

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.12 - phase: 0
(400 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
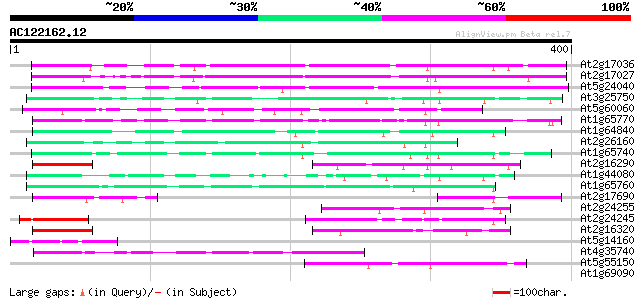
Score E
Sequences producing significant alignments: (bits) Value
At2g17036 F - Box like protein 160 9e-40
At2g17027 F - Box like protein 157 1e-38
At5g24040 putative protein 99 3e-21
At3g25750 hypothetical protein 86 4e-17
At5g60060 putative protein 84 2e-16
At1g65770 hypothetical protein 74 1e-13
At1g64840 hypothetical protein 67 2e-11
At2g26160 hypothetical protein 63 3e-10
At1g65740 hypothetical protein 60 3e-09
At2g16290 hypothetical protein 58 1e-08
At1g44080 hypothetical protein 55 7e-08
At1g65760 hypothetical protein 53 3e-07
At2g17690 hypothetical protein 52 6e-07
At2g24255 hypothetical protein 49 5e-06
At2g24245 unknown protein 49 6e-06
At2g16320 pseudogene; similar to MURA transposase of maize Muta... 47 1e-05
At5g14160 putative protein 46 4e-05
At4g35740 putative protein 43 4e-04
At5g55150 putative protein 42 5e-04
At1g69090 hypothetical protein 39 0.004
>At2g17036 F - Box like protein
Length = 404
Score = 160 bits (406), Expect = 9e-40
Identities = 130/417 (31%), Positives = 207/417 (49%), Gaps = 69/417 (16%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPN----DHNILPFQFPLLKYV 71
+DW+ LPKD++ LIS+ L+ DLI+FRSVCS+WRS+ P HN L +
Sbjct: 2 MDWATLPKDLLDLISKCLESSFDLIQFRSVCSSWRSAAGPKRLLWAHN--------LPFF 53
Query: 72 PTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN- 130
P+ D +N I+ ++ +S L+KP + Q + + W++++ N
Sbjct: 54 PSDDKPFLSNVIL-----------RVAHQSILLIKPNEPQCEADLF----GWIVKVWDNI 98
Query: 131 -SSGKTQLLKPPFLSLTSTAF-SHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSY- 187
S K LLKP LS + F HLP + D +KF+ + L + + D H++
Sbjct: 99 YVSRKMTLLKP--LSSSRNYFPQHLPRIFDMSKFTVRELCREVKLYHPDYYCVPGHTALE 156
Query: 188 LYPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSN-LSTAHGDICLFKGRF 246
L K + + K ++L L Y K +F D+ WT +++ + + D+ +F GRF
Sbjct: 157 LELGKTVVKYLNDDKFVLLTILEY--GKLAVFRSWDREWTVINDYIPSRCQDLIMFDGRF 214
Query: 247 YVVDQSGQTVTVGPDS-STELAAQPLYRRCVRGENRKLLVENEGELLLLDI--------- 296
+ +D +G+TV V S LAA PL + G ++K L+E+ GE+ L+DI
Sbjct: 215 FAIDYNGRTVVVDYSSFKLTLAANPL----IGGGDKKFLIESCGEMFLVDIEFCLNEKPE 270
Query: 297 ---------HQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL---C 344
++T + KFFKL E EK+WV+++ LG ++ F+G +FSAS D+ C
Sbjct: 271 FTGGFYSYFNETTVSYKFKFFKLVEREKRWVEVEDLGDKMFFLGDDSTFSASTADIIPRC 330
Query: 345 LPKGNCVIFI---DTSVLSSDNMAGGNRVFHLDRGQLSHVSEYPECENLFL-PPEWI 397
+ G+ V F ++ V+ D G VF G+ V++ PE LF PP WI
Sbjct: 331 VGTGSFVFFYTHEESLVVMDDRNLG---VFDFRSGKTELVNKLPEYAKLFWPPPPWI 384
>At2g17027 F - Box like protein
Length = 427
Score = 157 bits (397), Expect = 1e-38
Identities = 124/418 (29%), Positives = 199/418 (46%), Gaps = 65/418 (15%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRS-----SPIPNDHNILPFQFPLLKY 70
VDWS LPKD++ LIS+ L+ DLI+FRSVCS+WRS SP+P H P+L
Sbjct: 2 VDWSTLPKDLLDLISKSLESSFDLIQFRSVCSSWRSAAEPKSPLPTHH------LPIL-- 53
Query: 71 VPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQ- 129
PD N + +++ F LS+RS L+KP + + ++ WLI++ +
