

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.6 + phase: 0
(459 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
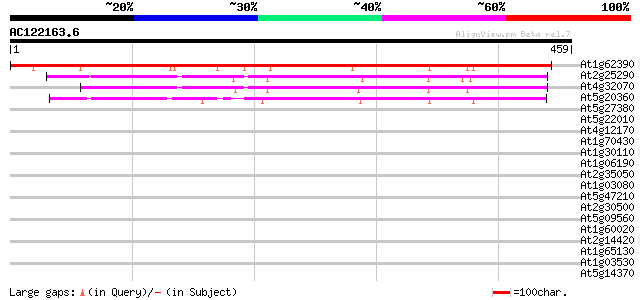
At1g62390 putative protein 419 e-117
At2g25290 unknown protein 302 2e-82
At4g32070 putative protein 298 4e-81
At5g20360 tetratricopeptide repeat protein 230 1e-60
At5g27380 glutathione synthetase gsh2 35 0.11
At5g22010 replication factor C large subunit-like protein 34 0.19
At4g12170 unknown protein (At4g12170) 33 0.25
At1g70430 hypothetical protein 32 0.56
At1g30110 unknown protein 32 0.56
At1g06190 unknown protein 32 0.56
At2g35050 putative protein kinase 32 0.73
At1g03080 unknown protein 32 0.73
At5g47210 unknown protein 32 0.96
At2g30500 unknown protein 32 0.96
At5g09560 putative protein 31 1.2
At1g60020 hypothetical protein 31 1.2
At2g14420 Mutator-like transposase 31 1.6
At1g65130 hypothetical protein 31 1.6
At1g03530 unknown protein 31 1.6
At5g14370 unknown protein 30 2.1
>At1g62390 putative protein
Length = 751
Score = 419 bits (1076), Expect = e-117
Identities = 247/534 (46%), Positives = 323/534 (60%), Gaps = 91/534 (17%)
Query: 1 MRRSGSRRKKGNVSQPNS-------PSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEH 53
+ + G G+VS PN+ P N E S K L+ P
Sbjct: 209 VHKKGVTSPVGSVSLPNASNGKVERPQVVNPVTENGGSVSKGQASRVVLKPVSHSPKGSK 268
Query: 54 LKE---------NQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRS 104
++E + ++ +I WR LK VYD DIRL QMP+NC F+ LR+IV +FP S++
Sbjct: 269 VEELGSSSVAVVGKVQEKRIRWRPLKFVYDHDIRLGQMPVNCRFKELREIVSSRFPSSKA 328
Query: 105 VLIKYKDNDDDLVTITSTEELRFAES---CVY-------KTDSVEILKLYIVEVSPEHEP 154
VLIKYKDND DLVTITST EL+ AES C+ K+DSV +L+L++V+VSPE EP
Sbjct: 329 VLIKYKDNDGDLVTITSTAELKLAESAADCILTKEPDTDKSDSVGMLRLHVVDVSPEQEP 388
Query: 155 PLLKEEKEEENNEK------QKPLDCVLDEKMCTE-CNKVVEN-------------LEID 194
LL+EE+EE + P + + + ++ TE +K VE LE+D
Sbjct: 389 MLLEEEEEEVEEKPVIEEVISSPTESLSETEINTEKTDKEVEKEKASSSEDPETKELEMD 448
Query: 195 DWLYEFAQLFRSRVGTDK--YIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVA 252
DWL++FA LFR+ VG D +ID H+LG E CS+ALEETVTS++AQ L DKA KFQEVA
Sbjct: 449 DWLFDFAHLFRTHVGIDPDAHIDLHELGMELCSEALEETVTSEKAQPLFDKASAKFQEVA 508
Query: 253 ALAFFNWGNVHMCAARKFVRMDENENE--VLVMNESEFDFVQEKYYLAREKYEQAVVIKP 310
ALAFFNWGNVHMCAARK + +DE+ + V ++ +++V+E+Y LA+EKYEQA+ IKP
Sbjct: 509 ALAFFNWGNVHMCAARKRIPLDESAGKEVVAAQLQTAYEWVKERYTLAKEKYEQALSIKP 568
Query: 311 DFYEGLLAIGQQQFELAKLNWSFGIANKMDLG----KETLRLFDVAEEKMTAANDAWENL 366
DFYEGLLA+GQQQFE+AKL+WS+ +A K+D+ ETL LFD AE KM A + WE L
