

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122163.2 + phase: 0
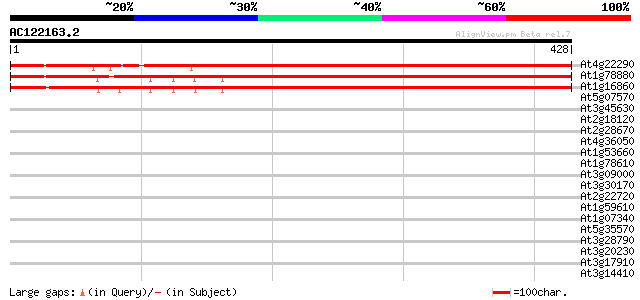
(428 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g22290 putative protein 555 e-158
At1g78880 unknown protein 433 e-121
At1g16860 unknown protein 432 e-121
At5g07570 glycine/proline-rich protein 38 0.012
At3g45630 unknown protein 35 0.079
At2g18120 unknown protein 34 0.14
At2g28670 putative disease resistance response protein 33 0.30
At4g36050 unknown protein 33 0.39
At1g53660 phosphate/phosphoenolpyruvate translocator precursor, ... 33 0.39
At1g78610 unknown protein 32 0.51
At3g09000 unknown protein 32 0.88
At3g30170 unknown protein 31 1.5
At2g22720 unknown protein 30 2.0
At1g59610 dynamin-like protein CF1 30 2.0
At1g07340 hexose transporter like protein 30 2.0
At5g35570 unknown protein 30 2.6
At3g28790 unknown protein 30 3.3
At3g20230 unknown protein 30 3.3
At3g17910 putative surfeit 1 protein 30 3.3
At3g14410 phosphate/phosphoenolpyruvate translocator protein like 30 3.3
>At4g22290 putative protein
Length = 445
Score = 555 bits (1429), Expect = e-158
Identities = 289/450 (64%), Positives = 355/450 (78%), Gaps = 27/450 (6%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
M RI SHQL +GLYVSG+ EQPKER PPTMA+R+VPYTGGD KKSGELG+M DI V+D
Sbjct: 1 MAGRIQSHQLPNGLYVSGKLEQPKER-PPTMAARAVPYTGGDIKKSGELGRMFDISVVDS 59
Query: 61 KS-----------HPSSSSSQLSTGP----ARSRPNSGQVGKNITGSGTLSRKSTGSGPI 105
S + S +S+L P + S PNSG V ++ SG++ + S GP+
Sbjct: 60 ASFQGPPPLIVGGNSSGGTSRLQAPPRVSGSSSNPNSGSV-RSGPNSGSVKKFS---GPL 115
Query: 106 A-LQPTGLITSGPVGS-GPV-GASRRSGQLEQSGS---MGKAVYGSAVTSLG-EEVKVGF 158
+ LQPTGLITSG +GS GP+ SRRSGQL+ S K YGS+VTSL + V+VGF
Sbjct: 116 SQLQPTGLITSGSLGSSGPILSGSRRSGQLDHQLSNLASSKPKYGSSVTSLNVDPVRVGF 175
Query: 159 RVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGGVIVPVLVLIIWNCVLGRKGLL 218
+V +++VW ++V AM LLVG FL VAVKK V++ A+ + P +V+++WNCV RKGLL
Sbjct: 176 KVPKAMVWAVLIVAAMGLLVGAFLTVAVKKPVVIAAVLAAVCPAIVVLVWNCVWRRKGLL 235
Query: 219 GFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESSYQRIPRCVYVSSELYEYKGWGGKS 278
F+K+YPDAELRGAIDGQ+VKVTGVVTCGSIPLESS+QR PRCVYVS+ELYEYKG+GGKS
Sbjct: 236 SFIKKYPDAELRGAIDGQFVKVTGVVTCGSIPLESSFQRTPRCVYVSTELYEYKGFGGKS 295
Query: 279 AHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVKAGYGNKVAPFVKPTTVVDVTKENR 338
A+PKHRCF+WGSR++EKY++DFYISDFQ+GLRALVKAGYG+KV+PFVKP TV +VT +N+
Sbjct: 296 ANPKHRCFSWGSRHAEKYVSDFYISDFQSGLRALVKAGYGSKVSPFVKPATVANVTTQNK 355