Sbjct: 54 ---PD---NGGSL---FPDSAVGF-RLSQRSILLIKPHEPCIESDSF----GWLIKVEED 99
Query: 130 -NSSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYL 188
N K LL P ++ + P V+D +KF + L +F + + S YL
Sbjct: 100 LNVPRKVTLL-DPLCDTRNSIPENFPRVLDMSKFKVRELGREFKLHYFNTVGDIVESLYL 158
Query: 189 YPQKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYV 248
+ + C VL T+ + S K +F D+ WT ++++ + + D+ LF GRF+
Sbjct: 159 EKAVVKYLDCDGDYKFVLLTI-HVSGKLAVFRSWDRAWTVINDMPSPYDDVMLFDGRFFA 217
Query: 249 VDQSGQTVTVGPDS-STELAAQPLYRRCVRGENRKLLVENEGELLLLDI----------- 296
VD +G+TV V S L A P++ G ++K L+E+ GE+LL+D+
Sbjct: 218 VDNNGRTVVVDYSSLKLTLVASPVF-----GGDKKFLIESCGEMLLVDMYLSLEAVEGDP 272
Query: 297 --------HQTFFQ----FSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDLC 344
H F+ K ++ E E+ WV + L ++LF+G +FSAS D+
Sbjct: 273 GFVEEIFEHPAFYMNERTVKFKVYRFVEREESWVDVYDLEDKMLFLGDDSTFSASASDIL 332
Query: 345 LPKGNCVIFIDTSVLSSDNMAGGN----RVFHLDRGQLSHVSEYPECENLFL-PPEWI 397
+F + +V + +++ VF G++ V + PE LF PP WI
Sbjct: 333 PLCDGSSVFFNGNVFNGEDLGAMQDRDLGVFDFRSGKIELVQKLPEYAKLFWPPPPWI 390
>At5g24040 putative protein
Length = 373
Score = 99.4 bits (246), Expect = 3e-21
Identities = 99/399 (24%), Positives = 180/399 (44%), Gaps = 46/399 (11%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTPD 75
V WSELP +I+ LIS ++D DLI FRSVCS WRSS + ++ + PL P
Sbjct: 2 VKWSELPPEILHLISLKIDNPFDLIHFRSVCSFWRSSSLLKFRHMTSLRCPL---PLDPG 58
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKT 135
++ I LS R + L P +++ Q W+ ++ +G+
Sbjct: 59 GCGDDCHI-------------LSSRVYLLKCPNRDRPQY--------WMFKLQAKENGEV 97
Query: 136 QLLKPPFLSLTSTAFSHL-PNV-VDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKI 193
+L FL S+A+ L P++ +D LA + + + F+ ++I
Sbjct: 98 -VLHSLFLRRNSSAYGCLYPSLSIDLLNCQIFELAQEHVACYSE-WFELFECISKCEERI 155
Query: 194 --LAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGD-ICLFKGRFYVVD 250
+ + + + ++LG LS+ + ++ D RWT + + + I FKG+FY +D
Sbjct: 156 GFMGLNREKNEYMILGKLSF--NGLAMYRSVDNRWTELEITPDSFFEGIVPFKGKFYAID 213
Query: 251 QSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLLLDIHQTFFQFSI----- 305
++G+T V P + E+ R C + R LL+ + +L+ ++ + F I
Sbjct: 214 RTGKTTVVDP--TLEVNTFQRSRPCDKTRKRWLLMTGDKLILVEMCTKSRYDFHIPNIRE 271
Query: 306 -----KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDLCLPKGNCVIFIDTSVLS 360
+ +L+E W +++ + GRVLF+ C+FS ++ + N +IF+D S
Sbjct: 272 KKIWFEISELNEERNDWDQVEDMDGRVLFLEHYCTFSCLATEIPGFRANSIIFMDLWGGS 331
Query: 361 SDNMAGGNRVFHLDRGQLSHVSEYPECENLF-LPPEWIL 398
+ V+ + + + + E LF PP W++
Sbjct: 332 NSYELESILVYEFNEKGVRSLRDKLEYIELFPFPPGWVI 370
>At3g25750 hypothetical protein
Length = 348
Score = 85.9 bits (211), Expect = 4e-17
Identities = 105/402 (26%), Positives = 160/402 (39%), Gaps = 78/402 (19%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M +WS+LP++++ LI+ R +D++R RS C +WRS+ + + QF +Y+P
Sbjct: 1 MEKTEWSDLPEELLDLIANRYSSNIDVLRIRSTCKSWRSAVAMSKERL---QFRFERYLP 57
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSS 132
T +N +I HLS +FF + P + WL+R Q S
Sbjct: 58 T-----SNKKIK----------AHLSPTTFFRITLPSS-------CPNKGWLVRTRQASK 95
Query: 133 --GKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYP 190
