

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122162.1 + phase: 0 /pseudo/partial
(517 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
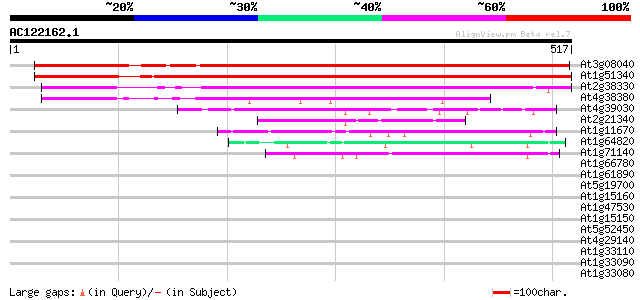
Score E
Sequences producing significant alignments: (bits) Value
At3g08040 putative integral membrane protein 582 e-166
At1g51340 putative protein 529 e-150
At2g38330 unknown protein 337 1e-92
At4g38380 putative protein 224 1e-58
At4g39030 enhanced disease susceptibility 5 gene (EDS5) 66 5e-11
At2g21340 unknown protein 56 4e-08
At1g11670 unknown protein 51 1e-06
At1g64820 hypothetical protein 44 2e-04
At1g71140 hypothetical protein 42 8e-04
At1g66780 hypothetical protein 41 0.001
At1g61890 unknown protein 40 0.002
At5g19700 putative protein 39 0.007
At1g15160 hypothetical protein 38 0.015
At1g47530 unknown protein 37 0.034
At1g15150 unknown protein (At1g15150) 37 0.034
At5g52450 unknown protein 36 0.059
At4g29140 unknown protein 36 0.059
At1g33110 Unknown protein (T9L6.1) 35 0.076
At1g33090 unknown protein 35 0.100
At1g33080 unknown protein 35 0.13
>At3g08040 putative integral membrane protein
Length = 526
Score = 582 bits (1499), Expect = e-166
Identities = 312/494 (63%), Positives = 379/494 (76%), Gaps = 17/494 (3%)
Query: 24 VFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRI 83
VF D+ +EILGIAFP+ALA+AADPIASLIDTAF+G LG V+LAA GVSIA+FNQASRI
Sbjct: 27 VFSRDTTGREILGIAFPAALALAADPIASLIDTAFVGRLGAVQLAAVGVSIAIFNQASRI 86
Query: 84 TIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQD-IEAGATKQ 142
TIFPLVS+TTSFVAEEDTM+++ +A K+N V + L+QD +E G +
Sbjct: 87 TIFPLVSLTTSFVAEEDTMEKMKEEAN----------KANLVHAETILVQDSLEKGISSP 136
Query: 143 DSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLI 202
S N + +NS NKS K +KR I +ASTA++ G +LGL+QA LI
Sbjct: 137 TS---NDTNQPQQPPAPDTKSNSGNKSNKK---EKRTIRTASTAMILGLILGLVQAIFLI 190
Query: 203 FAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIV 262
F++K LLG MG+K +SPML PA KYL +RALGAPA+LLSLAMQGIFRGFKDT TPL+ V
Sbjct: 191 FSSKLLLGVMGVKPNSPMLSPAHKYLSIRALGAPALLLSLAMQGIFRGFKDTKTPLFATV 250
Query: 263 SGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIF 322
+N+ +DP+ IF +LGI GAAI+HV+SQY M +L L KKV+L+PP+ DLQ
Sbjct: 251 VADVINIVLDPIFIFVLRLGIIGAAIAHVISQYFMTLILFVFLAKKVNLIPPNFGDLQFG 310
Query: 323 RFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQ 382
RFLKNG LLLAR IAVTFC TL+A++AARLG PMAAFQ CLQVW+TSSLL DGLAVA Q
Sbjct: 311 RFLKNGLLLLARTIAVTFCQTLAAAMAARLGTTPMAAFQICLQVWLTSSLLNDGLAVAGQ 370
Query: 383 AILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRL 442
AILACSFAEKDYNKVT A+R LQM FVLG+GLS+ VG GLYFGAGVFSK+ AVIHL+ +
Sbjct: 371 AILACSFAEKDYNKVTAVASRVLQMGFVLGLGLSVFVGLGLYFGAGVFSKDPAVIHLMAI 430
Query: 443 GLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWI 502
G+PF+AATQPINSLAFV DGVN+GASDFAY+AYS+V V+ S+ ++ ++ K+ GFIGIWI
Sbjct: 431 GIPFIAATQPINSLAFVLDGVNFGASDFAYTAYSMVGVAAISIAAVIYMAKTNGFIGIWI 490
Query: 503 ALTIYMSLRMFAGV 516
ALTIYM+LR G+
Sbjct: 491 ALTIYMALRAITGI 504
>At1g51340 putative protein
Length = 509
Score = 529 bits (1363), Expect = e-150