Query: 339 ELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTG 398
+LSP+FL WL+DR LS DDR+MRLKEG+IKEGSTVSVMG+VRRH+NVLMIVPP E VS+G
Sbjct: 356 DLSPSFLKWLSDRNLSADDRVMRLKEGYIKEGSTVSVMGMVRRHDNVLMIVPPAEAVSSG 415
Query: 399 CQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
C+W CL PT +GLIITC+DNQNADVI V
Sbjct: 416 CRWWHCLFPTYADGLIITCDDNQNADVIPV 445
>At1g78880 unknown protein
Length = 468
Score = 433 bits (1113), Expect = e-121
Identities = 235/472 (49%), Positives = 306/472 (64%), Gaps = 48/472 (10%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+R SHQLS+GL+VSGRPEQPKE+ PPTM+S ++PYTGGD KKSGELGKM DIP
Sbjct: 1 MGSRYASHQLSNGLFVSGRPEQPKEK-PPTMSSVAMPYTGGDIKKSGELGKMFDIPTDGT 59
Query: 61 KSHPS------SSSSQLSTGPARSRPNS-GQVGKNITGSGTLSRKSTGSGPIA------- 106
KS S SS S +GP PN+ G++ N+ +G+ S K T SGP++
Sbjct: 60 KSRKSGPITGGSSRSGAQSGPV---PNATGRMSGNLASAGSNSMKKTNSGPLSKHGEPLK 116
Query: 107 --------------------LQPTGLITSGPVGSGPV---GASRR-SGQLEQSGSMG--- 139
L TGLITSGP+ SGP+ GA R+ SG L+ SGSM
Sbjct: 117 KSSGPQSGGVTRQNSGPIPILPTTGLITSGPITSGPLNSSGAPRKISGPLDYSGSMKTHM 176
Query: 140 -KAVYGSAVTSLGEEVKVGFRVS--RSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALG 196
V+ AVT+L E S + V+W+ +++ M L G F++ AV ++L +
Sbjct: 177 PSVVHNQAVTTLAPEDDFSCMKSFPKPVLWLVILIFVMGFLAGGFILGAVHNAILLIVVA 236
Query: 197 GVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLESSYQ 256
+ V L IWN R+G+ F+ RYPDA+LR A +GQYVKVTGVVTCG++PLESS+
Sbjct: 237 VLFTVVAALFIWNISCERRGITDFIARYPDADLRTAKNGQYVKVTGVVTCGNVPLESSFH 296
Query: 257 RIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALVKAG 316
R+PRCVY S+ LYEY+GWG K A+ HR FTWG R +E+++ DFYISDFQ+GLRALVK G
Sbjct: 297 RVPRCVYTSTCLYEYRGWGSKPANASHRRFTWGLRSAERHVVDFYISDFQSGLRALVKTG 356
Query: 317 YGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTVSVM 376
G KV P V + V+D N + SP+F+ WL + L+ DDRIMRLKEG+IKEGSTVSV+
Sbjct: 357 NGAKVTPLVDDSVVIDFKPGNEQASPDFVRWLGKKNLTNDDRIMRLKEGYIKEGSTVSVI 416
Query: 377 GVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
GVV+R++NVLMIVP TEP++ G QW +C P +EG+++ CED+ N D I V
Sbjct: 417 GVVQRNDNVLMIVPTTEPLAAGWQWSKCTFPASLEGIVLRCEDSSNVDAIPV 468
>At1g16860 unknown protein
Length = 474
Score = 432 bits (1112), Expect = e-121
Identities = 233/475 (49%), Positives = 311/475 (65%), Gaps = 48/475 (10%)
Query: 1 MGTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDP 60
MG+R PSHQLS+GL+VSGRPEQPKER P TM++ ++PYTGGD K+SGELGKM DIP
Sbjct: 1 MGSRYPSHQLSNGLFVSGRPEQPKERAP-TMSAVAMPYTGGDIKRSGELGKMFDIPADGT 59