Sbjct: 569 DFYEGLLALGQQQFEMAKLHWSYLLAQKIDISGWDPSETLNLFDSAEAKMKDATEMWEKL 628
Query: 367 EKGKLGE------------------QGSVG-------------------MRSQIHLFWGN 389
E+ ++ + QG G MRSQIHLFWGN
Sbjct: 629 EEQRMDDLKNPNSNKKEEVSKRRKKQGGDGNEEVSETITAEEAAEQATAMRSQIHLFWGN 688
Query: 390 MLFERSQVEFKLGMSDWKKKLDASVERFKIAGASEADVSGILKKHCFNGNARDE 443
MLFERSQVE K+G W K LD++VERFK+AGASEAD++ ++K HC N A E
Sbjct: 689 MLFERSQVECKIGKDGWNKNLDSAVERFKLAGASEADIATVVKNHCSNEAAATE 742
>At2g25290 unknown protein
Length = 697
Score = 302 bits (774), Expect = 2e-82
Identities = 177/451 (39%), Positives = 268/451 (59%), Gaps = 47/451 (10%)
Query: 31 SPKKSDQISRRLREPKLGPTNEHLKENQ-KEDVKIAWRQLKLVYDDDIRLAQMPINCSFR 89
S +K I + E K+ ++ + ++ KED + R +KLV+ DDIR AQ+P++ S
Sbjct: 245 SGRKGKAIEEKKLEDKVAVMDKEVIASEIKEDATVT-RTVKLVHGDDIRWAQLPLDSSVV 303
Query: 90 LLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVEVS 149
L+RD++K++FP + LIKY+D++ DLVTIT+T+ELR A S K S +LYI EVS
Sbjct: 304 LVRDVIKDRFPALKGFLIKYRDSEGDLVTITTTDELRLAASTREKLGS---FRLYIAEVS 360
Query: 150 PEHEPPLLKEEKEEENNEKQKPLDCVLDEKMC---TECNKVVENLEIDDWLYEFAQLFRS 206
P EP + +E ++ K V D E K +LE W+++FAQLF++
Sbjct: 361 PNQEPTYDVIDNDESTDKFAKGSSSVADNGSVGDFVESEKASTSLE--HWIFQFAQLFKN 418
Query: 207 RVG--TDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGNVHM 264
VG +D Y++ H+LG + ++A+E+ VT ++AQ+L D A KFQE+AALA FNWGNVHM
Sbjct: 419 HVGFDSDSYLELHNLGMKLYTEAMEDIVTGEDAQELFDIAADKFQEMAALAMFNWGNVHM 478
Query: 265 CAARKFVRMDENENEVLVMNESE--FDFVQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQ 322
AR+ + E+ + ++ + E F++ + +Y A EKYE AV IK DFYE LLA+GQQ
Sbjct: 479 SKARRQIYFPEDGSRETILEKVEAGFEWAKNEYNKAAEKYEGAVKIKSDFYEALLALGQQ 538
Query: 323 QFELAKLNWSFGIANKMDL----GKETLRLFDVAEEKMTAANDAWENLEKG--------- 369
QFE AKL W ++ ++D+ ++ L+L++ AEE M WE +E+
Sbjct: 539 QFEQAKLCWYHALSGEVDIESDASQDVLKLYNKAEESMEKGMQIWEEMEERRLNGISNFD 598
Query: 370 -------KLGEQG-------------SVGMRSQIHLFWGNMLFERSQVEFKLGMSDWKKK 409
KLG G + M SQI+L WG++L+ERS VE+KLG+ W +
Sbjct: 599 KHKELLQKLGLDGIFSEASDEESAEQTANMSSQINLLWGSLLYERSIVEYKLGLPTWDEC 658
Query: 410 LDASVERFKIAGASEADVSGILKKHCFNGNA 440
L+ +VE+F++AGAS D++ ++K HC + NA
Sbjct: 659 LEVAVEKFELAGASATDIAVMVKNHCSSDNA 689
>At4g32070 putative protein
Length = 811
Score = 298 bits (763), Expect = 4e-81
Identities = 167/422 (39%), Positives = 253/422 (59%), Gaps = 45/422 (10%)
Query: 59 KEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVT 118
K++ R +KLV+ DDIR AQ+P++ + RL+RD+++++FP R LIKY+D + DLVT