Identities = 291/495 (58%), Positives = 352/495 (70%), Gaps = 20/495 (4%)
Query: 24 VFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRI 83
V K D L EI IA P+ALA+ ADPIASL+DTAFIG +GPVELAA GVSIA+FNQ SRI
Sbjct: 20 VLKFDELGLEIARIALPAALALTADPIASLVDTAFIGQIGPVELAAVGVSIALFNQVSRI 79
Query: 84 TIFPLVSITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQ- 142
IFPLVSITTSFVAEED S + DH + IE G
Sbjct: 80 AIFPLVSITTSFVAEEDA------------------CSSQQDTVRDHK-ECIEIGINNPT 120
Query: 143 DSTLKNGDDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLI 202
+ T++ + + + T+SS S SKP KKR+I SAS+AL+ G VLGL QA LI
Sbjct: 121 EETIELIPEKHKDSLSDEFKTSSSIFSISKPPAKKRNIPSASSALIIGGVLGLFQAVFLI 180
Query: 203 FAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIV 262
AAKPLL MG+K+DSPM+ P+ +YL LR+LGAPAVLLSLA QG+FRGFKDTTTPL+ V
Sbjct: 181 SAAKPLLSFMGVKHDSPMMRPSQRYLSLRSLGAPAVLLSLAAQGVFRGFKDTTTPLFATV 240
Query: 263 SGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIF 322
G N+ +DP+ IF F+LG+ GAA +HV+SQY+M +LL+ LM +VD+ S K LQ
Sbjct: 241 IGDVTNIILDPIFIFVFRLGVTGAATAHVISQYLMCGILLWKLMGQVDIFNMSTKHLQFC 300
Query: 323 RFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQ 382
RF+KNG LLL RVIAVTFCVTLSASLAAR G MAAFQ CLQVW+ +SLLADG AVA Q
Sbjct: 301 RFMKNGFLLLMRVIAVTFCVTLSASLAAREGSTSMAAFQVCLQVWLATSLLADGYAVAGQ 360
Query: 383 AILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRL 442
AILA +FA+KDY + A+R LQ+ VLG L++++G GL+FGA VF+K+ V+HLI +
Sbjct: 361 AILASAFAKKDYKRAAATASRVLQLGLVLGFVLAVILGAGLHFGARVFTKDDKVLHLISI 420
Query: 443 GLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWI 502
GLPFVA TQPIN+LAFVFDGVN+GASDF Y+A SLVMV+I S+ L FL + GFIG+W
Sbjct: 421 GLPFVAGTQPINALAFVFDGVNFGASDFGYAAASLVMVAIVSILCLLFLSSTHGFIGLWF 480
Query: 503 ALTIYMSLRMFAGVW 517
LTIYMSLR G W
Sbjct: 481 GLTIYMSLRAAVGFW 495
>At2g38330 unknown protein
Length = 521
Score = 337 bits (863), Expect = 1e-92
Identities = 194/490 (39%), Positives = 286/490 (57%), Gaps = 66/490 (13%)
Query: 30 LAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
+ EI+ IA P+ALA+AADPI SL+DTAF+GH+G ELAA GVS++VFN S++ PL+
Sbjct: 76 IGMEIMSIALPAALALAADPITSLVDTAFVGHIGSAELAAVGVSVSVFNLVSKLFNVPLL 135
Query: 90 SITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNG 149
++TTSFVAEE +A A K D
Sbjct: 136 NVTTSFVAEE------------------------------------QAIAAKDD------ 153
Query: 150 DDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
N +I S K+ + S ST+L+ +G+ +A L + L+
Sbjct: 154 ---NDSIETS-----------------KKVLPSVSTSLVLAAGVGIAEAIALSLGSDFLM 193
Query: 210 GAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNV 269
M + +DSPM +PA ++LRLRA GAP ++++LA QG FRGFKDTTTPLY +V+G LN
Sbjct: 194 DVMAIPFDSPMRIPAEQFLRLRAYGAPPIVVALAAQGAFRGFKDTTTPLYAVVAGNVLNA 253
Query: 270 AMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQIFRFLKNGG 329
+DP+LIF GI GAA + V+S+Y++A +LL+ L + V LL P +K + ++LK+GG
Sbjct: 254 VLDPILIFVLGFGISGAAAATVISEYLIAFILLWKLNENVVLLSPQIKVGRANQYLKSGG 313
Query: 330 LLLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSF 389
LL+ R +A+ TL+ SLAA+ GP MA Q L++W+ SLL D LA+A Q++LA ++
Sbjct: 314 LLIGRTVALLVPFTLATSLAAQNGPTQMAGHQIVLEIWLAVSLLTDALAIAAQSLLATTY 373