Query: 61 KSHPSS------SSSQLSTGPARSRPNS----GQVGKNITGSGTLSRKSTGSGPIA---- 106
KS S S S G A+S P + G++ ++ +G++S K T SGP++
Sbjct: 60 KSRKSGPIPGAPSRSGSFAGTAQSGPGAPMATGRMSGSLASAGSVSMKKTNSGPLSKHGE 119
Query: 107 -----------------------LQPTGLITSGPVGSGPV---GASRR-SGQLEQSGSMG 139
L TGLITSGP+ SGP+ GA R+ SG L+ SG M
Sbjct: 120 PLKKSSGPQSGGVTRQNSGSIPILPATGLITSGPITSGPLNSSGAPRKVSGPLDSSGLMK 179
Query: 140 K----AVYGSAVTSLGEEVKVGFRVS--RSVVWVFMVVVAMCLLVGVFLMVAVKKNVILF 193
V+ AVT+LG E S + V+W+ +++ M L G F++ AV ++L
Sbjct: 180 SHMPTVVHNQAVTTLGPEDDFSCLKSFPKPVLWLVVLIFIMGFLAGGFILGAVHNPILLV 239
Query: 194 ALGGVIVPVLVLIIWNCVLGRKGLLGFVKRYPDAELRGAIDGQYVKVTGVVTCGSIPLES 253
+ + V L IWN GR+G+ F+ RYPDA+LR A +GQ+VKVTGVVTCG++PLES
Sbjct: 240 VVAILFTVVAALFIWNICWGRRGITDFIARYPDADLRTAKNGQHVKVTGVVTCGNVPLES 299
Query: 254 SYQRIPRCVYVSSELYEYKGWGGKSAHPKHRCFTWGSRYSEKYIADFYISDFQTGLRALV 313
S+ R+PRCVY S+ LYEY+GWG K A+ HR FTWG R SE+++ DFYISDFQ+GLRALV
Sbjct: 300 SFHRVPRCVYTSTCLYEYRGWGSKPANSSHRHFTWGLRSSERHVVDFYISDFQSGLRALV 359
Query: 314 KAGYGNKVAPFVKPTTVVDVTKENRELSPNFLGWLADRKLSTDDRIMRLKEGHIKEGSTV 373
K G G KV P V + V+D + + ++SP+F+ WL + L++DDRIMRLKEG+IKEGSTV
Sbjct: 360 KTGSGAKVTPLVDDSVVIDFKQGSEQVSPDFVRWLGKKNLTSDDRIMRLKEGYIKEGSTV 419
Query: 374 SVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLPTGVEGLIITCEDNQNADVIAV 428
SV+GVV+R++NVLMIVP +EP++ G QW RC PT +EG+++ CED+ N D I V
Sbjct: 420 SVIGVVQRNDNVLMIVPSSEPLAAGWQWRRCTFPTSLEGIVLRCEDSSNVDAIPV 474
>At5g07570 glycine/proline-rich protein
Length = 1504
Score = 37.7 bits (86), Expect = 0.012
Identities = 27/107 (25%), Positives = 41/107 (38%), Gaps = 14/107 (13%)
Query: 28 PPTMASRSVPYTGGDPKKSGELG---KMLDIPV-----------LDPKSHPSSSSSQLST 73
PP + + DP +G LG +D PV L P P S
Sbjct: 633 PPNIEPSKADASDADPSNAGPLGISPLSIDPPVAGASDEGPLGKLPPDLSPPGKGSSGEG 692
Query: 74 GPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGS 120
S P+ G +G + G+G+L G P+ ++P G+ G + S
Sbjct: 693 PLVESPPSEGPLGVDPLGAGSLGISPLGKSPLRIRPLGISPFGKIPS 739
Score = 34.3 bits (77), Expect = 0.14
Identities = 25/88 (28%), Positives = 37/88 (41%), Gaps = 8/88 (9%)
Query: 41 GDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKST 100
G P G G + P++DP +S P P +GK +G L
Sbjct: 996 GGPPDVGPFG--ISPPIIDPPGEDASDGDPPGKFPLDISP----LGKCPSGVSPLGVGPP 1049
Query: 101 GSGPIALQPTGL--ITSGPVGSGPVGAS 126
G GP+ P G + +GP+G+G +G S
Sbjct: 1050 GVGPVIASPPGEGPLDAGPLGAGSLGIS 1077
Score = 32.0 bits (71), Expect = 0.67
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 7/70 (10%)
Query: 56 PVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITS 115
