

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.14 - phase: 0
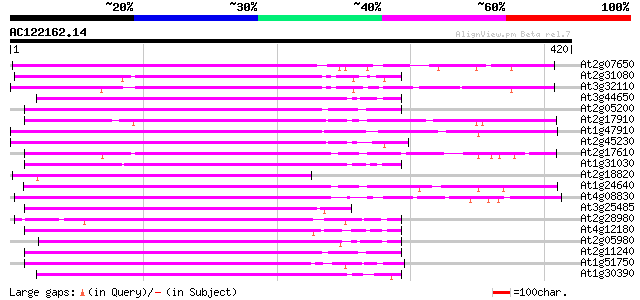
(420 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g07650 putative non-LTR retrolelement reverse transcriptase 162 4e-40
At2g31080 putative non-LTR retroelement reverse transcriptase 142 3e-34
At3g32110 non-LTR reverse transcriptase, putative 142 3e-34
At3g44650 putative protein 140 1e-33
At2g05200 putative non-LTR retroelement reverse transcriptase 140 1e-33
At2g17910 putative non-LTR retroelement reverse transcriptase 139 3e-33
At1g47910 reverse transcriptase, putative 137 1e-32
At2g45230 putative non-LTR retroelement reverse transcriptase 137 1e-32
At2g17610 putative non-LTR retroelement reverse transcriptase 135 5e-32
At1g31030 F17F8.5 134 1e-31
At2g18820 putative non-LTR retroelement reverse transcriptase 132 4e-31
At1g24640 hypothetical protein 131 6e-31
At4g08830 putative protein 130 1e-30
At3g25485 unknown protein 130 1e-30
At2g28980 putative non-LTR retroelement reverse transcriptase 127 9e-30
At4g12180 putative reverse transcriptase 127 1e-29
At2g05980 putative non-LTR retroelement reverse transcriptase 126 3e-29
At2g11240 pseudogene 125 4e-29
At1g51750 hypothetical protein 125 4e-29
At1g30390 hypothetical protein 124 7e-29
>At2g07650 putative non-LTR retrolelement reverse transcriptase
Length = 732
Score = 162 bits (409), Expect = 4e-40
Identities = 123/431 (28%), Positives = 196/431 (44%), Gaps = 55/431 (12%)
Query: 3 NAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNF 62
++++R +K + G +SH+ +ADD + EASV + ++ +L F +ASG KV+
Sbjct: 182 HSIARKDWKPISLSQGGPKLSHICFADDLILFAEASVAQIRVIRRVLERFCVASGQKVSL 241
Query: 63 YKSCLM-GINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKL 121
KS + NV + + D S KYLG+PV T+ +LE+L+ +L
Sbjct: 242 EKSKIFFSENVSRDLGKLISDESGISSTRELGKYLGMPVLQRRINKDTFGDILEKLTTRL 301
Query: 122 HSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKL 181
W +++SL GR+ L AV++SIP+ +S + +P + K+ FLWG + +K
Sbjct: 302 AGWKGRFLSLAGRVTLTKAVLSSIPVHTMSTIALPKSTLDGLDKVSRSFLWGSSVTQRKQ 361
Query: 182 CWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNL 241
+ W+ VC+PRS+GGLG+R + +N +LL+K WR++Q LW ++ CN
Sbjct: 362 HLISWKRVCKPRSEGGLGIRKAQDMNKALLSKVGWRLIQDYHSLWARIM-------RCNY 414
Query: 242 LVED--DGSWP---SYISRWWKDVAFLEDE---GGERWFNAEVVRKVGCGNSTSFWKDPW 293
V+D DG+W S S W+ VA E G W +G G FW D W
Sbjct: 415 RVQDVRDGAWTKVRSVCSSTWRSVALGMREVVIPGLSWV-------IGDGREILFWMDKW 467
Query: 294 RGDIPFSRKYPRLFAISNNKETLVEE---------CRQMNGVGGGWIFEWHRPLFVWEEE 344
+IP + E LV+E R + G GW P
Sbjct: 468 LTNIPLA-------------ELLVQEPPAGWKGMRARDLRRNGIGWDMATIAPYISSYNR 514
Query: 345 LLIS--LKEDLEGHRWVNDPDRWVKSCYGKLE-----RLLGGEVEWSLEELRVLESIWNS 397
L + + +D+ G R D W +S G+ + L + + R + +W+
Sbjct: 515 LQLQSFVLDDITGAR---DRISWGESQDGRFKVSTTYSFLTRDEAPRQDMKRFFDRVWSV 571
Query: 398 KAPLKVIAFSW 408
AP +V F W
Sbjct: 572 TAPERVRLFLW 582
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 142 bits (359), Expect = 3e-34
Identities = 99/301 (32%), Positives = 156/301 (50%), Gaps = 27/301 (8%)
Query: 4 AVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFY 63
+V + +K + G+ +SH+ +ADD + EASV + ++ +L F ASG KV+
Sbjct: 521 SVGKREWKPIAVSCGGSKLSHVCFADDLILFAEASVAQIRIIRRVLERFCEASGQKVSLE 580
Query: 64 KSCLM-GINVPSEFMTMACDF--LNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRK 120
KS + NV E + + + C++ + KYLG+P+ T+ +LE++S +
Sbjct: 581 KSKIFFSHNVSREMEQLISEESGIGCTKELG--KYLGMPILQKRMNKETFGEVLERVSAR 638