Query: 390 AEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAA 449
++ +Y + LQ+ G GL+ V+ + +F+ + V+ + G FVA
Sbjct: 374 SQGEYKQAREVLFGVLQVGLATGTGLAAVLFITFEPFSSLFTTDSEVLKIALSGTLFVAG 433
Query: 450 TQPINSLAFVFDGVNYGASDFAYSAYSLVMVSIASVTSLFFLYKSK--GFIGIWIALTIY 507
+QP+N+LAFV DG+ YG SDF ++AYS+V+V ++SLF L + G GIW L ++
Sbjct: 434 SQPVNALAFVLDGLYYGVSDFGFAAYSMVIVGF--ISSLFMLVAAPTFGLAGIWTGLFLF 491
Query: 508 MSLRMFAGVW 517
M+LR+ AG W
Sbjct: 492 MALRLVAGAW 501
>At4g38380 putative protein
Length = 479
Score = 224 bits (570), Expect = 1e-58
Identities = 154/463 (33%), Positives = 241/463 (51%), Gaps = 101/463 (21%)
Query: 30 LAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLV 89
+ +E++ ++ P+ A DP+ L++TA+IG LG VEL +AGVS+A+FN S++ PL+
Sbjct: 33 IKRELVMLSLPAIAGQAIDPLTLLMETAYIGRLGSVELGSAGVSMAIFNTISKLFNIPLL 92
Query: 90 SITTSFVAEEDTMDRINTKAAEKQFNESGKAKSNEVMPDDHLLQDIEAGATKQDSTLKNG 149
S+ TSFVAE+ I AA QD+ +
Sbjct: 93 SVATSFVAED-----IAKIAA----------------------QDLAS------------ 113
Query: 150 DDANSNISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAATLIFAAKPLL 209
+D+ S+I S+ + +++ ++S STAL+ +G+ +A L A+ P L
Sbjct: 114 EDSQSDIP-------------SQGLPERKQLSSVSTALVLAIGIGIFEALALSLASGPFL 160
Query: 210 GAMGLKYDSP------MLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVS 263
MG++ S M +PA ++L LRALGAPA ++SLA+QGIFRGFKDT TP+Y +V
Sbjct: 161 RLMGIQSVSSVQRMSEMFIPARQFLVLRALGAPAYVVSLALQGIFRGFKDTKTPVYCLVL 220
Query: 264 GYA----------LNVAMDPLLIFYFKLGIRGAAISHVLSQ-----------YIMATLLL 302
+ L V + PL I+ F++G+ GAAIS V+SQ Y +A L+L
Sbjct: 221 SFPNFHNSGIGNFLAVFLFPLFIYKFRMGVAGAAISSVISQMVLNPFPLIHRYTVAILML 280
Query: 303 FILMKKVDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASLAARLGPIPMAAFQT 362
+L K+V LLPP + L+ +LK+GG +L R ++V +T++ S+AAR G MAA Q
Sbjct: 281 ILLNKRVILLPPKIGSLKFGDYLKSGGFVLGRTLSVLVTMTVATSMAARQGVFAMAAHQI 340
Query: 363 CLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKV----------------------TTA 400
C+QVW+ SLL D LA + QA++A S +++D+ V
Sbjct: 341 CMQVWLAVSLLTDALASSGQALIASSASKRDFEGVKEFIFTFWGCYLISCYIYIYRERCN 400
Query: 401 ATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLG 443
+Q+ V G+ L++V+G A +N+ +H + G
Sbjct: 401 VFGVVQIGVVTGIALAIVLGMSFSSIADGGGRNIISVHAVCTG 443
>At4g39030 enhanced disease susceptibility 5 gene (EDS5)
Length = 543
Score = 65.9 bits (159), Expect = 5e-11
Identities = 87/369 (23%), Positives = 153/369 (40%), Gaps = 38/369 (10%)
Query: 155 NISKSSIVTNSSNKSESKPIRKKRHIASASTALLFGTVLGLIQAA-TLIFAAKPLLGAMG 213
+++ S++V S K + K + + S L G V GL+ T +F P
Sbjct: 156 SVATSNMVATSLAKQDKKEAQHQ-----ISVLLFIGLVCGLMMLLLTRLFG--PWAVTAF 208
Query: 214 LKYDSPMLVPAV-KYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMD 272
+ + +VPA KY+++R L P +L+ L Q G K++ PL + + +N D
Sbjct: 209 TRGKNIEIVPAANKYIQIRGLAWPFILVGLVAQSASLGMKNSWGPLKALAAATIINGLGD 268
Query: 273 PLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKK----VDLLPPSMKDLQIFRFLKNG 328
+L + GI GAA + SQ + A +++ L K+ PS ++L L
Sbjct: 269 TILCLFLGQGIAGAAWATTASQIVSAYMMMDSLNKEGYNAYSFAIPSPQELWKISALAAP 328
