

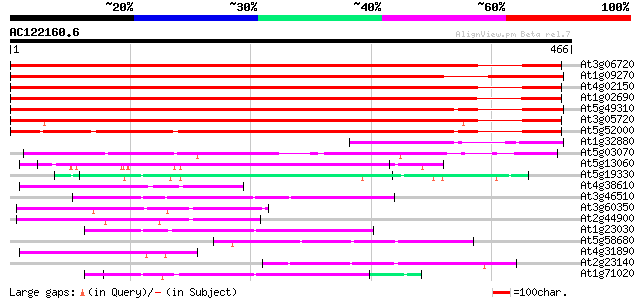
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.6 + phase: 0
(466 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g06720 importin alpha 591 e-169
At1g09270 putative importin beta binding domain 557 e-159
At4g02150 AtKAP alpha 540 e-154
At1g02690 importin alpha, putative 537 e-153
At5g49310 importin alpha 460 e-130
At3g05720 putative importin alpha 440 e-124
At5g52000 importin alpha subunit 360 e-100
At1g32880 hypothetical protein 146 2e-35
At5g03070 importin alpha - like protein 140 1e-33
At5g13060 unknown protein 75 8e-14
At5g19330 putative protein 65 6e-11
At4g38610 putative protein 58 1e-08
At3g46510 arm repeat containing protein homolog 58 1e-08
At3g60350 Arm repeat containing protein - like 55 1e-07
At2g44900 hypothetical protein 53 4e-07
At1g23030 unknown protein 52 9e-07
At5g58680 unknown protein 49 6e-06
At4g31890 unknown protein 47 2e-05
At2g23140 hypothetical protein 46 4e-05
At1g71020 unknown protein 46 4e-05
>At3g06720 importin alpha
Length = 532
Score = 591 bits (1523), Expect = e-169
Identities = 301/460 (65%), Positives = 359/460 (77%), Gaps = 38/460 (8%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MVA +WSDD + QLE+TT+FRK LS+++ PPI++VI +GVVPRFV+FL K D+P +Q EA
Sbjct: 77 MVAGVWSDDPALQLESTTQFRKLLSIERSPPIEEVISAGVVPRFVEFLKKEDYPAIQFEA 136
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+GTS++TKVV+DH AVP+FV+LL+SP DDVR QA WALGN+AGDSPR RDLV
Sbjct: 137 AWALTNIASGTSDHTKVVIDHNAVPIFVQLLASPSDDVREQAVWALGNVAGDSPRCRDLV 196
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
L GAL+ LL+QLNE L LRNA WTLSNFCRGKPQP +QV+ ALPAL+ L+ S DE
Sbjct: 197 LGCGALLPLLNQLNEHAKLSMLRNATWTLSNFCRGKPQPHFDQVKPALPALERLIHSDDE 256
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VLT+ACWALSYLSDGT+D IQ VI+AGV +LV++LL S S L PALRTVGNIV+GDD
Sbjct: 257 EVLTDACWALSYLSDGTNDKIQTVIQAGVVPKLVELLLHHSPSVLIPALRTVGNIVTGDD 316
Query: 240 MQTQAIVNHGLLPCLLSLLT-HPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
+QTQ ++N G LPCL +LLT + KKSIKKEAC T+SNITAGN++QIQ V+EA LI+PLV+
Sbjct: 317 IQTQCVINSGALPCLANLLTQNHKKSIKKEACWTISNITAGNKDQIQTVVEANLISPLVS 376
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
LLQNAEFDI KE A A++N TSGG+H+ IKYLV QGCIKPLCDLLVCPDPRI+TVCLEGL
Sbjct: 377 LLQNAEFDIKKEAAWAISNATSGGSHDQIKYLVEQGCIKPLCDLLVCPDPRIITVCLEGL 436
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN LKVGEAEK++ +TGD+N YAQ I D EG
Sbjct: 437 ENILKVGEAEKNLGHTGDMNYYAQLIDDAEG----------------------------- 467
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDE 458
LEKIENL SH++ EIYEKA KILE YWLE+ED+
Sbjct: 468 -------LEKIENLQSHDNNEIYEKAVKILETYWLEEEDD 500
>At1g09270 putative importin beta binding domain
Length = 538
Score = 557 bits (1435), Expect = e-159