Query: 121 LHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKK 180
L W + +SL GRI L AV++SIP+ +S + +P + + FLWG KK
Sbjct: 639 LAGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTLDTLDRYSRTFLWGSTMEKKK 698
Query: 181 LCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCN 240
+ WR +C+P+++GG+G+R R +N +L+AK WR+LQ +E LW V+ +KY +V
Sbjct: 699 QHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRKKY--KVGG 756
Query: 241 LLVEDDGSWPSYISRW---WKDVAFLEDEGGERWFNAEVVRK-----VGCGNSTSFWKDP 292
+ D SW RW W+ VA G R EVV K G G + FW D
Sbjct: 757 V---QDTSWLKPQPRWSSTWRSVAV-----GLR----EVVVKGVGWVPGDGCTIRFWLDR 804
Query: 293 W 293
W
Sbjct: 805 W 805
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 142 bits (358), Expect = 3e-34
Identities = 115/426 (26%), Positives = 185/426 (42%), Gaps = 41/426 (9%)
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
+ ++++ +K KI G +SH+ +ADD + EAS+D + L+ +L F ASG KV
Sbjct: 1194 IESSIAAKKWKPIKISQSGPRLSHICFADDLILFAEASIDQIRVLRGVLEKFCGASGQKV 1253
Query: 61 NFYKSCL---------MGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWE 111
+ KS + +G + E A L KYLG+P+ T+
Sbjct: 1254 SLEKSKIYFSKNVLRDLGKRISEESGMKATKDLG--------KYLGVPILQKRINKETFG 1305
Query: 112 PLLEQLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFL 171
++++ S +L W + +S GR+ L AV+ SI + +S +K+P + K+ FL
Sbjct: 1306 EVIKRFSSRLAGWKGRMLSFAGRLTLTKAVLTSILVHTMSTIKLPQSTLDGLDKVSRAFL 1365
Query: 172 WGGARGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLI 231
WG KK V W VC PR +GGLG+R +N +L+AK WRVL LW +V+
Sbjct: 1366 WGSTLEKKKQHLVAWTRVCLPRREGGLGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVR 1425
Query: 232 EKYGTRVCNLLVEDDGSWPSYISRW---WKDVAF-LEDEGGERWFNAEVVRKVGCGNSTS 287
KY +V ++ D +W S W W+ V L D W V +G G
Sbjct: 1426 SKY--KVGDV---HDRNWTVAKSNWSSTWRSVGVGLRD---VIWREQHWV--IGDGRQIR 1475
Query: 288 FWKDPWRGDIPFSRKYPRLFAISNNKETLVEECRQMNGVGGGWIFEWHRPLFVWEEELLI 347
FW D W + P + + + ++ R + G GW P FV + + L
Sbjct: 1476 FWTDRWLSETPIADD----SIVPLSLAQMLCTARDLWRDGTGWDMSQIAP-FVTDNKRLD 1530
Query: 348 SLKEDLEGHRWVNDPDRWVKSCYGKLE-----RLLGGEVEWSLEELRVLESIWNSKAPLK 402
L ++ +D W + G+ +L + + + +W +AP +
Sbjct: 1531 LLAVIVDSVTGAHDRLAWGMTSDGRFTVKSAFAMLTNDDSPRQDMSSLYGRVWKVQAPER 1590
Query: 403 VIAFSW 408
V F W
Sbjct: 1591 VRVFLW 1596
>At3g44650 putative protein
Length = 762
Score = 140 bits (354), Expect = 1e-33
Identities = 82/274 (29%), Positives = 141/274 (50%), Gaps = 10/274 (3%)
Query: 21 GISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGINVPSEFMTMA 80
G++HL +ADD + + + ++ + + F SGLK++ KS + + S
Sbjct: 250 GLTHLSFADDLMILTDGQCRSIEGIIEVFDLFSKWSGLKISMEKSTIFSAGLSSTSRAQL 309
Query: 81 CDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNA 140
G +P +YLGLP+ V + PL+EQ+ +++ SW ++++S GR L+++
Sbjct: 310 HTHFPFEVGELPIRYLGLPLVTKRLSSVDYAPLIEQIRKRIGSWSSRFLSFAGRFNLISS 369
Query: 141 VINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQPRSKGGLGV 200
+I S F+LS ++P ++I K+ FLW G K + W VC+P+S+GGLG+
Sbjct: 370 IIWSSCNFWLSAFQLPRACIQEIEKLCSSFLWSGTNLNSKKAKISWNQVCKPKSEGGLGL 429
Query: 201 RDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDV 260
R ++ N K WR++ + LW + + R +V+++ + S+I WK +
Sbjct: 430 RSLKEANDVCCLKLVWRIISHGDSLWVKWVEHNLLKREIFWIVKENANLGSWI---WKKI 486
Query: 261 AFLEDEG-GERWFNAEVVRKVGCGNSTSFWKDPW 293
L+ G +R+ AE VG G STSFW D W
Sbjct: 487 --LKYRGVAKRFCKAE----VGNGESTSFWFDDW 514
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 140 bits (354), Expect = 1e-33
