Gaps = 13/271 (4%)
Query: 5 PVVDMGKLNTE-ERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
P++DM L++ + ++ + AC+ WGFF+ VNH + + DK + ++ + ME+
Sbjct: 53 PIIDMSLLSSSTSMDSEIDKLDFACKEWGFFQLVNHGMDL---DKFKSDIQDFFNLPMEE 109
Query: 64 RFKEM-----VASKGLECVQSEINDLDWESTFFL--RHLPSSNISEIPDLDEDYRKTMKE 116
+ K + G V SE LDW FFL + +P P L +R T+
Sbjct: 110 KKKLWQQPGDIEGFGQAFVFSEEQKLDWADVFFLTMQPVPLRKPHLFPKLPLPFRDTLDT 169
Query: 117 FAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRA 176
++ +L+ +A+ L L L ++ ++K+F G +++ YPPCP+PD GL
Sbjct: 170 YSAELKSIAKVLFAKLASALKIKPEEMEKLFDDELGQRI--RMNYYPPCPEPDKAIGLTP 227
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRV 236
H+DA G+ +L Q ++V GLQ+ KD +W+ V P+P+++V+N+GD LE+ITNG Y+S+ HR
Sbjct: 228 HSDATGLTILLQVNEVEGLQIKKDGKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRG 287
Query: 237 IAQTDGARMSIASFYNPGNDAVISPASTLLK 267
+ ++ R+S+ASF+N G I P +L++
Sbjct: 288 VVNSEKERLSVASFHNTGFGKEIGPMRSLVE 318
>At1g17010 SRG1-like protein
Length = 361
Score = 164 bits (415), Expect = 6e-41
Identities = 88/281 (31%), Positives = 161/281 (56%), Gaps = 10/281 (3%)
Query: 1 MENFPVVDMGKL-NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK 59
+ P++DM +L ++ + +E + AC+ +GFF+ VNH I +DK++ ++ +
Sbjct: 51 ISEIPIIDMNRLCSSTAVDSEVEKLDFACKEYGFFQLVNHGIDPSFLDKIKSEIQDFFNL 110
Query: 60 CMEQRFK-----EMVASKGLECVQSEINDLDWESTFFLRHLPSSNISE--IPDLDEDYRK 112
ME++ K ++ G V SE LDW FFL P P L +R
Sbjct: 111 PMEEKKKLWQTPAVMEGFGQAFVVSEDQKLDWADLFFLIMQPVQLRKRHLFPKLPLPFRD 170
Query: 113 TMKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIK 172
T+ ++ +++ +A+ LL + + L ++ ++++F + +++ YPPCP+P+L+
Sbjct: 171 TLDMYSTRVKSIAKILLAKMAKALQIKPEEVEEIFGDDMMQSM--RMNYYPPCPQPNLVT 228
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSV 232
GL H+DA G+ +L Q ++V GLQ+ K+ +W V P+ ++ ++N+GD LE+ITNG Y+S+
Sbjct: 229 GLIPHSDAVGLTILLQVNEVDGLQIKKNGKWFFVKPLQNAFIVNVGDVLEIITNGTYRSI 288
Query: 233 MHRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSE 273
HR + + R+SIA+F+N G D I PA +L++ E ++
Sbjct: 289 EHRAMVNLEKERLSIATFHNTGMDKEIGPARSLVQRQEAAK 329
>At5g43450 1-aminocyclopropane-1-carboxylate oxidase
Length = 362
Score = 160 bits (404), Expect = 1e-39
Identities = 99/303 (32%), Positives = 159/303 (51%), Gaps = 22/303 (7%)
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKK--CME 62
P++D+G NT R + IKDA ENWGFF+ +NH + + +++++++ + +++ ++
Sbjct: 62 PIIDLGDRNTSSRNVVISKIKDAAENWGFFQVINHDVPLTVLEEIKESVRRFHEQDPVVK 121
Query: 63 QRF------KEMVASKGLECVQSEINDLDWESTFFLRHLPSS-NISEIPDLDEDYRKTMK 115
++ K V + + S + L+W +F P N EIP R +
Sbjct: 122 NQYLPTDNNKRFVYNNDFDLYHS--SPLNWRDSFTCYIAPDPPNPEEIPLA---CRSAVI 176
Query: 116 EFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLR 175
E+ + + +L L LL E LGL+ LK++ KG YPPCP+PDL G+
Sbjct: 177 EYTKHVMELGAVLFQLLSEALGLDSETLKRIDC-LKG--LFMLCHYYPPCPQPDLTLGIS 233
Query: 176 AHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHR 235
HTD + LL QD ++ GLQ+L +D W+DVPP+P ++V+N+GD +++ITN K+ SV HR
Sbjct: 234 KHTDNSFLTLLLQD-QIGGLQVLHEDYWVDVPPVPGALVVNIGDFMQLITNDKFLSVEHR 292