K LL P L+ + +D K + + I
Sbjct: 96 MYRKITLLCP----LSGERITRSHQTLDLLKVGVSEIRQSYEI----------------- 134
Query: 191 QKILAVTCPEKKPLVLGTLS-YCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVV 249
+I EK PL S Y + + + + WT + N DI L GR Y V
Sbjct: 135 -QIFDGLKDEKIPLDSEIFSNYIKNSDKIPSEGSRSWTKIKNQVEDFSDIILHMGRIYAV 193
Query: 250 DQSG--------QTVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLL---LDIHQ 298
D G Q V SST L Y C LVE G+L + L I +
Sbjct: 194 DLKGAIWWISLSQLTIVQQTSSTPLDYYK-YDSC----QDTRLVEYCGDLCIVHELSITR 248
Query: 299 TFFQFSI--KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFS--ASGWDLCLPKGNCVIFI 354
Q ++ K +K+DE KWV++ LG L V F+ AS + CL N + F
Sbjct: 249 NHIQRTVGFKVYKMDEDLAKWVEVSCLGDNTLIVACNSCFTVVASEYHGCLK--NSIYFS 306
Query: 355 DTSVLSSDNMAGGNRVFHLDRGQLSHVSEY--PECENLFLPP 394
V ++N+ +VF LD G ++ ++ C ++F P
Sbjct: 307 YYDVKKAENI----KVFKLDDGSITQRTDISSQSCFHMFSLP 344
>At5g60060 putative protein
Length = 374
Score = 83.6 bits (205), Expect = 2e-16
Identities = 95/359 (26%), Positives = 163/359 (44%), Gaps = 72/359 (20%)
Query: 10 TSSMASVDWSELPKDIIFLISQRLDVE----LDLIRFRSVCSTWRSS-PIPNDHNILPFQ 64
+SS+ WS+LP DI+ LIS RLD + + L+ RSVC+TWR S P+ N +N L +
Sbjct: 3 SSSLLPSQWSDLPLDILELISDRLDHDSSDTIHLLCLRSVCATWRLSLPLSNKNNRLS-K 61
Query: 65 FPLLKYVPTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWL 124
FP KY+P S S+ S+ F L + + + ++ P + R L
Sbjct: 62 FP--KYLPFWSSSSS-----------SSGFFTLKQSNVYKLEAP---------LNPRTCL 99
Query: 125 IRITQNSSGKTQLLKPPFLSLTSTAF--SHLPNVVDFTKFSPQHLATDFIIDKDDLTFQN 182
+++ + + G ++L S F + P+ +D +F + + + ++ + N
Sbjct: 100 VKLQETTPGIMRVLD--LFSNDRICFLPENFPSKIDLQEFHVRLVRRTYRME-----YAN 152
Query: 183 QHSSY------LYPQKILAVTCPEKKPLVL----GTLSYCSSKRVLFHDRDKRWTPVSNL 232
L+ K++ ++ E ++ G L + S D++W + N
Sbjct: 153 NGGGAVPCFWSLHSDKVVILSSGEDSAIIAIHSGGKLGFLKS------GNDEKWKILDNS 206
Query: 233 -STAHGDICLFK-GRFYVVDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVE-NEG 289
+ + DI L+K R VVD G+TV D A+ L G ++K LVE + G
Sbjct: 207 WNVIYEDIMLYKDNRCIVVDDKGKTVIYDVDFKVSDLAEGLVGG---GGHKKHLVECSGG 263
Query: 290 ELLLLD-----------IHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFS 337
E+ L+D I ++ +F + + L EK+W +++ LG LF+G CSFS
Sbjct: 264 EVFLVDKYVKTVWCKSDISKSVVEFRV--YNLKREEKRWEEVRDLGDVALFIGDDCSFS 320
>At1g65770 hypothetical protein
Length = 360
Score = 74.3 bits (181), Expect = 1e-13
Identities = 99/399 (24%), Positives = 166/399 (40%), Gaps = 66/399 (16%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQF-PLLKYVPTPD 75
DWS LP D++ +I+ RL ++L RFRS+C +WRSS +P PF+ PL+ P P
Sbjct: 3 DWSTLPVDLLNMIAGRLFSNIELKRFRSICRSWRSS-VPGAGKKNPFRTRPLILLNPNP- 60
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN-SSGK 134
N + D+ LS+ +FF V Q WLI+ + SSGK
Sbjct: 61 ----NKPLTDHRRRGE----FLSRSAFFRVTLSSSPSQ--------GWLIKSDVDVSSGK 104
Query: 135 TQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKIL 194
LL P L+ H VD ++F+ + + + ++ + + +++
Sbjct: 105 LHLLDP----LSRLPMEHSRKRVDLSEFTITEIREAYQVH----DWRTRKETRPIFKRVA 156
Query: 195 AVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQ 254
V E VLG S+ ++++ D K W DI + KG+ Y +D G