P++DP +S P + + +GK +G L G GP+ P G
Sbjct: 774 PIIDPPVEGASDGDP----PEKLSLDISPLGKGPSGVSLLGAGPPGVGPVIASPPG---E 826
Query: 116 GPVGSGPVGA 125
GP +GP+GA
Sbjct: 827 GPRDAGPLGA 836
Score = 29.6 bits (65), Expect = 3.3
Identities = 20/65 (30%), Positives = 25/65 (37%), Gaps = 3/65 (4%)
Query: 80 PNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSGQLEQSGSMG 139
P G + G L G GP + P G +GP G GPV AS +G +G
Sbjct: 1134 PGEGAFDGDPPGKLPLDISPVGKGPSGVSPPG---AGPPGDGPVIASPPGESPLDAGPLG 1190
Query: 140 KAVYG 144
G
Sbjct: 1191 AGSLG 1195
Score = 28.9 bits (63), Expect = 5.7
Identities = 23/88 (26%), Positives = 32/88 (36%), Gaps = 13/88 (14%)
Query: 41 GDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKST 100
G P G G + P++DP S P P S G
Sbjct: 881 GGPPDVGPFG--ISPPIIDPPGESESDGDPAGKLPLDISPLS---------KGPSCVSPL 929
Query: 101 GSGPIALQPTGL--ITSGPVGSGPVGAS 126
G GP+ P G + +GP+G+G +G S
Sbjct: 930 GVGPVIASPPGEGPLDAGPLGAGSLGIS 957
>At3g45630 unknown protein
Length = 989
Score = 35.0 bits (79), Expect = 0.079
Identities = 29/130 (22%), Positives = 54/130 (41%), Gaps = 9/130 (6%)
Query: 28 PPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRPNSGQVGK 87
PP + + + + G+P + +P S P SS S+G + + P + G
Sbjct: 256 PPPLDAYTSDSSTGNP--------IAKVPSSTSVSAPKSSPPSGSSGKSTALPAAASWGA 307
Query: 88 NITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGASRRSGQLEQSGSMGKAVYGSAV 147
+T +L+ + +G + Q +G + + V +G + S S+ KA +
Sbjct: 308 RLTNQHSLATSALSNGSLDNQ-RSTSENGTLATSTVVTKAANGPVSSSNSLQKAPLKEEI 366
Query: 148 TSLGEEVKVG 157
SL E+ K G
Sbjct: 367 QSLAEKSKPG 376
>At2g18120 unknown protein
Length = 222
Score = 34.3 bits (77), Expect = 0.14
Identities = 43/141 (30%), Positives = 61/141 (42%), Gaps = 25/141 (17%)
Query: 2 GTRIPSHQLSSGLYVSGRPEQPKERQPPTMASRSVPYTGGDPKKSGELGKMLDIPVLDPK 61
G P+H S+ + ++ R E+ ++ Q PT S P TGG SG +GK DI
Sbjct: 96 GLHCPTHVRSTWIPIAKRRERQQQLQTPT----SNP-TGG----SGRVGKYRDI-----N 141
Query: 62 SHPSSSSSQLSTGPAR--SRPNSGQVGKNITGSGTLSRKSTGSGPIALQPT----GLITS 115
H + SS L G R +S + + + SGT G G A Q T G +
Sbjct: 142 QHATLDSSGLEMGETRFPDEVSSDALFRCVRMSGT----DDGEGQYAYQTTVGIAGHLFK 197
Query: 116 GPV-GSGPVGASRRSGQLEQS 135
G + GP S RS Q ++
Sbjct: 198 GILYNQGPENKSMRSTQFYEN 218
>At2g28670 putative disease resistance response protein
Length = 447
Score = 33.1 bits (74), Expect = 0.30
Identities = 26/80 (32%), Positives = 36/80 (44%), Gaps = 3/80 (3%)
Query: 66 SSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGA 125
S S+ G SG G + G+GT S S+GSGP L PT G + G G+