Query: 329 GL--LLARVIAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILA 386
+ +++ +F + + S+ + +AA Q Q + ++ + L+ Q+ +
Sbjct: 329 VFISIFSKIAFYSFIIYCATSMGTHV----LAAHQVMAQTYRMCNVWGEPLSQTAQSFM- 383
Query: 387 CSFAEKDY--NKVTTAATRTLQMSFVLGVGLSLVVG------GGLYFGAGVFSKNVAVIH 438
E Y N+ A L+ ++G L LV+G GL+ G K V +
Sbjct: 384 ---PEMLYGANRNLPKARTLLKSLMIIGATLGLVLGVIGTAVPGLFPGVYTHDK-VIISE 439
Query: 439 LIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLVMVS---IASVTSLFFLYKSK 495
+ RL +PF A + + +G D + S VM S I +T +F
Sbjct: 440 MHRLLIPFFMALSAL-PMTVSLEGTLLAGRDLKF--VSSVMSSSFIIGCLTLMFVTRSGY 496
Query: 496 GFIGIWIAL 504
G +G W L
Sbjct: 497 GLLGCWFVL 505
Score = 34.3 bits (77), Expect = 0.17
Identities = 22/63 (34%), Positives = 32/63 (49%), Gaps = 2/63 (3%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVSI 91
KEI+ P+ P+ SLIDT IG +ELAA G + + S + +F +S+
Sbjct: 100 KEIVKFTGPAMGMWICGPLMSLIDTVVIGQGSSIELAALGPGTVLCDHMSYVFMF--LSV 157
Query: 92 TTS 94
TS
Sbjct: 158 ATS 160
>At2g21340 unknown protein
Length = 414
Score = 56.2 bits (134), Expect = 4e-08
Identities = 53/196 (27%), Positives = 88/196 (44%), Gaps = 8/196 (4%)
Query: 229 RLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAI 288
++R L PAVL+ Q G KD+ PL + A+N D +L + GI GAA
Sbjct: 183 QIRGLAWPAVLIGWVAQSASLGMKDSWGPLKALAVASAINGVGDVVLCTFLGYGIAGAAW 242
Query: 289 SHVLSQYIMATLLLFILMKK----VDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTL 344
+ ++SQ + A +++ L KK PS +L + F + + + V F TL
Sbjct: 243 ATMVSQVVAAYMMMDALNKKGYSAFSFCVPSPSEL-LTIFGLAAPVFITMMSKVLF-YTL 300
Query: 345 SASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRT 404
A +G +AA Q LQ++ S++ + L+ Q+ + N+ A
Sbjct: 301 LVYFATSMGTNIIAAHQVMLQIYTMSTVWGEPLSQTAQSFMPELLF--GINRNLPKARVL 358
Query: 405 LQMSFVLGVGLSLVVG 420
L+ ++G L +VVG
Sbjct: 359 LKSLVIIGATLGIVVG 374
Score = 34.3 bits (77), Expect = 0.17
Identities = 27/77 (35%), Positives = 38/77 (49%), Gaps = 10/77 (12%)
Query: 26 KMDSLA--------KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVF 77
K+D LA KEI+ P+A P+ SLIDTA IG +ELAA G + +
Sbjct: 99 KVDDLATQSIWGQMKEIVMFTGPAAGLWLCGPLMSLIDTAVIGQGSSLELAALGPATVIC 158
Query: 78 NQASRITIFPLVSITTS 94
+ +F +S+ TS
Sbjct: 159 DYLCYTFMF--LSVATS 173
>At1g11670 unknown protein
Length = 503
Score = 51.2 bits (121), Expect = 1e-06
Identities = 77/328 (23%), Positives = 134/328 (40%), Gaps = 23/328 (7%)
Query: 192 VLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGF 251
+ GL IF+ KPLL ++G D + Y + + A AV + +Q +
Sbjct: 141 ITGLPMTLLFIFS-KPLLISLGEPADVASVASVFVYGMIPMIFAYAV--NFPIQKFLQSQ 197
Query: 252 KDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDL 311
T Y+ + +++ + L +F F G+ G ++ H LS +I+ +L I+ K+
Sbjct: 198 SIVTPSAYISAATLVIHLILSWLSVFKFGWGLLGLSVVHSLSWWII--VLAQIIYIKIS- 254
Query: 312 LPPSMKDLQIFRFLKNGGLL----LARVIAVTFCVTLSAS----LAARLGPIPMAAFQT- 362
P + F + GL L+ AV C+ S L A L P A +
Sbjct: 255 -PRCRRTWDGFSWKAFDGLWDFFQLSAASAVMLCLESWYSQILVLLAGLLKDPELALDSL 313
Query: 363 --CLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVG 420
C+ + S +++ G A ++ + + T +SF+L + ++V+
Sbjct: 314 AICMSISAMSFMVSVGFNAAASVRVSNELGAGNPRSAAFSTAVTTGVSFLLSLFEAIVIL 373