Identities = 288/462 (62%), Positives = 350/462 (75%), Gaps = 38/462 (8%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MV ++SDD QLEATT+FRK LS+++ PPID+VI++GV+PRFV+FL + D PQLQ EA
Sbjct: 84 MVQGVYSDDPQAQLEATTQFRKLLSIERSPPIDEVIKAGVIPRFVEFLGRHDHPQLQFEA 143
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTN+A+GTS++T+VV++ GAVP+FVKLL+S DDVR QA WALGN+AGDSP R+LV
Sbjct: 144 AWALTNVASGTSDHTRVVIEQGAVPIFVKLLTSASDDVREQAVWALGNVAGDSPNCRNLV 203
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
L++GAL LL+QLNE L LRNA WTLSNFCRGKP EQV+ ALP L+ L++ DE
Sbjct: 204 LNYGALEPLLAQLNENSKLSMLRNATWTLSNFCRGKPPTPFEQVKPALPILRQLIYLNDE 263
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VLT+ACWALSYLSDG +D IQAVIEAGVC RLV++L S + L PALRTVGNIV+GDD
Sbjct: 264 EVLTDACWALSYLSDGPNDKIQAVIEAGVCPRLVELLGHQSPTVLIPALRTVGNIVTGDD 323
Query: 240 MQTQAIVNHGLLPCLLSLLT-HPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
QTQ I+ G+LP L +LLT + KKSIKKEAC T+SNITAGN+ QI+AV+ AG+I PLV+
Sbjct: 324 SQTQFIIESGVLPHLYNLLTQNHKKSIKKEACWTISNITAGNKLQIEAVVGAGIILPLVH 383
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
LLQNAEFDI KE A A++N TSGG+HE I+YLV+QGCIKPLCDLL+CPDPRIVTVCLEGL
Sbjct: 384 LLQNAEFDIKKEAAWAISNATSGGSHEQIQYLVTQGCIKPLCDLLICPDPRIVTVCLEGL 443
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN LKVGEA+K G VNLYAQ
Sbjct: 444 ENI------------------------------------LKVGEADKEMGLNSGVNLYAQ 467
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETL 460
+IE+ GL+K+ENL SH++ EIYEKA KILERYW E+E+E +
Sbjct: 468 IIEESDGLDKVENLQSHDNNEIYEKAVKILERYWAEEEEEQI 509
>At4g02150 AtKAP alpha
Length = 531
Score = 540 bits (1391), Expect = e-154
Identities = 279/460 (60%), Positives = 344/460 (74%), Gaps = 38/460 (8%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
MVA IWS+D++ QLEAT RK LS+++ PPI++V+QSGVVPR V+FL + DFP+LQ EA
Sbjct: 80 MVAGIWSEDSNSQLEATNLLRKLLSIEQNPPINEVVQSGVVPRVVKFLSRDDFPKLQFEA 139
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+GTSENT V+++ GAVP+F++LLSS +DVR QA WALGN+AGDSP+ RDLV
Sbjct: 140 AWALTNIASGTSENTNVIIESGAVPIFIQLLSSASEDVREQAVWALGNVAGDSPKCRDLV 199
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
LS+GA+ LLSQ NE L LRNA WTLSNFCRGKP PA EQ + ALP L+ LV S DE
Sbjct: 200 LSYGAMTPLLSQFNENTKLSMLRNATWTLSNFCRGKPPPAFEQTQPALPVLERLVQSMDE 259
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VLT+ACWALSYLSD ++D IQAVIEAGV RL+Q+L S S L PALRT+GNIV+GDD
Sbjct: 260 EVLTDACWALSYLSDNSNDKIQAVIEAGVVPRLIQLLGHSSPSVLIPALRTIGNIVTGDD 319
Query: 240 MQTQAIVNHGLLPCLLSLL-THPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
+QTQ +++ LPCLL+LL + KKSIKKEAC T+SNITAGN +QIQAVI+AG+I LV
Sbjct: 320 LQTQMVLDQQALPCLLNLLKNNYKKSIKKEACWTISNITAGNADQIQAVIDAGIIQSLVW 379
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
+LQ+AEF++ KE A ++N TSGGTH+ IK++VSQGCIKPLCDLL CPD ++VTVCLE L