Sbjct: 310 KKEGATVTRTIKLVHGDDIRWAQLPLDSTVRLVRDVIRDRFPALRGFLIKYRDTEGDLVT 369
Query: 119 ITSTEELRFAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDE 178
IT+T+ELR A S K S L+LYI EV+P+ EP E ++ K L + D
Sbjct: 370 ITTTDELRLAASTHDKLGS---LRLYIAEVNPDQEPTYDGMSNTESTDKVSKRLSSLADN 426
Query: 179 KMCTE---CNKVVENLEIDDWLYEFAQLFRSRVG--TDKYIDFHDLGTEFCSDALEETVT 233
E +K E +W+++FAQLF++ VG +D Y+D HDLG + ++A+E+ VT
Sbjct: 427 GSVGEYVGSDKASGCFE--NWIFQFAQLFKNHVGFDSDSYVDLHDLGMKLYTEAMEDAVT 484
Query: 234 SDEAQDLLDKAEFKFQEVAALAFFNWGNVHMCAARKFVRMDENENEVLVMN--ESEFDFV 291
++AQ+L A KFQE+ ALA NWGNVHM ARK V + E+ + ++ E+ F +
Sbjct: 485 GEDAQELFQIAADKFQEMGALALLNWGNVHMSKARKQVCIPEDASREAIIEAVEAAFVWT 544
Query: 292 QEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQFELAKLNWSFGIANKMDL----GKETLR 347
Q +Y A EKYE+A+ +KPDFYE LLA+GQ+QFE AKL W + +K+DL +E L+
Sbjct: 545 QNEYNKAAEKYEEAIKVKPDFYEALLALGQEQFEHAKLCWYHALKSKVDLESEASQEVLK 604
Query: 348 LFDVAEEKMTAANDAWENLEKGKLGE-----------------------------QGSVG 378
L++ AE+ M WE +E+ +L + +
Sbjct: 605 LYNKAEDSMERGMQIWEEMEECRLNGISKLDKHKNMLRKLELDELFSEASEEETVEQTAN 664
Query: 379 MRSQIHLFWGNMLFERSQVEFKLGMSDWKKKLDASVERFKIAGASEADVSGILKKHCFNG 438
M SQI+L WG++L+ERS VE+KLG+ W + L+ +VE+F++AGAS D++ ++K HC +
Sbjct: 665 MSSQINLLWGSLLYERSIVEYKLGLPTWDECLEVAVEKFELAGASATDIAVMVKNHCSSE 724
Query: 439 NA 440
+A
Sbjct: 725 SA 726
>At5g20360 tetratricopeptide repeat protein
Length = 809
Score = 230 bits (587), Expect = 1e-60
Identities = 152/445 (34%), Positives = 245/445 (54%), Gaps = 58/445 (13%)
Query: 33 KKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLR 92
K+SD+ S +E + E L EDV + +K VY DDIRLA++PINC+ LR
Sbjct: 329 KQSDKRSDTSKEQEKVIIEEELLVIGVEDVN---KDVKFVYSDDIRLAELPINCTLFKLR 385
Query: 93 DIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIVEVSPEH 152
++V E+FP R+V IKY+D + DLVTIT+ EELR +E + S ++ Y+VEVSPE
Sbjct: 386 EVVHERFPSLRAVHIKYRDQEGDLVTITTDEELRMSE---VSSRSQGTMRFYVVEVSPEQ 442
Query: 153 EPPL-----LKEEKEEENNEKQKPLDCVLDEKMCTECNKVVENLEIDDWLYEFAQLFR-- 205
+P +K+ K ++ K K V C +++DW+ EFA LF+
Sbjct: 443 DPFFGRLVEMKKLKITADSFKAK----VNGRGGC----------KVEDWMIEFAHLFKIQ 488
Query: 206 SRVGTDKYIDFHDLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGNVHMC 265
+R+ +D+ ++ +LG + S+A+EE VTSD AQ D+A +FQEVAA + N G VHM
Sbjct: 489 ARIDSDRCLNLQELGMKLNSEAMEEVVTSDAAQGPFDRAAQQFQEVAARSLLNLGYVHMS 548
Query: 266 AARKFVRMDENENEVLVMNE--SEFDFVQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQ 323
ARK + + + + V + + ++ ++++ A+EKYE+A+ IKP+ +E LA+G QQ