Sbjct: 157 LVKDKEGDNQVLGIR---STGKMMYWD-IKTW-KAKEEGYEFSDIIVHKGQTYALDSIGI 211
Query: 255 TVTVGPDSSTELAAQPLYRRCVRGENRKLLVENEGELLLLD-----------IHQTFFQF 303
+ D + PL G+ R LVE GE +++ T +++
Sbjct: 212 VYWIRSDLKF-IRFGPLVGDWT-GDRR--LVECCGEFYIVERLVGESTWKRKADDTGYEY 267
Query: 304 S----IKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDLCLPKGNCVIFIDTSVL 359
+ K +K D+ + K +++K LG + + + FS + N + F D +++
Sbjct: 268 AKTVGFKVYKFDDEQGKMMEVKSLGDKAFVIATDTCFSVLAHEFYGCLENAIYFTDDTMI 327
Query: 360 SSDNMAGGNRVFHLDRGQLSHVSE--YP---ECENLFLP 393
+VF LD G S + YP C +F+P
Sbjct: 328 ---------KVFKLDNGNGSSIETTIYPYAQSCFQMFVP 357
>At1g64840 hypothetical protein
Length = 384
Score = 66.6 bits (161), Expect = 2e-11
Identities = 87/358 (24%), Positives = 139/358 (38%), Gaps = 57/358 (15%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSS-PIPNDHNILPFQFPLLKYVPTPD 75
+WS+LP++++ LIS+ LD D++ RS+C +WRS+ P P+ + L + P P
Sbjct: 4 NWSQLPEELLNLISKNLDNCFDVVHARSICRSWRSAFPFPSSLSTLSYSLPTFAKFPL-- 61
Query: 76 SISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSSGKT 135
S L K FL + +L I Q+ S
Sbjct: 62 --------------VSKDLCTLKKIQIFLFRARNPAADIPEY-----FLGGIDQDQSNDH 102
Query: 136 QLLKPPFLSLTSTAFSHL-PNVVDFTKFSPQHLATDFI-IDKDDLTFQNQHSSYLYPQKI 193
L P F P +V+ + L +I I D + N + +
Sbjct: 103 MELPSPLQCSVKVKFPQSDPFLVNMLDYQIIPLGFQYIMIGWDPESLANGYVGVAF---- 158
Query: 194 LAVTCPEKK---PLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAH-GDICLFKGRFYVV 249
P KK + L Y + VL + RW V S A + F+GRFYV
Sbjct: 159 ----LPVKKNGGDEFVVLLRYRNHLLVL-RSSEMRWMKVKKTSIASCKGLVSFRGRFYVT 213
Query: 250 DQSGQTVTVGPDSSTE---LAAQPLYRRCVRGENRKLLVENEGELLLLDIHQTF------ 300
+G P S + + ++PL ++ L+ EL L++ F
Sbjct: 214 FLNGDIYVFDPYSLEQTLLMPSEPL------RSSKYLIPNGSDELFLVEKFNPFPEADVL 267
Query: 301 --FQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL---CLPKGNCVIF 353
+F+ + +LDE +WV++ LG RVLF+G + S +L C GN ++F
Sbjct: 268 DLSRFACRVSRLDEEAGQWVEVIDLGDRVLFIGHFGNVCCSAKELPDGCGVSGNSILF 325
>At2g26160 hypothetical protein
Length = 359
Score = 63.2 bits (152), Expect = 3e-10
Identities = 80/322 (24%), Positives = 124/322 (37%), Gaps = 53/322 (16%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M +WSELP D+I L + R D++R RS+C WRS+ P F
Sbjct: 1 MEKPEWSELPGDLINLTANRFSSISDVLRVRSICKPWRSAA------ATPKSFQC----- 49
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQ-NS 131
++ ++N++I+ + LS +FF V P + WLIR Q +
Sbjct: 50 ---NLPSSNKMIETV---------LSPTTFFRVTGPSS-------CSYKGWLIRTKQVSE 90
Query: 132 SGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQ 191
S K LL P F L + + +D KF + + I D P
Sbjct: 91 SSKINLLSPFFRQLLTPS----QQTLDLLKFEVSEIRQSYEIHIFDKYLIQGVIGKEGPS 146
Query: 192 KILA-VTCPEKKPLVLG---TLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFY 247
IL+ V + +G + C S + + WT + N DI L KG+ Y
Sbjct: 147 HILSRVVFLDNLIFAVGQDDKIWCCKSG----EESSRIWTKIKNQVEDFLDIILHKGQVY 202
Query: 248 VVDQSGQTVTVGPDSSTELAAQPLYRRCVRGEN--RKLLVENEGELLLLD--------IH 297
+D +G + + L P G + K LVE G+L ++ I
Sbjct: 203 ALDLTGAIWWISLSPLSLLQFTPSIPMDYDGYDSCNKRLVEYCGDLCIIHQLRLKKAYIR 262
Query: 298 QTFFQFSIKFFKLDEMEKKWVK 319
++ K +K+DE KWV+
Sbjct: 263 RSQRTVGFKVYKMDEYVAKWVE 284