Query: 421 GGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLV-- 478
+ + +F+ + AV + PF+A T +N + V GV G AY AY +
Sbjct: 374 SWRHVISYIFTDSPAVAEAVAELSPFLAITIVLNGVQPVLSGVAVGCGWQAYVAYVNIGC 433
Query: 479 --MVSIASVTSLFFLYKSKGFIGIWIAL 504
+V I L F Y G GIW +
Sbjct: 434 YYIVGIPIGYVLGFTY-DMGARGIWTGM 460
>At1g64820 hypothetical protein
Length = 502
Score = 44.3 bits (103), Expect = 2e-04
Identities = 80/327 (24%), Positives = 132/327 (39%), Gaps = 33/327 (10%)
Query: 202 IFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDT--TTPLY 259
IF PL+ + +Y S L+PA+ L +Q + R F+ T PL+
Sbjct: 138 IFHQDPLISQLACRY-SIWLIPALFGFTL-------------LQPMTRYFQSQGITLPLF 183
Query: 260 VIVSG-YALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKD 318
V G ++ LL++ K GI GAA+S S Y + LL+I M + L MK+
Sbjct: 184 VSSLGALCFHIPFCWLLVYKLKFGIVGAALSIGFS-YWLNVFLLWIFM-RYSALHREMKN 241
Query: 319 LQIFRFLKNGGLLLARVIAVTFCVTLS----ASLAARLGPIPMAAFQT-CLQVWMTSSLL 373
L + + + +A I + L L G +P + +T + + +T+S +
Sbjct: 242 LGLQELISSMKQFIALAIPSAMMICLEWWSFEILLLMSGLLPNSKLETSVISICLTTSAV 301
Query: 374 ADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLY----FGAGV 429
L AI A + + + AA + + LG +L+ LY V
Sbjct: 302 HFVLVNAIGASASTHVSNELGAGNHRAARAAVNSAIFLGGVGALITTITLYSYRKSWGYV 361
Query: 430 FSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAY----SLVMVSIASV 485
FS V+ P + + +NS V GV G+ Y S +V I
Sbjct: 362 FSNEREVVRYATQITPILCLSIFVNSFLAVLSGVARGSGWQRIGGYASLGSYYLVGIPLG 421
Query: 486 TSLFFLYKSKGFIGIWIALTIYMSLRM 512
L F+ K +G G+WI + I ++++
Sbjct: 422 WFLCFVMKLRG-KGLWIGILIASTIQL 447
>At1g71140 hypothetical protein
Length = 485
Score = 42.0 bits (97), Expect = 8e-04
Identities = 58/287 (20%), Positives = 117/287 (40%), Gaps = 19/287 (6%)
Query: 236 PAVLLSLAMQGIFRGFKDTTTPLYVI---VSGYALNVAMDPLLIFYFKLGIRGAAISHVL 292
PA+ +Q + R F+ + L ++ VS +++ + L+F F LG GAAI+ +
Sbjct: 155 PALFGYATLQPLVRFFQAQSLILPLVMSSVSSLCIHIVLCWSLVFKFGLGSLGAAIAIGV 214
Query: 293 SQYIMATLLLFIL------MKKVDLLPPSMKD--LQIFRF-LKNGGLLLARVIAVTFCVT 343
S ++ T+L + K + S+ + + FRF + + ++ + F V
Sbjct: 215 SYWLNVTVLGLYMTFSSSCSKSRATISMSLFEGMGEFFRFGIPSASMICLEWWSFEFLVL 274
Query: 344 LSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATR 403
LS L + + CL + + + L A +A + + A
Sbjct: 275 LSGILPN--PKLEASVLSVCLSTQSSLYQIPESLGAAASTRVANELGAGNPKQARMAVYT 332
Query: 404 TLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGV 463
+ ++ V + + +V G +FS V+ ++ P ++ + ++L GV
Sbjct: 333 AMVITGVESIMVGAIVFGARNVFGYLFSSETEVVDYVKSMAPLLSLSVIFDALHAALSGV 392
Query: 464 NYGASDFAYSAY----SLVMVSIASVTSLFFLYKSKGFIGIWIALTI 506
G+ AY + + I + L F +K +G G+WI +T+
Sbjct: 393 ARGSGRQDIGAYVNLAAYYLFGIPTAILLAFGFKMRG-RGLWIGITV 438
>At1g66780 hypothetical protein
Length = 456
Score = 41.2 bits (95), Expect = 0.001
Identities = 71/316 (22%), Positives = 127/316 (39%), Gaps = 42/316 (13%)
Query: 202 IFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGFKDTTTPLYVI 261
+F PL+ + +Y S L+PA L +VL S+ F + PL++
Sbjct: 144 LFHQDPLISQLACRY-SIWLIPA--------LFGYSVLQSMTR---FFQSQGLVLPLFLS 191
Query: 262 VSGYAL-NVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDLLPPSMKDLQ 320