Sbjct: 380 VLQSAEFEVKKEAAWGISNATSGGTHDQIKFMVSQGCIKPLCDLLTCPDLKVVTVCLEAL 439
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN L VGEAEK++ +TG+ NLYAQ I + EG
Sbjct: 440 ENILVVGEAEKNLGHTGEDNLYAQMIDEAEG----------------------------- 470
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDE 458
LEKIENL SH++ +IY+KA KILE +W ED +E
Sbjct: 471 -------LEKIENLQSHDNNDIYDKAVKILETFWTEDNEE 503
>At1g02690 importin alpha, putative
Length = 538
Score = 537 bits (1384), Expect = e-153
Identities = 278/460 (60%), Positives = 341/460 (73%), Gaps = 38/460 (8%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
M+A + S+D QLEAT FR+ LS+++ PPI++V+QSGVVP VQFL + DF QLQ EA
Sbjct: 81 MIAGVMSEDRDLQLEATASFRRLLSIERNPPINEVVQSGVVPHIVQFLSRDDFTQLQFEA 140
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+GTSENT+V++D GAVP+FVKLLSS ++VR QA WALGN+AGDSP+ RD V
Sbjct: 141 AWALTNIASGTSENTRVIIDSGAVPLFVKLLSSASEEVREQAVWALGNVAGDSPKCRDHV 200
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
LS A++ LL+Q +E L LRNA WTLSNFCRGKPQPA EQ ++ALPAL+ L+ S DE
Sbjct: 201 LSCEAMMSLLAQFHEHSKLSMLRNATWTLSNFCRGKPQPAFEQTKAALPALERLLHSTDE 260
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VLT+A WALSYLSDGT++ IQ VI+AGV RLVQ+L PS S L PALRT+GNIV+GDD
Sbjct: 261 EVLTDASWALSYLSDGTNEKIQTVIDAGVIPRLVQLLAHPSPSVLIPALRTIGNIVTGDD 320
Query: 240 MQTQAIVNHGLLPCLLSLLTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
+QTQA+++ LP LL+LL + KKSIKKEAC T+SNITAGN QIQ V +AG+I PL+N
Sbjct: 321 IQTQAVISSQALPGLLNLLKNTYKKSIKKEACWTISNITAGNTSQIQEVFQAGIIRPLIN 380
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
LL+ EF+I KE A++N TSGG H+ IK+LVSQGCI+PLCDLL CPDPR+VTV LEGL
Sbjct: 381 LLEIGEFEIKKEAVWAISNATSGGNHDQIKFLVSQGCIRPLCDLLPCPDPRVVTVTLEGL 440
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN LKVGEAEK++ NTG+ NLYAQ I+D +
Sbjct: 441 ENILKVGEAEKNLGNTGNDNLYAQMIEDAD------------------------------ 470
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDE 458
GL+KIENL SH++ EIYEKA KILE YW D++E
Sbjct: 471 ------GLDKIENLQSHDNNEIYEKAVKILESYWAADDEE 504
>At5g49310 importin alpha
Length = 519
Score = 460 bits (1183), Expect = e-130
Identities = 257/462 (55%), Positives = 309/462 (66%), Gaps = 41/462 (8%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEA 59
M+ ++SDD S QLE TT FR LS D+ PP D+VI+SGVVPRFV+FL K D P+LQ EA
Sbjct: 75 MITGVFSDDPSLQLEYTTRFRVVLSFDRSPPTDNVIKSGVVPRFVEFLKKDDNPKLQFEA 134
Query: 60 AWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLV 119
AWALTNIA+G SE+TKVV+DHG VP+FV+LL+SP DDVR QA W LGN+AGDS + RD V
Sbjct: 135 AWALTNIASGASEHTKVVIDHGVVPLFVQLLASPDDDVREQAIWGLGNVAGDSIQCRDFV 194
Query: 120 LSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
L+ GA I LL QLN L LRNA WTLSNF RGKP P + V+ LP LK LV+S DE
Sbjct: 195 LNSGAFIPLLHQLNNHATLSILRNATWTLSNFFRGKPSPPFDLVKHVLPVLKRLVYSDDE 254