>At1g65740 hypothetical protein
Length = 371
Score = 59.7 bits (143), Expect = 3e-09
Identities = 89/395 (22%), Positives = 152/395 (37%), Gaps = 63/395 (15%)
Query: 16 VDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPF-QFPLLKYVPTP 74
VDWS LP++++ I+ R ++ RF S+C +W SS N PF + PL+ + P
Sbjct: 2 VDWSTLPEELLHFIAARSFSLVEYKRFSSICVSWHSSVSGVKKN--PFHRRPLIDFNP-- 57
Query: 75 DSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNS-SG 133
I+ + ++++ + LS+ +FF V + W+I+ ++ SG
Sbjct: 58 --IAPSETLLEDHVFSCNPGAFLSRAAFFRVTLSSSPS--------KGWIIKSDMDTNSG 107
Query: 134 KTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKI 193
+ LL P L+ +D F+ + + + KD + + Y +
Sbjct: 108 RFHLLNP----LSRFPLRISSESLDLLDFTVSEIQESYAVLKD-AKGRLPNPGYQRSALV 162
Query: 194 LAVTCPEKKPLVLG-----TLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYV 248
+ +LG T++Y + + + DI + KG YV
Sbjct: 163 KVKEGDDHHHGILGIGRDGTINYWNGNVL---------NGFKQMGHHFSDIIVHKGVTYV 213
Query: 249 VDQSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLL--VENEGELLLLDI---------H 297
+D G + D L G R + V+ GEL ++
Sbjct: 214 LDSKGIVWCINSDLEMSRYETSLDENMTNGCRRYYMRFVDCCGELYVIKRLPKENSRKRK 273
Query: 298 QTFFQFS----IKFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL--CLPKGNCV 351
T FQFS K +K+D+ KWV++K LG + + FS + CLP N +
Sbjct: 274 STLFQFSRTAGFKVYKIDKELAKWVEVKTLGDNAFVMATDTCFSVLAHEYYGCLP--NSI 331
Query: 352 IFIDTSVLSSDNMAGGNRVFHLDRGQLSHVSEYPE 386
FI+ +VF LD G S ++ E
Sbjct: 332 YFIEDL---------EPKVFQLDNGNGSSITRKSE 357
>At2g16290 hypothetical protein
Length = 415
Score = 57.8 bits (138), Expect = 1e-08
Identities = 47/165 (28%), Positives = 81/165 (48%), Gaps = 21/165 (12%)
Query: 217 VLFHDRDKRWTPVSNL--STAHGDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLYRR 274
V++ R+ WT V + A+ D+ FKG+FY +D SG+ G EL+ +
Sbjct: 207 VMYRSRNMSWTQVVEHPETYAYQDLVAFKGKFYALDSSGR----GRVFVVELSLRVTEIP 262
Query: 275 CVRGE---NRKLLVENEGELLLLDI-------HQTFFQFSIKFFKLDEM--EKKWVKLKK 322
V+G +++ LV+ ELLL+ + + K F+LDE ++KWV++
Sbjct: 263 SVKGSQYCSKESLVQLGEELLLVQRFIPAGRRYDEYIYTWFKVFRLDEEGGKRKWVQVNY 322
Query: 323 LGGRVLFVGSGCSFSASGWDLCLPKGNCVIFI---DTSVLSSDNM 364
L RV+F+G+ S L NC++F+ D S+ +D++
Sbjct: 323 LNDRVIFLGARTKLCCSVHKLPGANENCIVFLASNDGSIFDNDSV 367
Score = 47.4 bits (111), Expect = 1e-05
Identities = 20/43 (46%), Positives = 28/43 (64%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHN 59
DWS LP D++ LI L+ +++ FRSVC +WRS P DH+
Sbjct: 3 DWSLLPNDLLELIVGHLETSFEIVLFRSVCRSWRSVVPPLDHS 45
>At1g44080 hypothetical protein
Length = 311
Score = 55.1 bits (131), Expect = 7e-08
Identities = 90/369 (24%), Positives = 141/369 (37%), Gaps = 113/369 (30%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVP 72
M V WS+L +D+I L++ L ++L+RFRS+C WRS+ V
Sbjct: 1 MGKVGWSDLHEDLIDLLANNLSSNINLLRFRSICKPWRST------------------VA 42
Query: 73 TPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQNSS 132
T + N+ E N+ +S +FF V P R + WLI+ Q S
Sbjct: 43 TKKRLHNHFE--RNLPTFKKKKTVVSPSTFFRVTLPSP-------CRNKGWLIKNRQVSE 93
Query: 133 GKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQK 192
+ S L NV F++D F+N+
Sbjct: 94 S---------IFADSDRVVFLDNVF-------------FVVD-----FKNE--------- 117