Identities = 84/283 (29%), Positives = 136/283 (47%), Gaps = 10/283 (3%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCL-MGI 70
G ++ +G ++HL +ADDT+ ++ ++ L +L + ASG +NF+KS +
Sbjct: 561 GVRVSINGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSINFHKSSVTFSS 620
Query: 71 NVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVS 130
P L + KYLGLP K + +++++ +K HSW ++++S
Sbjct: 621 KTPRSVKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQKSHSWASRFLS 680
Query: 131 LGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVC 190
G+ V+L AV+ S+P++ +S K+P+ + RKI + F W +K WV W +
Sbjct: 681 QAGKQVMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDVRKTSWVAWSKLT 740
Query: 191 QPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWP 250
P++ GGLG RDI N SLLAK WR+L E L +L+ KY C+ + P
Sbjct: 741 NPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKY----CHSSSFMECKLP 796
Query: 251 SYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW 293
S S W+ + G + + G S W DPW
Sbjct: 797 SQPSHGWRSII-----AGREILKEGLGWLITNGEKVSIWNDPW 834
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 139 bits (350), Expect = 3e-33
Identities = 107/413 (25%), Positives = 183/413 (43%), Gaps = 32/413 (7%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCL-MGI 70
G + ++ ++HL +ADDTL + +A+ L L + SG +N KS + G
Sbjct: 644 GIQFQDKKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMINLNKSAITFGK 703
Query: 71 NVPSEFMTMACDFLNCSEGVV----PFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGN 126
NV + D++ G+ KYLGLP + K + + E+L +L W
Sbjct: 704 NVDIQIK----DWIKSRSGISLEGGTGKYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYA 759
Query: 127 KYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKW 186
K +S GG+ VLL ++ ++P++ +S K+P + +K+ + DF W + +K+ W+ W
Sbjct: 760 KTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSW 819
Query: 187 RVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDD 246
+ + P+ +GG G +D++ N +LLAK WRVLQ + L+ V +Y + + L
Sbjct: 820 QRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVFQSRYFSN-SDFLSATR 878
Query: 247 GSWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRKYPRL 306
GS PSY W+ + F G + +G G T W D W D R R
Sbjct: 879 GSRPSYA---WRSILF-----GRELLMQGLRTVIGNGQKTFVWTDKWLHDGSNRRPLNRR 930
Query: 307 FAISNNKETLVEECRQMNGVGGGWIFEWHRPLFVWEE-ELLIS-----LKED----LEGH 356
I+ + ++ + ++ W R LF W++ E+++ KED L H
Sbjct: 931 RFINVD----LKVSQLIDPTSRNWNLNMLRDLFPWKDVEIILKQRPLFFKEDSFCWLHSH 986
Query: 357 RWVNDPDRWVKSCYGKLERLLGGEVEWSLEELRVLESIWNSKAPLKVIAFSWK 409
+ + ++ L E + + + IWN K+ F WK
Sbjct: 987 NGLYSVKTGYEFLSKQVHHRLYQEAKVKPSVNSLFDKIWNLHTAPKIRIFLWK 1039
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 137 bits (345), Expect = 1e-32
Identities = 110/420 (26%), Positives = 181/420 (42%), Gaps = 24/420 (5%)
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
+R A +NL G K+ +SHL +ADD+L +A+ + + +L+ +E SG ++
Sbjct: 439 IRKAERQNLITGIKVATPSPAVSHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQI 498
Query: 61 NFYKSCLM-GINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSR 119
NF KS + G V L YLGLP K + + ++L
Sbjct: 499 NFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQS 558
Query: 120 KLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGK 179
+++ W K++S GG+ V++ +V ++P + +S ++P + K+ F W +
Sbjct: 559 RINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWWSSNGDSR 618
Query: 180 KLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVC 239