Sbjct: 549 GARKRLSLLQGVSGESVSEQVKTAYECAKKEHANAKEKYEEAMKIKPECFEVFLALGLQQ 608
Query: 324 FELAKLNWSFGIANKMDLG----KETLRLFDVAEEKMTAANDAWENLEKGKLGEQGSVG- 378
FE A+L+W + + + +DL + ++ + AE + + + ENLE GK E G
Sbjct: 609 FEEARLSWYYVLVSHLDLKTWPYADVVQFYQSAESNIKKSMEVLENLETGKESEPSQAGK 668
Query: 379 ------------------------MRSQIHLFWGNMLFERSQVEFKLGMSDWKKKLDASV 414
++S I + +L+ERS +E+KL W++ L+A++
Sbjct: 669 TDCLTHEKDLGSSTQNNPAKEAGRLKSWIDILLCAVLYERSIMEYKLDQPFWRESLEAAM 728
Query: 415 ERFKIAGASEADVSGILKKHCFNGN 439
E+F++AG + DV I+ + GN
Sbjct: 729 EKFELAGTCKDDVVEIISEDYVAGN 753
>At5g27380 glutathione synthetase gsh2
Length = 539
Score = 34.7 bits (78), Expect = 0.11
Identities = 39/154 (25%), Positives = 73/154 (47%), Gaps = 25/154 (16%)
Query: 33 KKSDQISRRLREPKLGPTNEHLKENQKEDVKIAWRQLKLVYDDD----IRLAQMPINCSF 88
KK D + RL L ++ L+ N+KED+++ + + D++ +++ I+CSF
Sbjct: 162 KKVDVFTSRL----LDIHSKMLERNKKEDIRLGLHRFDYMLDEETNSLLQIEMNTISCSF 217
Query: 89 RLLRDIVKEKFPISRSVLIKYKDN---DDDLVTITSTEELRFAESCVYKTDSVEILKLYI 145
L +V + + +S+L Y D D + V I +T ++FA++ + K ++
Sbjct: 218 PGLSRLVSQ---LHQSLLRSYGDQIGIDSERVPI-NTSTIQFADA---------LAKAWL 264
Query: 146 VEVSPEHEPPLLKEEKEEENNEKQKPLDCVLDEK 179
E S ++ + EE N Q L +L EK
Sbjct: 265 -EYSNPRAVVMVIVQPEERNMYDQHLLSSILREK 297
>At5g22010 replication factor C large subunit-like protein
Length = 956
Score = 33.9 bits (76), Expect = 0.19
Identities = 37/131 (28%), Positives = 49/131 (37%), Gaps = 24/131 (18%)
Query: 9 KKGNVSQPNSPSSSN--IAFEPHPSPKKSDQISRRL----------------------RE 44
+KGN S P S SS + +P KS+Q S L +E
Sbjct: 13 EKGNGSAPKSTSSKAGPVKNAAETAPIKSEQASEDLETADRRKTSKYFGKDKTKVKDEKE 72
Query: 45 PKLGPTNEHLKENQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLRDIVKEKFPISRS 104
+ P LK + VK R++ V DDD +PI+ R K K R
Sbjct: 73 VEAIPAKRKLKTESDDLVKPRPRKVTKVVDDDDDDFDVPISRKTRDTTPSKKLKSGSGRG 132
Query: 105 VLIKYKDNDDD 115
+ K DNDDD
Sbjct: 133 IASKTVDNDDD 143
>At4g12170 unknown protein (At4g12170)
Length = 128
Score = 33.5 bits (75), Expect = 0.25
Identities = 23/84 (27%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Query: 135 TDSVEILKLYIVEVSPEHEPPLLKEEKEEENN---EKQKPLDCVLDEKMCTECNKVVENL 191
+DS +K Y+ P H+ + E N+ + + P+ V K C EC ++ L
Sbjct: 5 SDSDPFIKAYVSSSLPSHDGLVQSLSASEWNSLVIQSKVPVIVVFIAKDCAECGSLMPEL 64
Query: 192 EIDDWLYEFAQLFRSRVGTDKYID 215
E D YE+ F + V TD+ ++
Sbjct: 65 EFLDSEYEYMLKFYT-VDTDEELE 87
>At1g70430 hypothetical protein
Length = 594
Score = 32.3 bits (72), Expect = 0.56
Identities = 30/118 (25%), Positives = 52/118 (43%), Gaps = 12/118 (10%)