G +V LL++ + GI GAA+S S ++ V LL M+D
Sbjct: 192 SLGALFFHVPFSWLLVYKLRFGIVGAALSIGFSYWL-----------NVGLLWAFMRDSA 240
Query: 321 IFRFLKNGGLLLARV-IAVTFCVTLSASLAARLGPIPMAAFQTCLQVWMTSSLLADGLAV 379
++R KN L + +++ +TL+ IP A TCL + ++ + +
Sbjct: 241 LYR--KNWNLRAQEIFLSMKQFITLA---------IP-TAMMTCLTMSSLHYVIVNAIGA 288
Query: 380 AIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHL 439
A ++ + +AA + + + +S+ + A +FS V
Sbjct: 289 AASTHVSNKLGAGNPKAARSAANSAIFLGMIDAAIVSISLYSYRRNWAYIFSNESEVADY 348
Query: 440 IRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAY----SLVMVSIASVTSLFFLYKSK 495
+ PF+ + ++S V GV G AY S +V I + L F+ K +
Sbjct: 349 VTQITPFLCLSIGVDSFLAVLSGVARGTGWQHIGAYANIGSYYLVGIPVGSILCFVVKLR 408
Query: 496 GFIGIWIALTIYMSLR 511
G G+WI + + +L+
Sbjct: 409 G-KGLWIGILVGSTLQ 423
>At1g61890 unknown protein
Length = 501
Score = 40.4 bits (93), Expect = 0.002
Identities = 67/328 (20%), Positives = 126/328 (37%), Gaps = 22/328 (6%)
Query: 192 VLGLIQAATLIFAAKPLLGAMGLKYDSPMLVPAVKYLRLRALGAPAVLLSLAMQGIFRGF 251
+L + + L + P+L A+G L Y + + A AV + +Q +
Sbjct: 137 ILTCLPMSFLFLFSNPILTALGEPEQVATLASVFVYGMIPVIFAYAV--NFPIQKFLQSQ 194
Query: 252 KDTTTPLYVIVSGYALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMKKVDL 311
T Y+ + +++ + + ++ G+ ++ H S +I+ + + +
Sbjct: 195 SIVTPSAYISAATLVIHLILSWIAVYRLGYGLLALSLIHSFSWWIIVVAQIVY----IKM 250
Query: 312 LPPSMKDLQIFRFLKNGGLL----LARVIAVTFCVTLSAS----LAARLGPIPMAAFQT- 362
P + + F + GL L+ AV C+ S L A L P A +
Sbjct: 251 SPRCRRTWEGFSWKAFEGLWDFFRLSAASAVMLCLESWYSQILVLLAGLLKNPELALDSL 310
Query: 363 --CLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVG 420
C+ + S +++ G A ++ + + T +SF+L V ++VV
Sbjct: 311 AICMSISAISFMVSVGFNAAASVRVSNELGAGNPRAAAFSTVVTTGVSFLLSVFEAIVVL 370
Query: 421 GGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAYSLV-- 478
+ + F+ + AV + PF+A T +N + V GV G A+ AY +
Sbjct: 371 SWRHVISYAFTDSPAVAEAVADLSPFLAITIVLNGIQPVLSGVAVGCGWQAFVAYVNIGC 430
Query: 479 --MVSIASVTSLFFLYKSKGFIGIWIAL 504
+V I L F Y G GIW +
Sbjct: 431 YYVVGIPVGFVLGFTY-DMGAKGIWTGM 457
Score = 28.9 bits (63), Expect = 7.2
Identities = 15/47 (31%), Positives = 24/47 (50%)
Query: 32 KEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFN 78
K + +A P+ + S++ F GH+G ELAAA + + FN
Sbjct: 49 KFLFHLAAPAIFVYVINNGMSILTRIFAGHVGSFELAAASLGNSGFN 95
>At5g19700 putative protein
Length = 508
Score = 38.9 bits (89), Expect = 0.007
Identities = 19/55 (34%), Positives = 31/55 (55%)
Query: 24 VFKMDSLAKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFN 78
+ ++ S A+ + +AFP+ LA S I F+GH+G +ELA ++IA N
Sbjct: 30 ITELKSEARSLFSLAFPTILAALILYARSAISMLFLGHIGELELAGGSLAIAFAN 84
Score = 38.1 bits (87), Expect = 0.012
Identities = 35/155 (22%), Positives = 61/155 (38%), Gaps = 9/155 (5%)
Query: 356 PMAAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGL 415
P+A+ +Q + L +A+ + NK +A + + V+G+
Sbjct: 295 PVASMGILIQTTSLLYIFPSSLGLAVSTRVGNELGSNRPNKARLSAIVAVSFAGVMGLTA 354
Query: 416 SLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYG------ASD 469
S G +F+ +VA+I L LP + + N V GV G A++