+ W+ W +C +S GGLG R++ N +LLAK WR++ + L+ +V +Y R
Sbjct: 619 GMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRY-FRKS 677
Query: 240 NLLVEDDGSWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW-RGDIP 298
N L PSY R L +G ++++VG G S S W DPW P
Sbjct: 678 NPLDSIKSYSPSYGWRSMISARSLVYKG--------LIKRVGSGASISVWNDPWIPAQFP 729
Query: 299 FSRKYPRLFAISNNKETLVEECRQMNGVGGGWIFEWHRPLFVWEEELLISL-------KE 351
KY + K + + R W + + LF E+ LIS E
Sbjct: 730 RPAKYGGSIVDPSLKVKSLIDSR-----SNFWNIDLLKELFDPEDVPLISALPIGNPNME 784
Query: 352 DLEGHRWVNDPDRWVKSCYGKLERLLGGEVEWSLEELRVLES-IWNSKAPLKVIAFSWKL 410
D G + + VKS Y L +L L++ IW + P K+ F W++
Sbjct: 785 DTLGWHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQCPPKLRHFLWQI 844
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 137 bits (344), Expect = 1e-32
Identities = 84/305 (27%), Positives = 153/305 (49%), Gaps = 22/305 (7%)
Query: 1 MRNAVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKV 60
+++A +N G K+ ISHL +ADD++ + + + L + ++ + +ASG +V
Sbjct: 655 LQSAEQKNQITGLKVARGAPPISHLLFADDSMFYCKVNDEALGQIIRIIEEYSLASGQRV 714
Query: 61 NFYKSCL-MGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSR 119
N+ KS + G ++ E + L YLGLP K+ T L ++L +
Sbjct: 715 NYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKDRLGK 774
Query: 120 KLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGK 179
K+ W + ++S GG+ +LL AV ++P + +S K+P + ++I + +F W + G+
Sbjct: 775 KVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAEFWWKNKKEGR 834
Query: 180 KLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVC 239
L W W + +P++ GGLG ++I N++LL K WR++ ++ L +V +Y ++
Sbjct: 835 GLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVFKSRYFSK-S 893
Query: 240 NLLVEDDGSWPSYISRWWKDVAFLEDEGGERWFNAEVVRK------VGCGNSTSFWKDPW 293
+ L GS PS+ WK + + A+V+ K +G G + + W DPW
Sbjct: 894 DPLNAPLGSRPSFA---WKSI-----------YEAQVLIKQGIRAVIGNGETINVWTDPW 939
Query: 294 RGDIP 298
G P
Sbjct: 940 IGAKP 944
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 135 bits (339), Expect = 5e-32
Identities = 116/426 (27%), Positives = 184/426 (42%), Gaps = 58/426 (13%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLM--- 68
GFK +G ISHL +A D+L +A+++ TL +L+ +E ASG VNF KS ++
Sbjct: 121 GFKASRNGPAISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAILFGK 180
Query: 69 GINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKY 128
G++ + + +EG +YLGLP K + + + + +K+ +W NK
Sbjct: 181 GLDFRTSEQLSQLLGIYKTEGFG--RYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKL 238
Query: 129 VSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRV 188
+S G+ VL+ +++ +IP + +S +P ++ +I F W + K+ WV W
Sbjct: 239 LSPAGKEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSK 298
Query: 189 VCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGS 248
+ P+ GGL +RD++ N++LLAK WR+LQ L V KY + L D
Sbjct: 299 LNDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLL----DAK 354
Query: 249 WPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRKYPRLFA 308
S S WK + G + + + G GN+ WKD W +P + P +
Sbjct: 355 ATSQSSYAWKSILH-----GTKLISRGLKYIAGNGNNIQLWKDNW---LPLNPPRPPVGT 406
Query: 309 ISNNKETLVEECRQMNGVGGGWIFEWHRPLFVWEEELLISL--KEDLEGHRWV------- 359
+ L + G W E+LL L + D+ R +
Sbjct: 407 CDSIYSQLKVSDLLIEG--------------RWNEDLLCKLIHQNDIPHIRAIRPSITGA 452
Query: 360 NDPDRW---------VKSCYGKLERL-------LGGEVEWSLEELRVLESIWNSKAPLKV 403
ND W VKS Y L +L L E S + V +IW AP K+