Sbjct: 58 SGSTGFGFGAGSGSSGSGSTGSGL-GAGTGSIPSSGSGP-GLLPTASSVPGSLAGGGSGS 115
Query: 126 SRRSGQLEQSGS-MGKAVYG 144
+G +G+ G A+ G
Sbjct: 116 LPTTGSATGAGAGTGSALGG 135
>At4g36050 unknown protein
Length = 609
Score = 32.7 bits (73), Expect = 0.39
Identities = 36/134 (26%), Positives = 60/134 (43%), Gaps = 18/134 (13%)
Query: 20 PEQPKERQPPTMASRSVPYTGG---------DPKKSGELGKMLDIPVLDP-KSHPSSSSS 69
P+ P+ PP +ASR +P G +++ E K +++ +S+ SS
Sbjct: 335 PDIPEHSTPP-LASRYLPMIYGFQQTLVSVFKKRRANEEAKAIEVSCSSSTQSNTSSICG 393
Query: 70 QLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIA--LQPTGLITSGPVGSGPVGASR 127
+STGP R N G +G ++ S + KST A + TG I + + G +S
Sbjct: 394 DISTGPLR---NCGSMGISLEKSCSFENKSTSGVTEAETVAATGSIDN--LSDGIRASSV 448
Query: 128 RSGQLEQSGSMGKA 141
R+ + + G KA
Sbjct: 449 RALNISRDGDRKKA 462
>At1g53660 phosphate/phosphoenolpyruvate translocator precursor,
putative
Length = 316
Score = 32.7 bits (73), Expect = 0.39
Identities = 22/80 (27%), Positives = 42/80 (52%), Gaps = 2/80 (2%)
Query: 138 MGKAVYGSAVTSLGEEVKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGG 197
M KA+ AV LG V VG + + + M V++ +LV + + + +++ +GG
Sbjct: 101 MLKAIMPVAVFILG--VCVGLEIMSCKMLLIMSVISFGVLVSSYGELNINWVGVVYQMGG 158
Query: 198 VIVPVLVLIIWNCVLGRKGL 217
++ L LI+ ++ RKG+
Sbjct: 159 IVSEALRLILMEILVKRKGI 178
>At1g78610 unknown protein
Length = 856
Score = 32.3 bits (72), Expect = 0.51
Identities = 35/159 (22%), Positives = 68/159 (42%), Gaps = 22/159 (13%)
Query: 46 SGELGKMLDIPVLDPKSH---PSSSSSQLSTG-------PARSRPNSGQVGKNITGSGTL 95
SGE+ +D+ + + +S P S S ++ST +RS N+ G+ + SG
Sbjct: 99 SGEICLDMDLGMDELQSRGLTPVSESPRVSTKRDPVGRRDSRSNTNNNDDGEVVKCSGNN 158
Query: 96 SRKSTGSGPIALQPTGLITSGPVGSG--PVGASRRSGQLEQSGSMGKAVYGSAVTSLGEE 153
+ S + T S P P A +SG++ +SG M +G + + GEE
Sbjct: 159 APIQRSSSTLLKMRTRSRLSDPPTPQLPPQTADMKSGRIPKSGQMKSGFFGKSPKTQGEE 218
Query: 154 ----------VKVGFRVSRSVVWVFMVVVAMCLLVGVFL 182
+ +R + +W+ + +++ L++ F+
Sbjct: 219 EEDDPFAAEDLPEEYRKDKLSLWIVLEWLSLILIIAGFV 257
>At3g09000 unknown protein
Length = 541
Score = 31.6 bits (70), Expect = 0.88
Identities = 41/151 (27%), Positives = 57/151 (37%), Gaps = 20/151 (13%)
Query: 17 SGRPEQPKE------RQPP----TMASRSVPYTGGDPKKSGELGKMLDIPVLD--PKSHP 64
S RP +P E PP T+A R V + G P + G P S
Sbjct: 291 SSRPWKPPEMPGFSLEAPPNLRTTLADRPVSASRGRPGVASAPGSRSGSIERGGGPTSGG 350
Query: 65 SSSSSQLSTGPARSRPNSGQVGKNITG-SGTLSRKSTGSGPIALQPTGL---ITSGPVGS 120
S ++ + S P+R R G ++TG G + GSG L P + + V
Sbjct: 351 SGNARRQSCSPSRGRAPIGNTNGSLTGVRGRAKASNGGSGCDNLSPVAMGNKMVERVVNM 410