Sbjct: 355 SAFAWGVSDVWGWIFTNDVAIIKLTAAALPILGLCELGNCPQTVGCGVVRGTARPSMAAN 414
Query: 470 FAYSAYSLVMVSIASVTSLFFLYKSKGFIGIWIAL 504
A+ LV +A + + Y GF G+W+ L
Sbjct: 415 INLGAFYLVGTPVAVGLTFWAAY---GFCGLWVGL 446
>At1g15160 hypothetical protein
Length = 487
Score = 37.7 bits (86), Expect = 0.015
Identities = 72/292 (24%), Positives = 114/292 (38%), Gaps = 19/292 (6%)
Query: 229 RLRALGAPAVLLSLAMQGIFRGFKDTT--TPLYVIVSG-YALNVAMDPLLIFYFKLGIRG 285
R A P + +Q + R FK+ + TPL + + L+V + LL++ L G
Sbjct: 153 RFAAWLIPGLFAYAVLQPLTRYFKNQSLITPLLITSCVVFCLHVPLCWLLVYKSGLDHIG 212
Query: 286 AAISHVLSQYIMATLL----LFILMKKVDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFC 341
A++ LS ++ A L F P +M+ + R L A ++ + +
Sbjct: 213 GALALSLSYWLYAIFLGSFMYFSSACSETRAPLTMEIFEGVREFIKYALPSAAMLCLEWW 272
Query: 342 VTLSASLAARLGPIPM---AAFQTCLQVW-MTSSL-LADGLAVAIQAILACSFAEKDYNK 396
L + L P P + CLQ MT S+ LA A + +
Sbjct: 273 SYELIILLSGLLPNPQLETSVLSVCLQTLSMTYSIPLAIAAAASTRISNELGAGNSRAAH 332
Query: 397 VTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSL 456
+ A +L + L VG SL+ G L VFS + I + P V+ + ++SL
Sbjct: 333 IVVYAAMSLAVVDALMVGTSLLAGKNLL--GQVFSSDKNTIDYVAKMAPLVSISLILDSL 390
Query: 457 AFVFDGVNYGASDFAYSAY----SLVMVSIASVTSLFFLYKSKGFIGIWIAL 504
V GV G AY + + I SL F KG +G+WI +
Sbjct: 391 QGVLSGVASGCGWQHIGAYINFGAFYLWGIPIAASLAFWVHLKG-VGLWIGI 441
>At1g47530 unknown protein
Length = 484
Score = 36.6 bits (83), Expect = 0.034
Identities = 56/214 (26%), Positives = 92/214 (42%), Gaps = 18/214 (8%)
Query: 274 LLIFYFKLGIRGAAISHVLSQY-IMATLLLFILMKKVDLLPPSMKDL---QIFRFLKNGG 329
L I YFK G+ GAAI+ S + I+ LL+IL+ K D L ++ F+K
Sbjct: 205 LFILYFKWGLVGAAITLNTSWWLIVIGQLLYILITKSDGAWTGFSMLAFRDLYGFVKL-S 263
Query: 330 LLLARVIAVTFCVTLSASLAARLGP---IPMAAFQTCLQVWMTSSLLADGLAVAIQA--- 383
L A ++ + F + + L P IP+ A C+ + +++++ G AI
Sbjct: 264 LASALMLCLEFWYLMVLVVVTGLLPNPLIPVDAISICMNIEGWTAMISIGFNAAISVRVS 323
Query: 384 --ILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIR 441
+ A + A ++ + + T TL +G+ +VV +F+ + AV
Sbjct: 324 NELGAGNAALAKFSVIVVSITSTL-----IGIVCMIVVLATKDSFPYLFTSSEAVAAETT 378
Query: 442 LGLPFVAATQPINSLAFVFDGVNYGASDFAYSAY 475
+ T +NSL V GV GA A AY
Sbjct: 379 RIAVLLGFTVLLNSLQPVLSGVAVGAGWQALVAY 412
>At1g15150 unknown protein (At1g15150)
Length = 487
Score = 36.6 bits (83), Expect = 0.034
Identities = 63/285 (22%), Positives = 113/285 (39%), Gaps = 19/285 (6%)
Query: 236 PAVLLSLAMQGIFRGFKDTT--TPLYVIVSG-YALNVAMDPLLIFYFKLGIRGAAISHVL 292
P + +Q + R FK+ + TPL V S + ++V + LL++ LG G A++ L
Sbjct: 160 PGLFAYAVLQPLIRYFKNQSLITPLLVTSSVVFCIHVPLCWLLVYKSGLGHIGGALALSL 219
Query: 293 SQYIMATLL----LFILMKKVDLLPPSMKDLQIFRFLKNGGLLLARVIAVTFCVTLSASL 348
S ++ A L + P +M+ + R L A ++ + + L
Sbjct: 220 SYWLYAIFLGSFMYYSSACSETRAPLTMEIFEGVREFIKYALPSAAMLCLEWWSYELIIL 279
Query: 349 AARLGPIPM---AAFQTCLQVWMTSSLLADGLAVAIQAILACSFAEKDYNK--VTTAATR 403
+ L P P + C + + + +A A ++ + + A
Sbjct: 280 LSGLLPNPQLETSVLSICFETLSITYSIPLAIAAAASTRISNELGAGNSRAAHIVVYAAM 339
Query: 404 TLQMSFVLGVGLSLVVGGGLYFGAGVFSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGV 463
+L + L V +SL+ G ++ VFS + I + P V+ + ++SL V GV