Query: 1 MRRSGS----RRKKGNVSQPNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEHLKE 56
+R++GS + K G P + SS ++ EP K+ + +PK G H+
Sbjct: 439 IRKAGSDQQEKPKNGYADSPVNRESSTLSKEPLADTKQVRKPGNEQEKPKNGYIVSHVNR 498
Query: 57 NQKEDVKIAWRQLKLVYDDDIRLAQMPINCSFRLLR---DIVKEKFPISRSVLIKYKD 111
+I L+ +DI+ AQ+ RL+R K + PIS++ ++ KD
Sbjct: 499 ESSTSEEILPLLQSLLVQNDIQRAQV-----IRLIRFFDRTAKTENPISKTEGVQEKD 551
>At1g30110 unknown protein
Length = 175
Score = 32.3 bits (72), Expect = 0.56
Identities = 22/64 (34%), Positives = 34/64 (52%), Gaps = 5/64 (7%)
Query: 259 WGNVHMCAARKF----VRMDENENEV-LVMNESEFDFVQEKYYLAREKYEQAVVIKPDFY 313
WG A+K+ +R DE+E E+ L NE++ +F + K+ E EQAV K Y
Sbjct: 91 WGGEWHGQAQKWYLVRLRNDEDEKEINLANNEADSEFAEWKWAKPEEVVEQAVDYKRPTY 150
Query: 314 EGLL 317
E ++
Sbjct: 151 EEVI 154
>At1g06190 unknown protein
Length = 401
Score = 32.3 bits (72), Expect = 0.56
Identities = 35/138 (25%), Positives = 57/138 (40%), Gaps = 14/138 (10%)
Query: 122 TEELRFAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEENNEKQKPLDCVL--DEK 179
+ E F +S Y + + PEHEP + E E EN + P+ +L D +
Sbjct: 249 SSEATFDQSSSYSVTWTQKKDTVELHDEPEHEP-AYEHEHEPENESEPGPVTTMLEPDSE 307
Query: 180 MCTECNKVVENLEIDDWLYEFAQLFRSRVGTDKYIDFHDLGTEFCSDALEETVTSDEAQD 239
+ E + + E DD ++ D +D E DA E+ SDEA++
Sbjct: 308 LKPESSSFYQEEEDDDVTFDVLS------QDDGILDVLSDDDESLDDADED---SDEAEE 358
Query: 240 --LLDKAEFKFQEVAALA 255
+ D +E K E+ +A
Sbjct: 359 EAVKDLSELKLVELRGIA 376
>At2g35050 putative protein kinase
Length = 1257
Score = 32.0 bits (71), Expect = 0.73
Identities = 27/103 (26%), Positives = 50/103 (48%), Gaps = 4/103 (3%)
Query: 67 RQLKLVY-DDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLIKYKDND-DDLVTITSTEE 124
R KL Y + R+ ++ SF+ L +KE FP +R++ + D D LV+++S E+
Sbjct: 192 RDQKLRYVGGETRIIRISKTISFQELMHKMKEIFPEARTIKYQLPGEDLDALVSVSSDED 251
Query: 125 LR--FAESCVYKTDSVEILKLYIVEVSPEHEPPLLKEEKEEEN 165
L+ E V+ E ++++ S E + E E ++
Sbjct: 252 LQNMMEECIVFGNGGSEKPRMFLFSSSDIEEAQFVMEHAEGDS 294
>At1g03080 unknown protein
Length = 1744
Score = 32.0 bits (71), Expect = 0.73
Identities = 39/160 (24%), Positives = 68/160 (42%), Gaps = 17/160 (10%)
Query: 48 GPTNEHLKENQ---KEDVKIAWRQLKLVYDDDIRLAQMPINCSFR-LLRDIVKEKFP--- 100
G N+ L E K +++ + KL D + L++ + LL D++ EK
Sbjct: 1086 GDNNKTLDEKAYLTKSTLQLEEEKCKLEDDISLLLSETIYQSNLIILLEDVILEKLSGAM 1145
Query: 101 -----ISRSVLIKYKDNDDDLVT---ITSTEELRFAESCVYKTDSVEILKLYIVEVSPEH 152
+ R ++K K ++ + S + F V + + E+L V EH
Sbjct: 1146 KLNEDLDRLSIVKCKLEEEVRELGDKLKSADIANFQLQVVLEKSNAELLSARSANVHLEH 1205