Query: 193 ILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHG--DICLFKGRFYVVD 250
+ C C S H WT ++N A G DI L KG+ Y +D
Sbjct: 118 ---IWC-------------CKSGEETRH-----WTRINN-EEAKGFLDIILHKGKIYALD 155
Query: 251 QSGQTVTVGPDSSTELA------AQPLYRRCVRGENRKLLVENEGELLLLDIHQTFFQFS 304
+G + S +EL+ + P+ + K LVE GEL + +H+ + +F
Sbjct: 156 LTGAIWWI---SLSELSIYQYGPSTPVDFYEIDNCKEKRLVEYCGELCV--VHRFYKKFC 210
Query: 305 I-----------KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSF--SASGWDLCLPKGNCV 351
+ K +K+D+ +WV++ LG + L V + F AS + CL N +
Sbjct: 211 VKRVLTERTVCFKVYKMDKNLVEWVEVSSLGDKALIVATDNCFLVLASEYYGCLE--NAI 268
Query: 352 IFIDTSVLS 360
F D +S
Sbjct: 269 YFNDREDVS 277
>At1g65760 hypothetical protein
Length = 362
Score = 52.8 bits (125), Expect = 3e-07
Identities = 76/342 (22%), Positives = 135/342 (39%), Gaps = 32/342 (9%)
Query: 13 MASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPI-PNDHNILPFQFPLLKYV 71
M DWS LP++++ +I L ++L RFRS+C +WRSS N +N P + P + +
Sbjct: 1 MVDCDWSNLPEELLHMIVLLLFSVVELKRFRSICRSWRSSTSGVNRNNPFPSR-PRIHF- 58
Query: 72 PTPDSISNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQN- 130
D I + + + S LS+ +FF V + W+I+ +
Sbjct: 59 ---DPIDPSETLSSDDSYISRPGAFLSRAAFFRVTLSSSSS--------KGWMIKSDMDI 107
Query: 131 SSGKTQLLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYP 190
+SG+ +LL L+ +D +F+ + + + L + S+
Sbjct: 108 NSGRFRLLN----VLSRYPLRLYSESIDLLEFTVSEIREAYTV----LNTAKRRSATKGY 159
Query: 191 QKILAVTCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLSTAHGDICLFKGRFYVVD 250
Q+ V E + G L + + D + + + + DI + KG
Sbjct: 160 QRSAPVKVKEGEGHRDGVLGIGRDGNINYWDGNV-LSELKQMGDHFSDIIVHKGLINSDL 218
Query: 251 QSGQTVTVGPDSSTELAAQPLYRRCVRGENRKLLVE----NEGELLLLDIHQTFFQFSIK 306
+ + T ++ T+ L R V G +VE N + D K
Sbjct: 219 EISRFGTSLDENITDGCWGDL--RFVEGCGELYIVERLPKNTRKRKAGDFFHNSRTVGFK 276
Query: 307 FFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL--CLP 346
+K+DE KW+++K LG +G+ FS ++ CLP
Sbjct: 277 AYKMDEELAKWIEIKTLGDNAFLMGTDTCFSVLAHEVYGCLP 318
>At2g17690 hypothetical protein
Length = 421
Score = 52.0 bits (123), Expect = 6e-07
Identities = 32/94 (34%), Positives = 54/94 (57%), Gaps = 13/94 (13%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSP--IPNDHNILPFQFPLLKYVPTP 74
DWS+LP++++ LI+ RL ++LIRFRS+C +WRSS + +H++ PL+ + P
Sbjct: 3 DWSKLPEELLGLIALRLYSVIELIRFRSICKSWRSSASGVNKNHSL---SSPLIYFKPLQ 59
Query: 75 DSISN---NNEIIDNIENTSTSFGHLSKRSFFLV 105
++ N +I+ T LS+ +FF V
Sbjct: 60 IILARAQANGQILSKYHGTV-----LSRATFFRV 88
Score = 43.9 bits (102), Expect = 2e-04
Identities = 28/91 (30%), Positives = 45/91 (48%), Gaps = 12/91 (13%)
Query: 306 KFFKLDEMEKKWVKLKKLGGRVLFVGSGCSFSASGWDL--CLPKGNCVIFIDTSVLSSDN 363
K +K DE KWV++K LG + + + + FS S + CLP S+ +D
Sbjct: 337 KVYKNDEELLKWVEVKSLGDKAIVIATDACFSVSAHEFYGCLP---------NSIYFTDK 387
Query: 364 MAGGNRVFHLDRGQLSHVSEYPE-CENLFLP 393
+VF LD G ++ +SE + C +F+P
Sbjct: 388 KEEEVKVFKLDDGSITTMSESEQSCFQMFVP 418
>At2g24255 hypothetical protein
Length = 336
Score = 48.9 bits (115), Expect = 5e-06
Identities = 48/150 (32%), Positives = 70/150 (46%), Gaps = 25/150 (16%)