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
VL +ACWALS LSD +++NIQ+VIEAGV RLV++L S L PALR +GNIVSG+
Sbjct: 255 QVLIDACWALSNLSDASNENIQSVIEAGVVPRLVELLQHASPVVLVPALRCIGNIVSGNS 314
Query: 240 MQTQAIVNHGLLPCLLSLLT-HPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVN 298
QT ++N G+LP L LLT + + I++EAC T+SNITAG EQIQ+VI+A LI LVN
Sbjct: 315 QQTHCVINCGVLPVLADLLTQNHMRGIRREACWTISNITAGLEEQIQSVIDANLIPSLVN 374
Query: 299 LLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEGL 358
L Q+AEFDI KE A++N + GG+ IKYLV Q CIK LCD+LVCPD RI+ V L GL
Sbjct: 375 LAQHAEFDIKKEAIWAISNASVGGSPNQIKYLVEQNCIKALCDILVCPDLRIILVSLGGL 434
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
E L GE +K N DVN Y+Q I+D EG
Sbjct: 435 EMILIAGEVDK---NLRDVNCYSQMIEDAEG----------------------------- 462
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETL 460
LEKIENL H + EIYEKA KIL+ Y L +ED L
Sbjct: 463 -------LEKIENLQHHGNNEIYEKAVKILQTYGLVEEDGRL 497
>At3g05720 putative importin alpha
Length = 528
Score = 440 bits (1132), Expect = e-124
Identities = 242/468 (51%), Positives = 308/468 (65%), Gaps = 46/468 (9%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVD--KPPIDDVIQSGVVPRFVQFLDKGDFPQLQLE 58
+++ IWSD+ +EATT+ R L + +++VIQ+G+VPRFV+FL D PQLQ E
Sbjct: 64 LISCIWSDERDLLIEATTQIRTLLCGEMFNVRVEEVIQAGLVPRFVEFLTWDDSPQLQFE 123
Query: 59 AAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDL 118
AAWALTNIA+GTSENT+VV+DHGAV + V+LL+SP D VR Q WALGNI+GDSPR RD+
Sbjct: 124 AAWALTNIASGTSENTEVVIDHGAVAILVRLLNSPYDVVREQVVWALGNISGDSPRCRDI 183
Query: 119 VLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKD 178
VL H AL LL QLN L L NA WTLSN CRGKPQP +QV +ALPAL L+ D
Sbjct: 184 VLGHAALPSLLLQLNHGAKLSMLVNAAWTLSNLCRGKPQPPFDQVSAALPALAQLIRLDD 243
Query: 179 EVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGD 238
+ +L CWAL YLSDG+++ IQAVIEA VC RL+ + + S S + PALRT+GNIV+G+
Sbjct: 244 KELLAYTCWALVYLSDGSNEKIQAVIEANVCARLIGLSIHRSPSVITPALRTIGNIVTGN 303
Query: 239 DMQTQAIVNHGLLPCLLSLLTHP-KKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLV 297
D QTQ I++ LPCL++LL K+I+KEAC T+SNITAG + QIQAV +A + LV
Sbjct: 304 DSQTQHIIDLQALPCLVNLLRGSYNKTIRKEACWTVSNITAGCQSQIQAVFDADICPALV 363
Query: 298 NLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEG 357
NLLQN+E D+ KE A A+ N +GG+++ I +LV Q CIKPLCDLL C D ++V VCLE
Sbjct: 364 NLLQNSEGDVKKEAAWAICNAIAGGSYKQIMFLVKQECIKPLCDLLTCSDTQLVMVCLEA 423
Query: 358 LENFLKVGEAEKSISNTG-------DVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNT 410
L+ LKVGE S G +VN +AQ I++ EG
Sbjct: 424 LKKILKVGEVFSSRHAEGIYQCPQTNVNPHAQLIEEAEG--------------------- 462
Query: 411 GDVNLYAQMIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDE 458
LEKIE L SHE+ +IYE A KILE YW+E+E+E
Sbjct: 463 ---------------LEKIEGLQSHENNDIYETAVKILETYWMEEEEE 495