Sbjct: 453 NDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASLPSPNEVSAQ--TVFTNIWKQNAPPKI 510
Query: 404 IAFSWK 409
F W+
Sbjct: 511 KHFWWR 516
>At1g31030 F17F8.5
Length = 872
Score = 134 bits (336), Expect = 1e-31
Identities = 82/283 (28%), Positives = 142/283 (49%), Gaps = 10/283 (3%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGIN 71
GF K G++HL +ADD + + + ++ + + F SGL+++ KS L
Sbjct: 335 GFHPKCQRLGLTHLSFADDLMVLSDGKTRSIEGILEVFDEFCKRSGLRISLEKSTLYMAG 394
Query: 72 V-PSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVS 130
V P +A FL G +P +YLGLP+ + PLLEQ+ +++ +W ++ S
Sbjct: 395 VSPIIKQEIAAKFL-FDVGQLPVRYLGLPLVTKRLTSADYSPLLEQIKKRIATWTFRFFS 453
Query: 131 LGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVC 190
GR L+ +V+ SI F+L+ ++P + R+I K+ FLW G+ + W +VC
Sbjct: 454 FAGRFNLIKSVLWSICNFWLAAFRLPRQCIREIDKLCSSFLWSGSEMSSHKAKISWDIVC 513
Query: 191 QPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWP 250
+P+++GGLG+R+++ N K WR++ LW + + E + ++ S
Sbjct: 514 KPKAEGGLGLRNLKEANDVSCLKLVWRIISNSNSLWTKWVAEYLIRKKSIWSLKQSTSMG 573
Query: 251 SYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW 293
S+I W+ + + D + F+ +VG G S SFW D W
Sbjct: 574 SWI---WRKILKIRDVA--KSFSRV---EVGNGESASFWYDHW 608
>At2g18820 putative non-LTR retroelement reverse transcriptase
Length = 1412
Score = 132 bits (332), Expect = 4e-31
Identities = 76/231 (32%), Positives = 117/231 (49%), Gaps = 7/231 (3%)
Query: 3 NAVSRNLFKGFKIKNDG-------TGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMA 55
N +S L KG K G G++HL +ADD + S +L + A+ + F
Sbjct: 927 NVLSAMLDKGAVEKRFGYHPRCRNMGLTHLCFADDIMVFSAGSAHSLEGVLAIFKDFAAF 986
Query: 56 SGLKVNFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLE 115
SGL ++ KS L ++ SE G +P +YLGLP+ L PLLE
Sbjct: 987 SGLNISLEKSTLFMASISSETCASILARFPFDSGSLPVRYLGLPLMTKRMTLADCLPLLE 1046
Query: 116 QLSRKLHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGA 175
++ ++ SW N+++S GR+ LLN+VI+S+ F++S ++P R+I +I FLW G
Sbjct: 1047 KIRSRISSWKNRFLSYAGRLQLLNSVISSLTKFWISAFRLPRACIREIEQISAAFLWSGT 1106
Query: 176 RGGKKLCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLW 226
V W VC+P+S+GGLG+R + N K WR++ + LW
Sbjct: 1107 DLNPHKAKVAWHDVCKPKSEGGLGLRSLVDANKICCFKLIWRLVSAKHSLW 1157
>At1g24640 hypothetical protein
Length = 1270
Score = 131 bits (330), Expect = 6e-31
Identities = 110/413 (26%), Positives = 176/413 (41%), Gaps = 28/413 (6%)
Query: 11 KGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCL-MG 69
+G + +G + HL +ADD+L + +AS + L+ +L+ + A+G +N KS + G
Sbjct: 628 EGIQFSENGPMVHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQTINLNKSSITFG 687
Query: 70 INVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYV 129
V + L YLGLP + K+ L ++L KL W + +
Sbjct: 688 EKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLKEKLDVWFTRCL 747
Query: 130 SLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVV 189
S GG+ VLL +V ++P+F +S K+P + F W +K+ W W +
Sbjct: 748 SQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWWDSCDHSRKIHWQSWERL 807
Query: 190 CQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSW 249
C P+ GGLG RDI+ N +LLAK WR+L + L +L +Y L D +
Sbjct: 808 CLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKSRYFDATDFL----DAAL 863
Query: 250 PSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRKYPR--LF 307
S W+ + F G + + ++VG G S W DPW D F + + ++
Sbjct: 864 SQRPSFGWRSILF-----GRELLSKGLQKRVGDGASLFVWIDPWIDDNGFRAPWRKNLIY 918
Query: 308 AISNNKETLVEECRQMNGVGGGWIFEWHRPLFVWEEELLISL------KEDLEGHRWVND 361