Query: 153 EPPLLKEEKEEENNEKQKPLDCVLDEKMCTECNKVVENLE 192
E +K +KE+E E + + +EK +E +K VE LE
Sbjct: 1206 EIANVKVQKEKELLEAMLMISIMQNEK--SELSKAVEGLE 1243
>At5g47210 unknown protein
Length = 357
Score = 31.6 bits (70), Expect = 0.96
Identities = 31/108 (28%), Positives = 51/108 (46%), Gaps = 10/108 (9%)
Query: 149 SPEHEPPLLKEEKEEENNEKQKPLDCVLD--EKMCTECNKV-----VENLEIDDWLYEFA 201
+PE + L EEK ++ E+ + + L+ EK+ E K VE ++D ++E
Sbjct: 206 TPEAKKELTAEEKAQKEAEEAEAREMTLEEYEKILEEKKKALQATKVEERKVDTKVFESM 265
Query: 202 QLFRSRVGTDKYIDFHDLGT--EFCSDALEETVTSDEAQDLLDKAEFK 247
Q ++ TD+ I F LG+ E DA E+ S + L A+ K
Sbjct: 266 QQLSNKKNTDEEI-FIKLGSDKEKRKDATEKAKKSLSINEFLKPADGK 312
>At2g30500 unknown protein
Length = 517
Score = 31.6 bits (70), Expect = 0.96
Identities = 31/159 (19%), Positives = 67/159 (41%), Gaps = 9/159 (5%)
Query: 259 WGNVHMCA------ARKFVRMDENENEVLVMNESEFDFVQEKYYLAREKYEQAVVIKPDF 312
W + H C A +MD+ N +L + E + D +K + +K + + + +F
Sbjct: 24 WWDSHNCPKNSKWLAENLEKMDDRVNHMLKLIEEDADSFAKKAQMYFQKRPELIQLVEEF 83
Query: 313 YEGLLAIGQQQFELAKLNWSFGIANKMDLGKETLRLFDVAEEKMTAANDAW-ENLEKGKL 371
Y A+ ++++ A +++ + +L + +EK++ + E + L
Sbjct: 84 YRMYRALA-ERYDQASGELQKNHTSEIQ-SQSSLEISSPTKEKLSRRQSSHKEEEDSSSL 141
Query: 372 GEQGSVGMRSQIHLFWGNMLFERSQVEFKLGMSDWKKKL 410
+ GS S + G+ R E +L + + K+KL
Sbjct: 142 TDSGSDSDHSSANDEDGDEALIRRMAELELELQETKQKL 180
>At5g09560 putative protein
Length = 567
Score = 31.2 bits (69), Expect = 1.2
Identities = 28/117 (23%), Positives = 54/117 (45%), Gaps = 17/117 (14%)
Query: 27 EPHPSPKKSDQIS----------RRLREPKLG----PTNEH-LKENQKEDVKIAWRQLKL 71
+PH S + +IS R + +P++ P+ +H L + ++ + +Q
Sbjct: 248 DPHGSLHRHVEISQEDALVRPFFRTITQPRIDYLPHPSYDHRLITSASKNPPVTIKQPLQ 307
Query: 72 VYDDDIRLAQMPINCSFRLLRDIVKEKFPISRSVLI--KYKDNDDDLVTITSTEELR 126
DDIR + I CS ++K + S+ + ++ D D+ LVTIT+ E+ +
Sbjct: 308 ASKDDIRQVDLKILCSNESASVVIKTRSVTDASISVGDRHPDCDERLVTITAFEKTK 364
>At1g60020 hypothetical protein
Length = 1194
Score = 31.2 bits (69), Expect = 1.2
Identities = 22/69 (31%), Positives = 33/69 (46%), Gaps = 6/69 (8%)
Query: 4 SGSRRKKGNVSQPNSPSSSNIAFEPHPSPKKSDQISRRLREPKLGPTNEH-LKENQKEDV 62
S S + N + PNSPSSS+ + P PSP S P P N+H ++ K ++
Sbjct: 825 SPSHSNQPNKTPPNSPSSSSSSPTPIPSPSPQSSNS-----PPPPPQNQHSMRTRAKNNI 879
Query: 63 KIAWRQLKL 71
++L L
Sbjct: 880 TKPIKKLTL 888
>At2g14420 Mutator-like transposase
Length = 241
Score = 30.8 bits (68), Expect = 1.6
Identities = 15/55 (27%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query: 132 VYKTDSVEILKLYIVEVSPEHEPPLL--KEEKEEENNEKQKPLDCVLDEKMCTEC 184