>At5g52000 importin alpha subunit
Length = 441
Score = 360 bits (924), Expect = e-100
Identities = 214/460 (46%), Positives = 279/460 (60%), Gaps = 49/460 (10%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAA 60
++ +WSDD QLE+ T+ R+ S + I VI+SGVVPR VQ L FP+LQ E A
Sbjct: 14 IIDGLWSDDPPLQLESVTKIRRITS--QRDISCVIRSGVVPRLVQLLKNQVFPKLQYEVA 71
Query: 61 WALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVL 120
WALTNIA +N VVV++ AVP+ ++L++SP D VR QA W L N+AG S RD VL
Sbjct: 72 WALTNIAV---DNPGVVVNNNAVPVLIQLIASPKDYVREQAIWTLSNVAGHSIHYRDFVL 128
Query: 121 SHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEV 180
+ G L+ LL L + LR A W L N CRGKP PA +QV+ ALPAL+ L+ S DE
Sbjct: 129 NSGVLMPLLRLLYKDT---TLRIATWALRNLCRGKPHPAFDQVKPALPALEILLHSHDED 185
Query: 181 VLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDM 240
VL AC AL +LS+G+ D IQ+VIEAG +LVQIL PS L PAL T+G + +G+
Sbjct: 186 VLKNACMALCHLSEGSEDGIQSVIEAGFVPKLVQILQLPSPVVLVPALLTIGAMTAGNHQ 245
Query: 241 QTQAIVNHGLLPCLLSLLT-HPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNL 299
QTQ ++N G LP + ++LT + + IKK AC +SNITAG +EQIQ+VI+A LI LVNL
Sbjct: 246 QTQCVINSGALPIISNMLTRNHENKIKKCACWVISNITAGTKEQIQSVIDANLIPILVNL 305
Query: 300 LQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDLLVCPDPR-IVTVCLEGL 358
Q+ +F + KE A++N+ G+H+ IKY+ Q CIK LCD+LV D R + CL+GL
Sbjct: 306 AQDTDFYMKKEAVWAISNMALNGSHDQIKYMAEQSCIKQLCDILVYSDERTTILKCLDGL 365
Query: 359 ENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYAQ 418
EN LK GEAEK N+ DVN Y I+D EG
Sbjct: 366 ENMLKAGEAEK---NSEDVNPYCLLIEDAEG----------------------------- 393
Query: 419 MIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDE 458
LEKI L +++ +IYEKA KIL W E++DE
Sbjct: 394 -------LEKISKLQMNKNDDIYEKAYKILVTNWFEEDDE 426
>At1g32880 hypothetical protein
Length = 183
Score = 146 bits (369), Expect = 2e-35
Identities = 81/178 (45%), Positives = 107/178 (59%), Gaps = 39/178 (21%)
Query: 283 QIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCDL 342
++Q+VI+A LI LV L QNAEFD+ KE A++N T G+H+ IKY+V Q CIKPLCD+
Sbjct: 40 KLQSVIDANLIPTLVKLTQNAEFDMKKESVCAISNATLLGSHDQIKYMVEQSCIKPLCDI 99
Query: 343 LVCPDPRIVTVCLEGLENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGE 402
L CPD + + CL+G+EN LKVGEAEK N GD
Sbjct: 100 LFCPDVKTILKCLDGMENTLKVGEAEK---NAGD-------------------------- 130
Query: 403 AEKRFGNTGDVNLYAQMIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLEDEDETL 460
DV+ Y ++IE +GL+KI NL HE+ EIY+KA KIL+ YWLE++DE +
Sbjct: 131 ---------DVSWYTRLIE-AEGLDKILNLQRHENIEIYDKALKILQTYWLEEDDEDI 178
Score = 37.0 bits (84), Expect = 0.023
Identities = 24/78 (30%), Positives = 36/78 (45%), Gaps = 19/78 (24%)
Query: 30 PIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKL 89
PI+ VI+SG V R +QFL FP+LQ V+D +P VKL