Sbjct: 340 SLAVMDALMVSMSLLAGRHVF--GHVFSSDKKTIEYVAKMAPLVSISIILDSLQGVLSGV 397
Query: 464 NYGASDFAYSAY----SLVMVSIASVTSLFFLYKSKGFIGIWIAL 504
G AY + + I SL F KG +G+WI +
Sbjct: 398 ASGCGWQHIGAYINFGAFYLWGIPIAASLAFWVHLKG-VGLWIGI 441
>At5g52450 unknown protein
Length = 486
Score = 35.8 bits (81), Expect = 0.059
Identities = 63/271 (23%), Positives = 114/271 (41%), Gaps = 36/271 (13%)
Query: 266 ALNVAMDPLLIFYFKLGIRGAAISHVLSQYIMATLLLFILMK--------KVDLLPPSMK 317
+L+V + +L+F LG +GAA+++ +S Y + +LLF +K +++
Sbjct: 188 SLHVLLCWVLVFKSGLGFQGAALANSIS-YWLNVVLLFCYVKFSPSCSLTWTGFSKEALR 246
Query: 318 DLQIFRFLKNGGLLLARVIAVTFCVTL-SASLAARL-GPIPMAAFQT-----CLQVWMTS 370
D+ F L LA A+ C+ + S L L G +P +T CL T
Sbjct: 247 DILPF-------LRLAVPSALMVCLEMWSFELLVLLSGLLPNPVLETSVLSICLNTSGTM 299
Query: 371 SLLADGLAVAIQAILACSFAEKDYNKVTTAATRTLQMSFVLGVGLSLVVGGGLYFGAGV- 429
++ GL+ A ++ + KV A R + + V S+V+G L +
Sbjct: 300 WMIPFGLSGAASTRISNELGAGN-PKVAKLAVRVV---ICIAVAESIVIGSVLILIRNIW 355
Query: 430 ---FSKNVAVIHLIRLGLPFVAATQPINSLAFVFDGVNYGASDFAYSAY----SLVMVSI 482
+S + V+ + +P +A ++SL V GV G A S +V +
Sbjct: 356 GLAYSSELEVVSYVASMMPILALGNFLDSLQCVLSGVARGCGWQKIGAIINLGSYYLVGV 415
Query: 483 ASVTSLFFLYKSKGFIGIWIALTIYMSLRMF 513
S L F + G G+W+ + + +++F
Sbjct: 416 PSGLLLAFHFHVGG-RGLWLGIICALVVQVF 445
>At4g29140 unknown protein
Length = 532
Score = 35.8 bits (81), Expect = 0.059
Identities = 18/48 (37%), Positives = 27/48 (55%)
Query: 31 AKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFN 78
AK + +AFP A+ + S + F+G LG +ELAA ++IA N
Sbjct: 57 AKSLFTLAFPIAVTALVLYLRSAVSMFFLGQLGDLELAAGSLAIAFAN 104
>At1g33110 Unknown protein (T9L6.1)
Length = 494
Score = 35.4 bits (80), Expect = 0.076
Identities = 19/60 (31%), Positives = 33/60 (54%)
Query: 31 AKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVS 90
+K++ +A P+ + S+I +FIGHLGP+ELAA ++ V + S + + S
Sbjct: 37 SKKLWIVAAPAIFTRFSTFGVSIISQSFIGHLGPIELAAYSITFTVLLRFSNGILLGMAS 96
>At1g33090 unknown protein
Length = 494
Score = 35.0 bits (79), Expect = 0.100
Identities = 18/60 (30%), Positives = 31/60 (51%)
Query: 31 AKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVS 90
+K++ +A PS + SL+ F+GH+GP ELAA ++ V + S + + S
Sbjct: 37 SKKLWVVAAPSIFTKFSTYGVSLVTQGFVGHIGPTELAAYSITFTVLLRFSNGILLGMAS 96
>At1g33080 unknown protein
Length = 494
Score = 34.7 bits (78), Expect = 0.13
Identities = 20/60 (33%), Positives = 32/60 (53%)
Query: 31 AKEILGIAFPSALAVAADPIASLIDTAFIGHLGPVELAAAGVSIAVFNQASRITIFPLVS 90
+K++ +A P+ + SLI AFIGHLG ELAA +++ V + S + + S
Sbjct: 37 SKKLWVVAGPAIFTRFSTSGLSLISQAFIGHLGSTELAAYSITLTVLLRFSNGILLGMAS 96
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,409,800
Number of Sequences: 26719
Number of extensions: 357514
Number of successful extensions: 1715
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1655
Number of HSP's gapped (non-prelim): 84
length of query: 517
length of database: 11,318,596
effective HSP length: 104
effective length of query: 413
effective length of database: 8,539,820
effective search space: 3526945660
effective search space used: 3526945660
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC122162.1