++ + L +N G W E LF+ E+ L I + D +
Sbjct: 919 DVTLKVKAL------LNPRTGFWDEEVLHDLFLPEDILRIKAIKPVISQADFFVWKLNKS 972
Query: 362 PDRWVKS----CYGKLERLLGGEVEWSLEELRVLESIWNSKAPLKVIAFSWKL 410
D VKS Y + L EV L + +WN + K+ F WK+
Sbjct: 973 GDFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTDPKIKIFLWKV 1025
>At4g08830 putative protein
Length = 947
Score = 130 bits (328), Expect = 1e-30
Identities = 112/419 (26%), Positives = 180/419 (42%), Gaps = 47/419 (11%)
Query: 4 AVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFY 63
AV+ +K + G ISH+ +ADD + EASV + ++ +L F +ASG KV+
Sbjct: 335 AVAVKEWKSIGLSQGGPKISHICFADDLILFAEASVSQIRVIRRILETFCIASGQKVSLD 394
Query: 64 KSCLM-GINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLH 122
KS + NV + + KYLG+P+ T+ +LE++S +L
Sbjct: 395 KSKIFFSKNVSRDLEKLISKESGIKSTRELGKYLGMPILQRRINKDTFGEVLERVSSRLA 454
Query: 123 SWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLC 182
W + +S GR+ L +V++ IPI +S + +P + K+ FL G + KKL
Sbjct: 455 GWKGRSLSFAGRLTLTKSVLSLIPIHTMSTISLPQSTLEGLDKLARVFLLGSSAEKKKLH 514
Query: 183 WVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLL 242
V W VC P+S+GGLG+R + +N +L++K WR++ LW +L KY + +
Sbjct: 515 LVAWDRVCLPKSEGGLGIRTSKCMNKALVSKVGWRLINDRYSLWARILRSKYRVGLREV- 573
Query: 243 VEDDGSWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRGDIPFSRK 302
+SR G RW VG G FW D W +
Sbjct: 574 ----------VSR------------GSRWV-------VGNGRDILFWSDNWLSHEALINR 604
Query: 303 YPRLFAISNNKETLVEECRQMNGVGGGWIFEWHRPLFVWEE--ELLISLKEDLEGHR--- 357
+ I N+++ L + NG+ GW + P + EL + + + G R
Sbjct: 605 --AVIEIPNSEKELRVKDLWANGL--GWKLDKIEPYISYHTRLELAAVVVDSVTGARDRL 660
Query: 358 -WVNDPDR--WVKSCYGKLERLLGGEVEWSLEELRVLESIWNSKAPLKVIAFSWKLDES 413
W D VKS Y RLL + + + +W A +V F W + ++
Sbjct: 661 SWGYSADGVFTVKSAY----RLLTEDHDPRPNMAAFFDRLWRVVALERVKTFLWHIGDT 715
>At3g25485 unknown protein
Length = 979
Score = 130 bits (328), Expect = 1e-30
Identities = 70/248 (28%), Positives = 129/248 (51%), Gaps = 5/248 (2%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGIN 71
G+ + G++HL +ADD + + + V ++ + ++ F SGLK++ KS +
Sbjct: 604 GYHPRCQTIGLTHLTFADDLMVLSDGKVRSVEGIVSVFDEFAKKSGLKISMEKSTIYLAG 663
Query: 72 VPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSL 131
+ + + + G +P +YLGLP+ F + PL+EQ+ RK+ +W +Y+S
Sbjct: 664 SANMYRREIEERFKFAVGSLPVRYLGLPLVTKRFSSADYRPLVEQIKRKIGTWTARYLSY 723
Query: 132 GGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQ 191
GR+ L+++VI SI F+++ ++P + R+I K+ FLW G K V W +C+
Sbjct: 724 AGRLNLISSVIWSICNFWIAAFRLPRECIREIDKLCSAFLWSGPELNSKRAKVNWEAICK 783
Query: 192 PRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKY---GTRVCNLLVEDDGS 248
P+ +GGLG+R ++ N K WR + + LW+++L KY + C + + + G
Sbjct: 784 PKREGGLGLRSLKEANDVSCLKLIWRWVSRGDSLWRKLL--KYRDTAKQFCKVDIRNGGR 841
Query: 249 WPSYISRW 256
+ W
Sbjct: 842 TSFWFDNW 849
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 127 bits (320), Expect = 9e-30
Identities = 86/298 (28%), Positives = 140/298 (46%), Gaps = 26/298 (8%)
Query: 4 AVSRNLFKGFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEM---ASGLKV 60
AV RN+ G+ K + G++HL +ADD + + + W+++ ++ F+ SGL++
Sbjct: 979 AVHRNI--GYHPKCEKIGLTHLCFADDLMVFVDG---HQWSIEGVINVFKEFAGRSGLQI 1033
Query: 61 NFYKSCLMGINVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRK 120
+ KS + V + + G +P +YLGLP+ + PL+E + K
Sbjct: 1034 SLEKSTIYLAGVSASDRVQTLSSFPFANGQLPVRYLGLPLLTKQMTTADYSPLIEAVKTK 1093