+ T +E + LY+V V + +L +EE+EEE + +P+D + + + C
Sbjct: 29 INNTYELESMNLYVVRVDDPYLDDVLVGEEEEEEEPKNEDEPIDLLFQDAVSCTC 83
>At1g65130 hypothetical protein
Length = 1088
Score = 30.8 bits (68), Expect = 1.6
Identities = 21/82 (25%), Positives = 42/82 (50%), Gaps = 5/82 (6%)
Query: 87 SFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTITSTEELRFAESCVYKTDSVEILKLYIV 146
S LL+ +V K P+ S ++ +++ I+ ++L Y++ ++ +KLY+
Sbjct: 626 SINLLKSVVTYKVPLVDSKILLVENS-----RISLLKDLVSLSVFDYRSYILQPVKLYLE 680
Query: 147 EVSPEHEPPLLKEEKEEENNEK 168
E + PP L EK++E +K
Sbjct: 681 EELEDAAPPELLSEKKQEKAKK 702
>At1g03530 unknown protein
Length = 801
Score = 30.8 bits (68), Expect = 1.6
Identities = 44/168 (26%), Positives = 70/168 (41%), Gaps = 24/168 (14%)
Query: 218 DLGTEFCSDALEETVTSDEAQDLLDKAEFKFQEVAALAFFNWGNVHMCAARKFVRMDENE 277
++G E S A+++ SDEA+ +D AE + + ++ A + D +
Sbjct: 219 EVGLEKVSLAVDDDEKSDEAKGEMDSAESESETSSSSASSS---------------DSSS 263
Query: 278 NEVLVMNESEFDFVQEKYYLAREKYEQAVVIKPDFYEGLLAIGQQQFELAKLNWSFGIAN 337
+E +E E D + K EK+E VV K D G L G+ + L + N I +
Sbjct: 264 SEEEESDEDESDKEENK---KEEKFEHMVVGKEDDLAGELEEGEIE-NLDEENGDDDIED 319
Query: 338 KMDLGKETLRLFDVAEEKMTAANDAWENLEKGKLGEQGSVGMRSQIHL 385
+ D + D E + AW N E LG Q +RS+ L
Sbjct: 320 EDDDDDDDDDDDDDVNEMV-----AWSNDEDDDLGLQTKEPIRSKNEL 362
>At5g14370 unknown protein
Length = 339
Score = 30.4 bits (67), Expect = 2.1
Identities = 37/144 (25%), Positives = 59/144 (40%), Gaps = 11/144 (7%)
Query: 86 CSFRLLRDIVKEKFPISRSVLIKYKDNDDDLVTIT--STEELRFAESCVYKTDSVEILKL 143
CS + + +EK S IK + N + T+ S+ L + KT + L L
Sbjct: 2 CSNKASPVVGEEKQSTRSSKRIKKRKNREATTTMEDKSSSNLDASRKIRTKTKKPKFLSL 61
Query: 144 YIVEVSPEHE---PPLLKEEKEEENNEKQ----KPLDCVLDEKMCTECNKVVENLEIDDW 196
+ E++ HE P K+ K++ NN+KQ +P EK E + E ++
Sbjct: 62 KL-ELNTSHEINENPRSKKSKKKNNNKKQSKKKEPDTTPFKEKKRAETTTTLGGGEKEEE 120
Query: 197 LYEFAQLFRSRVGTDKYI-DFHDL 219
Y+ + TD I HDL
Sbjct: 121 QYDTVAAYLFNSATDSTISSIHDL 144
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,658,653
Number of Sequences: 26719
Number of extensions: 485846
Number of successful extensions: 1860
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1822
Number of HSP's gapped (non-prelim): 51
length of query: 459
length of database: 11,318,596
effective HSP length: 103
effective length of query: 356
effective length of database: 8,566,539
effective search space: 3049687884
effective search space used: 3049687884
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC122163.6