Sbjct: 16 PIETVIRSGAVHRLIQFLLDESFPKLQ-------------------SVIDANLIPTLVKL 56
Query: 90 LSSPCDDVRGQAAWALGN 107
+ D++ ++ A+ N
Sbjct: 57 TQNAEFDMKKESVCAISN 74
>At5g03070 importin alpha - like protein
Length = 483
Score = 140 bits (353), Expect = 1e-33
Identities = 125/458 (27%), Positives = 198/458 (42%), Gaps = 92/458 (20%)
Query: 12 QQLEATTEFRKRLSVDK-PPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGT 70
+++ A E R+ LS + PP++ +++G +P VQ L G + LE+AW LTNIAAG
Sbjct: 99 KRVTALRELRRLLSKSEFPPVEAALRAGAIPLLVQCLSFGSPDEQLLESAWCLTNIAAGK 158
Query: 71 SENTKVVVDHGAVPMFVKLLSSPCD-DVRGQAAWALGNIAGDSPRGRDLVLSHGALILLL 129
E TK ++ A+P+ + L V Q AWA+GN+AG+ R+++LS GAL L
Sbjct: 159 PEETKALLP--ALPLLIAHLGEKSSAPVAEQCAWAIGNVAGEGEDLRNVLLSQGALPPLA 216
Query: 130 SQLNEQEGLYRLRNAVWTLSNFCRG---KPQPALEQVRSALPALKCLVFSKDEVVLTEAC 186
+ +G +R A W LSN +G K L ++ L A+ + DE TE
Sbjct: 217 RMIFPDKG-STVRTAAWALSNLIKGPESKAAAQLVKIDGILDAILRHLKKTDEETATEIA 275
Query: 187 WALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIV 246
W + YLS + +++ G+ L+ L S L
Sbjct: 276 WIIVYLSALSDIATSMLLKGGILQLLIDRLATSSSLQL---------------------- 313
Query: 247 NHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFD 306
L+PCL S + +KKEA LSNI AG+ E + + ++ L+ +L + FD
Sbjct: 314 ---LIPCLRS----EHRVLKKEAAWVLSNIAAGSIEHKRMIHSTEVMPLLLRILSTSPFD 366
Query: 307 ITKEVAQALTNVTSGGT---------HEHIKYLVSQGCIKPLCDLLVCPDPRIVTVCLEG 357
I KEVA L N+ EH+ +VS GC++ +L+ PD + L+
Sbjct: 367 IRKEVAYVLGNLCVESAEGDRKPRIIQEHLVSIVSGGCLRGFIELVRSPDIEAARLGLQF 426
Query: 358 LENFLKVGEAEKSISNTGDVNLYAQTIKDVEGLEKGLEKFLKVGEAEKRFGNTGDVNLYA 417
+E L+ G N GE K
Sbjct: 427 IELVLR-----------GMPN----------------------GEGPK------------ 441
Query: 418 QMIEDVKGLEKIENLDSHEDREIYEKAAKILERYWLED 455
++E G++ +E HE+ E+ A ++++Y+ ED
Sbjct: 442 -LVEGEDGIDAMERFQFHENEELRVMANSLVDKYFGED 478
>At5g13060 unknown protein
Length = 709
Score = 75.1 bits (183), Expect = 8e-14
Identities = 74/315 (23%), Positives = 136/315 (42%), Gaps = 26/315 (8%)
Query: 24 LSVDKPPIDDVIQSGVVPRFVQFLDKG----DFPQLQLEAAWALTNIAAGTSENTKVVVD 79
+ +D PID V+ + R V+ L+ DF ++ A A A EN +++V+
Sbjct: 36 IDIDDEPIDLVV---AIRRHVEVLNSSFSDPDFDHEAVKEAAADIADLAKIDENVEIIVE 92
Query: 80 HGAVPMFVKLLSSP---CDDV--------RGQAAWALGNIAGDSPRGRDLVLSHGALILL 128
+GA+P V+ L SP C +V A ALG IA P + L++ GA++
Sbjct: 93 NGAIPALVRYLESPLVVCGNVPKSCEHKLEKDCALALGLIAAIQPGYQQLIVDAGAIVPT 152
Query: 129 LSQLNEQ----EGLYR---LRNAVWTLSNFCRGKPQPALE-QVRSALPALKCLVFSKDEV 180
+ L + E ++ +R A ++N P+ +V + L L+ D
Sbjct: 153 VKLLKRRGECGECMFANAVIRRAADIITNIAHDNPRIKTNIRVEGGIAPLVELLNFPDVK 212
Query: 181 VLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDM 240
V A AL +S +N ++E LV +L + A+ +GN+V
Sbjct: 213 VQRAAAGALRTVSFRNDENKSQIVELNALPTLVLMLQSQDSTVHGEAIGAIGNLVHSSPD 272
Query: 241 QTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLL 300