Query: 236 VIAQTDGARMSIASFYNPG---NDAVISPASTLLKEDETSEIYPKFIFDDYMKLYMGLKF 292
V D R+S+A F++ N V P LL DE Y +Y ++ F
Sbjct: 293 VRPNKDRPRISVACFFSSSLSPNSTVYGPIKDLL-SDENPAKYKDITIPEYTAGFLASIF 351
Query: 293 QAK 295
K
Sbjct: 352 DEK 354
>At5g08640 flavonol synthase (FLS) (sp|Q96330)
Length = 336
Score = 160 bits (404), Expect = 1e-39
Identities = 97/290 (33%), Positives = 157/290 (53%), Gaps = 19/290 (6%)
Query: 5 PVVDMGKLNTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQR 64
PVVD L+ + ++ + A E WG F+ VNH I EL+ +++ + ++ ++ +
Sbjct: 44 PVVD---LSDPDEESVRRAVVKASEEWGLFQVVNHGIPTELIRRLQDVGRKFFE--LPSS 98
Query: 65 FKEMVA----SKGLECVQSEIND-----LDWESTFFLRHLPSS--NISEIPDLDEDYRKT 113
KE VA SK +E +++ W F R P S N P +YR+
Sbjct: 99 EKESVAKPEDSKDIEGYGTKLQKDPEGKKAWVDHLFHRIWPPSCVNYRFWPKNPPEYREV 158
Query: 114 MKEFAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKG 173
+E+A ++KL+E LL +L + LGL++ LK+ G + + K++ YPPCP+PDL G
Sbjct: 159 NEEYAVHVKKLSETLLGILSDGLGLKRDALKEGL-GGEMAEYMMKINYYPPCPRPDLALG 217
Query: 174 LRAHTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVM 233
+ AHTD GI LL ++ V GLQ+ KDD W D +P ++++++GDQ+ ++NG+YK+V+
Sbjct: 218 VPAHTDLSGITLLVPNE-VPGLQVFKDDHWFDAEYIPSAVIVHIGDQILRLSNGRYKNVL 276
Query: 234 HRVIAQTDGARMSIASFYNPGNDAVISPASTLLKEDETSEIYPKFIFDDY 283
HR + RMS F P + ++ P L +D + P F F DY
Sbjct: 277 HRTTVDKEKTRMSWPVFLEPPREKIVGPLPELTGDDNPPKFKP-FAFKDY 325
>At4g25300 SRG1-like protein
Length = 356
Score = 159 bits (401), Expect = 2e-39
Identities = 84/276 (30%), Positives = 155/276 (55%), Gaps = 10/276 (3%)
Query: 5 PVVDMGKL-NTEERKATMEMIKDACENWGFFECVNHSISIELMDKVEKLTKEHYKKCMEQ 63
P++DM L ++ + ++ + AC+ WGFF+ VNH + ++KV+ ++ + ME+
Sbjct: 53 PIIDMSLLCSSTSMDSEIDKLDSACKEWGFFQLVNHGMESSFLNKVKSEVQDFFNLPMEE 112
Query: 64 RFK-----EMVASKGLECVQSEINDLDWESTFFLRHLPSSNISE--IPDLDEDYRKTMKE 116
+ + + G V SE LDW FFL P P L +R T+
Sbjct: 113 KKNLWQQPDEIEGFGQVFVVSEEQKLDWADMFFLTMQPVRLRKPHLFPKLPLPFRDTLDM 172
Query: 117 FAEKLEKLAEELLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGLRA 176
++ +++ +A+ LL + L ++ + K+F G +++ YP CP+PD + GL
Sbjct: 173 YSAEVKSIAKILLGKIAVALKIKPEEMDKLFDDELGQRI--RLNYYPRCPEPDKVIGLTP 230
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDDQWIDVPPMPHSIVINLGDQLEVITNGKYKSVMHRV 236
H+D+ G+ +L Q ++V GLQ+ K+ +W+ V P+P+++V+N+GD LE+ITNG Y+S+ HR
Sbjct: 231 HSDSTGLTILLQANEVEGLQIKKNAKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRG 290
Query: 237 IAQTDGARMSIASFYNPGNDAVISPASTLLKEDETS 272
+ ++ R+S+A+F+N G I P +L++ + +
Sbjct: 291 VVNSEKERLSVAAFHNIGLGKEIGPMRSLVERHKAA 326
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,509,189
Number of Sequences: 26719
Number of extensions: 334839
Number of successful extensions: 1423
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 1106
Number of HSP's gapped (non-prelim): 136
length of query: 317
length of database: 11,318,596
effective HSP length: 99
effective length of query: 218
effective length of database: 8,673,415
effective search space: 1890804470
effective search space used: 1890804470
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122163.7