Query: 121 LHSWGNKYVSLGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKK 180
+ SW + +S GR+ LLN+VI SI F++S ++P R+I K+ FLW G K
Sbjct: 1094 ISSWTARSLSYAGRLALLNSVIVSIANFWMSAYRLPAGCIREIEKLCSAFLWSGPVLNPK 1153
Query: 181 LCWVKWRVVCQPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCN 240
+ W +CQP+ +GGLG++ + N K WR+L + LW T +
Sbjct: 1154 KAKIAWSSICQPKKEGGLGIKSLAEANKVSCLKLIWRLLSTQPSLWV--------TWIWT 1205
Query: 241 LLVEDDGSWP----SYISRW-WKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW 293
++ W S + W WK + E + EV G+STSFW D W
Sbjct: 1206 FIIRKGTFWSANERSSLGSWMWKKLLKYR-ELAKSMHKVEVRN----GSSTSFWYDHW 1258
>At4g12180 putative reverse transcriptase
Length = 662
Score = 127 bits (319), Expect = 1e-29
Identities = 78/286 (27%), Positives = 138/286 (47%), Gaps = 17/286 (5%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGIN 71
G+ K G++HL +ADD + + + + +L + + F SGLK++ K+ +
Sbjct: 206 GYHPKCKNLGLTHLSFADDIMVLTDGKLRSLEGIVEVFDSFAKQSGLKISMAKTTIYFAG 265
Query: 72 VPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSL 131
+ D + + G +P +YL LP+ F + PLLEQ+ R++ +W +++S
Sbjct: 266 ISKSVCKEFEDQFHFAVGRLPVRYLCLPLVTKRFTSQDYSPLLEQIKRRIGTWTARFLSY 325
Query: 132 GGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQ 191
GR+ L+++V+ SI F+LS ++P + R+I K+ FLW G + W VC+
Sbjct: 326 AGRLNLVSSVLWSICNFWLSAFRLPRECVREIDKLCSAFLWSGPELSTNKAKIAWETVCR 385
Query: 192 PRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLW----KEVLIEKYGTRVCNLLVEDDG 247
P+ +GGLG++ I+ N K WR++ + LW + L+++ + G
Sbjct: 386 PKREGGLGLQSIKEANDVCCLKLIWRIVSQGDSLWVQWIRTYLLKR--NTFWSFRSASQG 443
Query: 248 SWPSYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW 293
SW WK + D + F+ +R G + SFW D W
Sbjct: 444 SW------MWKKLLKYRDTA--KAFSKVDIRN---GETASFWYDDW 478
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 126 bits (316), Expect = 3e-29
Identities = 77/275 (28%), Positives = 133/275 (48%), Gaps = 14/275 (5%)
Query: 22 ISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGINVPSEFMTMAC 81
++HL +ADD + + + ++ A+ F S LK++ KS + + T
Sbjct: 845 LTHLCFADDIMVFSDGTSKSIQGTLAIFEKFAAMSWLKISLEKSTIFMAGISPNAKTSIL 904
Query: 82 DFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNAV 141
G +P KYLGLP+ + PL+E++ ++ SW N+++S GR+ L+ +V
Sbjct: 905 QQFPFELGTLPVKYLGLPLLTKRMTQSDYLPLVEKIRARITSWTNRFLSFAGRLQLIKSV 964
Query: 142 INSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQPRSKGGLGVR 201
++SI F+LS ++P ++I K+ FLW G K + W VC+ + +GGLG++
Sbjct: 965 LSSITNFWLSVFRLPKACLQEIEKMFSAFLWSGPDLNTKKAKIAWSEVCKLKEEGGLGLK 1024
Query: 202 DIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDD---GSWPSYISRWWK 258
++ N L K WR+L + LW + + + + V+++ GSW W+
Sbjct: 1025 PLKEANEVSLLKLIWRILSARDSLWVKWVNKHLIRKETFWSVKENTGLGSW------LWR 1078
Query: 259 DVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW 293
+ L+ R F+ VR G TSFW D W
Sbjct: 1079 KI--LKQRDKARLFHRMEVRS---GTFTSFWHDHW 1108
>At2g11240 pseudogene
Length = 1044
Score = 125 bits (314), Expect = 4e-29
Identities = 74/283 (26%), Positives = 135/283 (47%), Gaps = 10/283 (3%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCL-MGI 70
G ++ ++HL +ADDT+ + + + T +L+ +E ASG +N KS +
Sbjct: 492 GLRVSKGNPRVNHLLFADDTIFFCRSDLKSCKTFLCILKKYEEASGQMINKSKSAITFSR 551
Query: 71 NVPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVS 130
P T A L KYLGLP K + +++++ ++ SW ++++S
Sbjct: 552 KTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGRKKRDLFNQIVDRIRQRSLSWSSRFLS 611
Query: 131 LGGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVC 190