+ ++ G L ++ LL+ ++EA + A + + + + G I PL+ +L
Sbjct: 273 IKKEVIRAGALQPVIGLLSSTCLETQREAALLIGQFAAPDSDCKVHIAQRGAITPLIKML 332
Query: 301 QNAEFDITKEVAQAL 315
++++ + + A AL
Sbjct: 333 ESSDEQVVEMSAFAL 347
Score = 70.5 bits (171), Expect = 2e-12
Identities = 88/373 (23%), Positives = 157/373 (41%), Gaps = 26/373 (6%)
Query: 9 DNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDK-----GDFPQ-----LQLE 58
D+ EA + +D+ ++ ++++G +P V++L+ G+ P+ L+ +
Sbjct: 66 DHEAVKEAAADIADLAKIDEN-VEIIVENGAIPALVRYLESPLVVCGNVPKSCEHKLEKD 124
Query: 59 AAWALTNIAAGTSENTKVVVDHGAVPMFVKLLS--SPCDD------VRGQAAWALGNIAG 110
A AL IAA +++VD GA+ VKLL C + V +AA + NIA
Sbjct: 125 CALALGLIAAIQPGYQQLIVDAGAIVPTVKLLKRRGECGECMFANAVIRRAADIITNIAH 184
Query: 111 DSPRGRDLVLSHGALILLLSQLNEQE-GLYRLRNAVWTLSNFCRGKPQPALEQVRSALPA 169
D+PR + + G + L+ LN + + R +F + + + ++ +ALP
Sbjct: 185 DNPRIKTNIRVEGGIAPLVELLNFPDVKVQRAAAGALRTVSFRNDENKSQIVEL-NALPT 243
Query: 170 LKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALR 229
L ++ S+D V EA A+ L + D + VI AG ++ +L L A
Sbjct: 244 LVLMLQSQDSTVHGEAIGAIGNLVHSSPDIKKEVIRAGALQPVIGLLSSTCLETQREAAL 303
Query: 230 TVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIE 289
+G + D I G + L+ +L + + + + L + Q
Sbjct: 304 LIGQFAAPDSDCKVHIAQRGAITPLIKMLESSDEQVVEMSAFALGRLAQDAHNQAGIAHR 363
Query: 290 AGLIAPLVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKPLCD--LLVCPD 347
G+I+ L+NLL + A AL + E++ + G I+ L D V P
Sbjct: 364 GGIIS-LLNLLDVKTGSVQHNAAFALYGLAD--NEENVADFIKAGGIQKLQDDNFTVQPT 420
Query: 348 PRIVTVCLEGLEN 360
V L+ L+N
Sbjct: 421 RDCVVRTLKRLQN 433
>At5g19330 putative protein
Length = 636
Score = 65.5 bits (158), Expect = 6e-11
Identities = 88/407 (21%), Positives = 163/407 (39%), Gaps = 38/407 (9%)
Query: 59 AAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPC------------DDVRGQAAWALG 106
A L +A + V+VD GAVP + L +P +V +A+ALG
Sbjct: 74 ATQVLAELAKNAEDLVNVIVDGGAVPALMTHLQAPPYNDGDLAEKPYEHEVEKGSAFALG 133
Query: 107 NIAGDSPRGRDLVLSHGALILLLSQL--NEQEGLYR-----LRNAVWTLSNFCRGKPQPA 159
+A P + L++ GAL L++ L N+ R +R A ++N
Sbjct: 134 LLA-IKPEYQKLIVDKGALPHLVNLLKRNKDGSSSRAVNSVIRRAADAITNLAHENSSIK 192
Query: 160 LE-QVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLE 218
+V +P L L+ D V A AL L+ DN ++E L+ +L
Sbjct: 193 TRVRVEGGIPPLVELLEFSDSKVQRAAAGALRTLAFKNDDNKNQIVECNALPTLILMLGS 252
Query: 219 PSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITA 278
+ + A+ +GN+V + ++ G L ++ LL+ ++EA L +
Sbjct: 253 EDAAIHYEAVGVIGNLVHSSPHIKKEVLTAGALQPVIGLLSSCCPESQREAALLLGQFAS 312
Query: 279 GNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQGCIKP 338
+ + +++ G + PL+ +LQ+ + + + A AL + ++ + G + P
Sbjct: 313 TDSDCKVHIVQRGAVRPLIEMLQSPDVQLKEMSAFALGRLAQDAHNQ--AGIAHSGGLGP 370
Query: 339 LCDLLVCPDPRI---VTVCLEGL-------ENFLKVGEAEKSISNTGDVNLYAQTIKDVE 388