Query: 121 GPVGASRRSGQLEQSGSMGKAVYGSAVTSLG 151
+G R L ++G G SA SLG
Sbjct: 411 RKLGPPR----LTENGGRGSGKSSSAFNSLG 437
Score = 30.8 bits (68), Expect = 1.5
Identities = 34/114 (29%), Positives = 47/114 (40%), Gaps = 13/114 (11%)
Query: 2 GTRIPSHQLSSGLYVSGRPEQPKERQ--PPTMASRSVPYTGGDPKKSGELGKML------ 53
G R PS SS + RP P R P T SR V + + S +
Sbjct: 143 GLRRPSSSGSSRS--TSRPATPTRRSTTPTTSTSRPVTTRASNSRSSTPTSRATLTAARA 200
Query: 54 DIPVLDPKSHPSSSSSQLSTGPARS--RPNSGQVGKNITGSGTLSRK-STGSGP 104
P++ +SS S S P RS RP+S K ++ T +R+ ST +GP
Sbjct: 201 TTSTAAPRTTTTSSGSARSATPTRSNPRPSSASSKKPVSRPATPTRRPSTPTGP 254
>At3g30170 unknown protein
Length = 995
Score = 30.8 bits (68), Expect = 1.5
Identities = 24/93 (25%), Positives = 39/93 (41%), Gaps = 7/93 (7%)
Query: 60 PKSHPSSSSSQLSTGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVG 119
P+SH S + S GP+ + G G + G G S+ + +G GL + GP
Sbjct: 807 PQSHQGSQADLESQGPSAAGRGLGSQGPSAAGRGLGSQGPSAAG------RGLGSQGPSA 860
Query: 120 SG-PVGASRRSGQLEQSGSMGKAVYGSAVTSLG 151
+G +G+ S G+ G + G + G
Sbjct: 861 AGRGLGSQEPSAAGRGRGAQGPSAAGRGRVAQG 893
>At2g22720 unknown protein
Length = 340
Score = 30.4 bits (67), Expect = 2.0
Identities = 29/86 (33%), Positives = 39/86 (44%), Gaps = 2/86 (2%)
Query: 62 SHPSSSSSQLS-TGPARSRPNSGQVGKNITGSGTLSRKSTGSGPIALQPTGLITSGPVGS 120
S PSSS S+++ + PA S +GS SR +GSG A + + S P S
Sbjct: 4 SRPSSSGSKMNHSRPATSGSQMPNSRPASSGSQMQSRAVSGSGRPASSGSQMQNSRPQNS 63
Query: 121 GPVGA-SRRSGQLEQSGSMGKAVYGS 145
P A S+ + SGS A GS
Sbjct: 64 RPASAGSQMQQRPASSGSQRPASSGS 89
>At1g59610 dynamin-like protein CF1
Length = 920
Score = 30.4 bits (67), Expect = 2.0
Identities = 28/93 (30%), Positives = 43/93 (46%), Gaps = 7/93 (7%)
Query: 44 KKSGELGKML-DIPVLDPKSHPSSSSSQLSTGPARSRPNSGQVGK------NITGSGTLS 96
K+S L K+ + + D ++ +SS S S + R N G G+ N SG S
Sbjct: 810 KQSSLLSKLTRQLSIHDNRAAAASSWSDNSGTESSPRTNGGSSGEDWMNAFNAAASGPDS 869
Query: 97 RKSTGSGPIALQPTGLITSGPVGSGPVGASRRS 129
K GSG + + + +G SG G+SRR+
Sbjct: 870 LKRYGSGGHSRRYSDPAQNGEDSSGSGGSSRRT 902
>At1g07340 hexose transporter like protein
Length = 383
Score = 30.4 bits (67), Expect = 2.0
Identities = 15/58 (25%), Positives = 30/58 (50%)
Query: 150 LGEEVKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGGVIVPVLVLII 207
+ E +R +V++ F++ + + V + + KN ++LGG VP L+L+I
Sbjct: 157 ISEIAPARYRGGLNVMFQFLITIGILAASYVNYLTSTLKNGWRYSLGGAAVPALILLI 214
>At5g35570 unknown protein
Length = 652
Score = 30.0 bits (66), Expect = 2.6
Identities = 28/89 (31%), Positives = 37/89 (41%), Gaps = 19/89 (21%)