Query: 223 DKRWTPVSNLS-TAHGDICLFKGRFYVVDQSGQTVTVGPDS--STELAAQPLYRRCVRGE 279
+ RW + N+ T DI F+GRFY + SGQ V + P S T L + P R E
Sbjct: 161 EMRWKQLENVPYTICVDIVTFRGRFYALFLSGQIVVIDPYSLEVTLLLSSPQDPR----E 216
Query: 280 NRKLLVENEGELLLL------DIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFVGS- 332
R L+ + EL L+ D+ F + + + ++DE KWV++ LG VL +G+
Sbjct: 217 LRSLVPYGKDELFLVEVIIPNDVVIDFSRLARRVRRVDEKAGKWVEVGDLGDCVLLLGNL 276
Query: 333 GCSFSASGWDLCLPKG-----NCVIFIDTS 357
CS P+G N V+F D S
Sbjct: 277 SCSAKE------FPRGFGLTENAVLFGDQS 300
>At2g24245 unknown protein
Length = 374
Score = 48.5 bits (114), Expect = 6e-06
Identities = 43/147 (29%), Positives = 68/147 (46%), Gaps = 15/147 (10%)
Query: 212 CSSKRVLFHDRDKRWTPVSNLSTAH-GDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQP 270
C+ ++ + RW + STA ++ F+GRFY +G T + P S L A P
Sbjct: 185 CTDLFLVLRSTEMRWIRLEKPSTASCKELFTFRGRFYATFFNGDTFVIDPSS---LEATP 241
Query: 271 LYRRCVRGENRKLLVENEGELLLLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLGGRVLFV 330
L + LV + E L L + F + + +LDE +WV++ LG RVLF+
Sbjct: 242 LTPHI----DSNFLVPSGNEELFL-VKTDFLRCRVS--RLDEEAAEWVEVSDLGDRVLFL 294
Query: 331 GSGC-SFSASGWDL---CLPKGNCVIF 353
G +F S +L C G+ ++F
Sbjct: 295 GGHLGNFYCSAKELPHGCGLTGDSILF 321
Score = 42.4 bits (98), Expect = 5e-04
Identities = 22/51 (43%), Positives = 35/51 (68%), Gaps = 3/51 (5%)
Query: 8 VATSSMASVDWSELPKDIIFLISQRL-DVELDLIRFRSVCSTWRSS-PIPN 56
++TSS+ DWS+LP++++ +IS L D D + RSVC +WRS+ P P+
Sbjct: 7 ISTSSIMP-DWSQLPEELLHIISTHLEDHYFDAVHARSVCRSWRSTFPFPS 56
>At2g16320 pseudogene; similar to MURA transposase of maize
Mutator transposon
Length = 467
Score = 47.4 bits (111), Expect = 1e-05
Identities = 20/43 (46%), Positives = 28/43 (64%)
Query: 17 DWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHN 59
DWS LP D++ LI L+ +++ FRSVCS+WRS P D +
Sbjct: 3 DWSLLPNDLLELIVGHLETSFEIVLFRSVCSSWRSVVPPQDQS 45
Score = 45.8 bits (107), Expect = 4e-05
Identities = 40/146 (27%), Positives = 65/146 (44%), Gaps = 16/146 (10%)
Query: 217 VLFHDRDKRWTPVSNLST--AHGDICLFKGRFYVVDQSGQTVTVGPDSSTELAAQPLYRR 274
V++ R+ WT V A+ D+ FKG+FY +D SG+ + S E+ P R
Sbjct: 203 VMYRSRNMSWTQVVEHPEKYAYQDLVAFKGKFYALDSSGRGRVFVVELSLEVMEIPSVRG 262
Query: 275 CVRGENRKLLVENEGELLLLDIHQTFFQFSIKFFKLDEMEKKWVKLKKLG---GRVLFVG 331
+ L++ E ELLL+ +F+ + DE W ++ +L GR
Sbjct: 263 SQQSSKENLVLSGE-ELLLVQ------RFTPEVRHYDEYLYTWFRVFRLDEEEGRETQTN 315
Query: 332 SGCSFSASGWDLCLPKGNCVIFIDTS 357
CS L K NC++FI+++
Sbjct: 316 LCCSVH----KLPCAKENCIVFIESN 337
Score = 40.4 bits (93), Expect = 0.002
Identities = 20/53 (37%), Positives = 29/53 (53%), Gaps = 6/53 (11%)
Query: 18 WSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKY 70
WS L D++ LI + +++ RSVCS+WRS P D+ FPL +Y
Sbjct: 402 WSLLLNDLLKLIVGHFETSFEIVNLRSVCSSWRSVVPPLDN------FPLFRY 448
>At5g14160 putative protein
Length = 352
Score = 45.8 bits (107), Expect = 4e-05
Identities = 32/77 (41%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 1 MIYFHTAVATSSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNI 60
M H A +S VDWSELP+D+I L+ +RL + D R R+VCSTW + D
Sbjct: 1 MALIHKRQALNSRG-VDWSELPEDVIRLVLRRLRLS-DFHRARAVCSTW--CRVWGDCVS 56