G+ +L +V+ S+P + +S K+ + ++I F W + KK+CW+ W +
Sbjct: 612 TAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQSALTHFWWDSSADKKKMCWIAWSKMA 671
Query: 191 QPRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWP 250
+ + +GGLG +DI N +LLAK WR++Q + +L+ KY C D S
Sbjct: 672 KNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSCVLVRILLGKY----CRTSSFLDCSVT 727
Query: 251 SYISRWWKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPW 293
+ S W+ + G+ +++ + +G G T W +PW
Sbjct: 728 AASSHGWRGIC-----TGKDLIKSQLGKVIGSGLDTLVWNEPW 765
>At1g51750 hypothetical protein
Length = 629
Score = 125 bits (314), Expect = 4e-29
Identities = 82/289 (28%), Positives = 137/289 (47%), Gaps = 18/289 (6%)
Query: 12 GFKIKNDGTGISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGIN 71
GF K G++HL +ADD + + + V ++ + ++ F SGL++N K+ L
Sbjct: 93 GFHPKCKNLGLTHLCFADDLMILTDGKVRSVDGIVEVMNLFAKRSGLQINMEKTTLYTAG 152
Query: 72 VPSEFMTMACDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSL 131
V M G +P +YLGLP+ PL EQ+ ++ +W ++Y+S
Sbjct: 153 VSDHNRYMMISRYPFGLGQLPVRYLGLPLVTKRLTKEDLSPLFEQIRNRIGTWTSRYLSF 212
Query: 132 GGRIVLLNAVINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQ 191
GR+ L+++V+ S F++S ++P+ ++I I FLW G ++ V W +C+
Sbjct: 213 AGRLNLISSVLWSTMNFWMSAFRLPSACLKEINSICSAFLWSGPELHRRKAKVSWDDICK 272
Query: 192 PRSKGGLGVRDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWP- 250
P+ +GGLG+R + N+ + K WRV ++ LW V K L++ + W
Sbjct: 273 PKQEGGLGLRSLTEANVVSVLKLIWRVTSNDDSLW--VKWSKMN------LLKQESFWSL 324
Query: 251 ---SYISRW-WKDVAFLEDEGGERWFNAEVVRKVGCGNSTSFWKDPWRG 295
S + W WK + E + + E V G TSFW D W G
Sbjct: 325 TPNSSLGSWMWKKMLKYR-ETAKPFSRVE----VNNGARTSFWFDNWSG 368
>At1g30390 hypothetical protein
Length = 504
Score = 124 bits (312), Expect = 7e-29
Identities = 72/275 (26%), Positives = 133/275 (48%), Gaps = 12/275 (4%)
Query: 21 GISHLQYADDTLCIGEASVDNLWTLKALLRGFEMASGLKVNFYKSCLMGINVPSEFMTMA 80
G++HL +ADD + + + V ++ + + F S LK++ KS + +
Sbjct: 27 GLTHLSFADDLMVLSDGKVRSIEGIVDVFDTFAKCSDLKISMEKSTVYLAGLSHTTRQEV 86
Query: 81 CDFLNCSEGVVPFKYLGLPVGANSFKLVTWEPLLEQLSRKLHSWGNKYVSLGGRIVLLNA 140
D + + G +P +YLGLP+ F + PL++ + +K+ SW +++S GR+ L+++
Sbjct: 87 IDRFSFAVGTLPVRYLGLPLVTKQFSSTDYLPLIDHIKQKICSWSARFLSYTGRLNLISS 146
Query: 141 VINSIPIFYLSFLKMPNKVWRKIVKIQCDFLWGGARGGKKLCWVKWRVVCQPRSKGGLGV 200
++ SI F++ ++P R+I K+ +LW G + W VC+P+ +GGLG+
Sbjct: 147 ILWSICNFWMGAFRLPRDCIREIDKMCSAYLWSGGELNTSKAKIAWAFVCKPKEEGGLGL 206
Query: 201 RDIRVVNLSLLAKWRWRVLQGEEGLWKEVLIEKYGTRVCNLLVEDDGSWPSYISRWWKDV 260
R ++ N K WR++ + LW + + +V V ++ S S++ W+ +
Sbjct: 207 RSLKEANDVCCLKLIWRIISHADSLWVKWIQSSLLKKVFFWAVRENTSLGSWM---WRKI 263
Query: 261 AFLEDEGGERWFNAEVVRKVGCGN--STSFWKDPW 293
D A + KV N TSFW D W
Sbjct: 264 LKFRD-------IARTLCKVEINNGAQTSFWYDDW 291
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.141 0.472
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,001,162
Number of Sequences: 26719
Number of extensions: 516120
Number of successful extensions: 1190
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 62
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1017
Number of HSP's gapped (non-prelim): 99
length of query: 420
length of database: 11,318,596
effective HSP length: 102
effective length of query: 318
effective length of database: 8,593,258
effective search space: 2732656044
effective search space used: 2732656044
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122162.14