Query: 109 PTGLITSGPVGSGPVGA--------SRRSGQLEQSGSMG--KAVYGSAVT--SLGEEVKV 156
P I SG GS P G + GQL + G + GS V+ SL E K+
Sbjct: 79 PRSEIVSGSSGSDPSGGFDSALNRKHQTYGQLRERVVKGLLRKPMGSVVSDFSLRERKKL 138
Query: 157 GFRVSRSVVWVFMVVVAMCLLVGVFLMVA 185
G W+F +CL +GVF + A
Sbjct: 139 GH-------WMFFAFCGVCLFLGVFKICA 160
>At3g28790 unknown protein
Length = 608
Score = 29.6 bits (65), Expect = 3.3
Identities = 30/110 (27%), Positives = 45/110 (40%), Gaps = 4/110 (3%)
Query: 20 PEQPKERQP-PTMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARS 78
P P P P+ + S P G +K E M +S S++S +S +
Sbjct: 298 PSTPTPSTPTPSTPAPSTPAAGKTSEKGSESASMKKESNSKSESE-SAASGSVSKTKETN 356
Query: 79 RPNSGQVGKNITG--SGTLSRKSTGSGPIALQPTGLITSGPVGSGPVGAS 126
+ +SG K+ TG SG+ S +GS + G +S S GAS
Sbjct: 357 KGSSGDTYKDTTGTSSGSPSGSPSGSPTPSTSTDGKASSKGSASASAGAS 406
>At3g20230 unknown protein
Length = 187
Score = 29.6 bits (65), Expect = 3.3
Identities = 15/52 (28%), Positives = 26/52 (49%)
Query: 30 TMASRSVPYTGGDPKKSGELGKMLDIPVLDPKSHPSSSSSQLSTGPARSRPN 81
T +V + PK S E+G ++ P++ S ++ ++ S G A RPN
Sbjct: 25 TKLGSAVSWQSSFPKLSIEIGSVISSPIVKKDSFVQAAWTRRSRGEAAKRPN 76
>At3g17910 putative surfeit 1 protein
Length = 354
Score = 29.6 bits (65), Expect = 3.3
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 2/42 (4%)
Query: 366 HIKEGSTVSVMGVVRRHENVLMIVPPTEPVSTGCQWMRCLLP 407
HI V V+GV+R EN + VP +P STG QW +P
Sbjct: 239 HISAVKPVEVVGVIRGGENPSIFVPSNDP-STG-QWFYVDVP 278
>At3g14410 phosphate/phosphoenolpyruvate translocator protein like
Length = 340
Score = 29.6 bits (65), Expect = 3.3
Identities = 21/80 (26%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Query: 138 MGKAVYGSAVTSLGEEVKVGFRVSRSVVWVFMVVVAMCLLVGVFLMVAVKKNVILFALGG 197
M KA+ AV LG V G + + + M +++ +LV + + + +++ +GG
Sbjct: 115 MLKAIMPVAVFILG--VAAGLEMMSCRMLLIMSIISFGVLVASYGELNINWIGVVYQMGG 172
Query: 198 VIVPVLVLIIWNCVLGRKGL 217
V+ L LI ++ RKG+
Sbjct: 173 VVGEALRLIFMELLVKRKGI 192
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,036,979
Number of Sequences: 26719
Number of extensions: 451984
Number of successful extensions: 1415
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1370
Number of HSP's gapped (non-prelim): 63
length of query: 428
length of database: 11,318,596
effective HSP length: 102
effective length of query: 326
effective length of database: 8,593,258
effective search space: 2801402108
effective search space used: 2801402108
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122163.2