Query: 61 LPFQFPLLKYVPTPDSI 77
P Q P L P P I
Sbjct: 57 KPNQVPWLILFPDPAQI 73
>At4g35740 putative protein
Length = 926
Score = 42.7 bits (99), Expect = 4e-04
Identities = 51/238 (21%), Positives = 98/238 (40%), Gaps = 46/238 (19%)
Query: 18 WSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFPLLKYVPTPDSI 77
WS+LP +++ I+ L +++L+RFRS+C T+RS+ + +D N L D +
Sbjct: 7 WSDLPGELLDHIANGLFSKVELLRFRSICKTFRSA-VDSDKNFL-------------DHL 52
Query: 78 SNNNEIIDNIENTSTSFGHLSKRSFFLVKPPQEQQQQETLIRRRPWLIRITQ-NSSGKTQ 136
N + + ST+F + S+ + WLI++ S + Q
Sbjct: 53 KRNRRRL--LSPYSTAFYRVVLSSY----------------PDKGWLIKLQDAYVSSQKQ 94
Query: 137 LLKPPFLSLTSTAFSHLPNVVDFTKFSPQHLATDFIIDKDDLTFQNQHSSYLYPQKILAV 196
LL P L+ + +D +F+ + + D + L + + +S+ + + +LA
Sbjct: 95 LLSP----LSRFSIKSSGKTLDLLEFTVSEIHQSY--DVEYLYYNSTRASFNFARVVLA- 147
Query: 197 TCPEKKPLVLGTLSYCSSKRVLFHDRDKRWTPVSNLST-AHGDICLFKGRFYVVDQSG 253
+ V +Y ++ D W + + DI KG Y +D +G
Sbjct: 148 -----EDFVFIVDNYKKIWLCNSNESDSHWVRIMDEEVKLFSDIVFHKGYMYALDLTG 200
>At5g55150 putative protein
Length = 340
Score = 42.4 bits (98), Expect = 5e-04
Identities = 38/170 (22%), Positives = 77/170 (44%), Gaps = 19/170 (11%)
Query: 211 YCSSKRVLFHDR-DKRWTPVSNLSTAHGDICLFKGRFYVVDQSGQ----TVTVGPDSSTE 265
Y + +++ F R DK+WT + +++++ DI G F+ +D+ G+ ++ +T
Sbjct: 140 YNTDRKLAFCRRGDKQWTDLESVASSVDDIVFCNGVFFAIDRLGEIYHCELSANNPKATP 199
Query: 266 LAAQPLYRRCVRGENRKLLVENEGELLLLDIHQ------TFFQFSIKFFKLDEMEKKWVK 319
L + +R +K L E++ + L + + + F+ S + ++ + +W K
Sbjct: 200 LCSTSPFR---YDSCKKYLAESDYDELWVVLKKLELNDDCDFETSFEIYEFNRETNEWTK 256
Query: 320 LKKLGGRVLFVG-SGCSFSASGWDLCLPKGNCVIFIDTSVLSSDNMAGGN 368
+ L G+ LF+ G + + K N V FID D GG+
Sbjct: 257 VMSLRGKALFLSPQGRCIAVLAGERGFFKDNSVYFID----GDDPSVGGS 302
Score = 30.8 bits (68), Expect = 1.4
Identities = 15/56 (26%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 12 SMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWR--SSPIPNDHNILPFQF 65
++ S WSE +++ + L+ D++ +VCS+W+ SS + PF F
Sbjct: 2 ALPSSSWSEFLPELLNTVFHNLNDARDILNCATVCSSWKDSSSAVYYSRTFSPFLF 57
>At1g69090 hypothetical protein
Length = 401
Score = 39.3 bits (90), Expect = 0.004
Identities = 35/120 (29%), Positives = 57/120 (47%), Gaps = 13/120 (10%)
Query: 7 AVATSSMASVDWSELPKDIIFLISQRLDVELDLIRFRSVCSTWRSSPIPNDHNILPFQFP 66
AV+ S WS+LP D++ L+ +RL LD R +SVCS+W+ + N Q P
Sbjct: 18 AVSVSQRNPHCWSKLPLDLMQLVFERL-AFLDFERAKSVCSSWQFGSKQSKPN---NQIP 73
Query: 67 LLKYVPTPDS--ISNNNEIIDNIENTSTSFGHLSK-------RSFFLVKPPQEQQQQETL 117
+ PT + + N E + + T +K RS+ L++P E+ + +TL
Sbjct: 74 WMILFPTDKNYCLLFNPEDKEKLYKTQHLGDDFAKSIVLATYRSWLLMQPRYEELEDQTL 133
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,725,833
Number of Sequences: 26719
Number of extensions: 438901
Number of successful extensions: 1085
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 1014
Number of HSP's gapped (non-prelim): 81
length of query: 400
length of database: 11,318,596
effective HSP length: 101
effective length of query: 299
effective length of database: 8,619,977
effective search space: 2577373123
effective search space used: 2577373123
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122162.12