L LL + + L GL +F++VG +K V + + +
Sbjct: 371 LLKLLDSRNGSLQHNAAFALYGLADNEDNVSDFIRVGGIQKLQDGEFIVQVLRHLLYLMR 430
Query: 389 GLEKGLEKFLKVGEA----EKRFGNTGDVNLYAQMIEDVKGLEKIEN 431
EK +++ + + A E G G +N Q ++ L K+ N
Sbjct: 431 ISEKSIQRRVALALAHLWLELLLGLLGSLNT-KQQLDGAAALYKLAN 476
Score = 50.4 bits (119), Expect = 2e-06
Identities = 66/295 (22%), Positives = 113/295 (37%), Gaps = 60/295 (20%)
Query: 38 GVVPRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDV 97
G +P V+ L+ D ++Q AA AL +A +N +V+ A+P + +L S +
Sbjct: 199 GGIPPLVELLEFSD-SKVQRAAAGALRTLAFKNDDNKNQIVECNALPTLILMLGSEDAAI 257
Query: 98 RGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQ 157
+A +GN+ SP + VL+ GAL Q
Sbjct: 258 HYEAVGVIGNLVHSSPHIKKEVLTAGAL-------------------------------Q 286
Query: 158 PALEQVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILL 217
P + + S P + EA L + SD +++ G L+++L
Sbjct: 287 PVIGLLSSCCPESQ-----------REAALLLGQFASTDSDCKVHIVQRGAVRPLIEMLQ 335
Query: 218 EPSLSALFPALRTVGNIVSGDDMQTQAIVNH-GLLPCLLSLLTHPKKSIKKEACRTLSNI 276
P + + +G + D QA + H G L LL LL S++ A L +
Sbjct: 336 SPDVQLKEMSAFALGRL--AQDAHNQAGIAHSGGLGPLLKLLDSRNGSLQHNAAFALYGL 393
Query: 277 TAGNREQIQAVIEAG-------------LIAPLVNLLQNAEFDITKEVAQALTNV 318
A N + + I G ++ L+ L++ +E I + VA AL ++
Sbjct: 394 -ADNEDNVSDFIRVGGIQKLQDGEFIVQVLRHLLYLMRISEKSIQRRVALALAHL 447
>At4g38610 putative protein
Length = 1083
Score = 58.2 bits (139), Expect = 1e-08
Identities = 56/189 (29%), Positives = 91/189 (47%), Gaps = 8/189 (4%)
Query: 9 DNSQQLEATTEFRKRLSVD-KPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIA 67
+ +Q+EA T+ + LS+ + + VP V L+ P + L AA ALT++
Sbjct: 207 EEGKQVEALTQLCEMLSIGTEDSLSTFSVDSFVPVLVGLLNHESNPDIMLLAARALTHLC 266
Query: 68 AGTSENTKVVVDHGAVPMFV-KLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALI 126
+ VV +GAV V +LL+ D+ Q+ AL I+ + P L GAL+
Sbjct: 267 DVLPSSCAAVVHYGAVSCLVARLLTIEYMDLAEQSLQALKKISQEHPTA---CLRAGALM 323
Query: 127 LLLSQLNE-QEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEA 185
+LS L+ G+ R+ A+ T +N C+ P A + V A+P L L+ D VL A
Sbjct: 324 AVLSYLDFFSTGVQRV--ALSTAANMCKKLPSDASDYVMEAVPLLTNLLQYHDSKVLEYA 381
Query: 186 CWALSYLSD 194
L+ +++
Sbjct: 382 SICLTRIAE 390
>At3g46510 arm repeat containing protein homolog
Length = 660
Score = 57.8 bits (138), Expect = 1e-08
Identities = 61/267 (22%), Positives = 118/267 (43%), Gaps = 4/267 (1%)
Query: 53 PQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDS 112
P+ Q AA + +A ++N + + GA+P+ V LLS+P ++ + AL N++
Sbjct: 366 PEDQRSAAGEIRLLAKRNADNRVAIAEAGAIPLLVGLLSTPDSRIQEHSVTALLNLS-IC 424
Query: 113 PRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKC 172
+ ++S GA+ ++ Q+ ++ + NA TL + A+P L
Sbjct: 425 ENNKGAIVSAGAIPGIV-QVLKKGSMEARENAAATLFSLSVIDENKVTIGALGAIPPLVV 483
Query: 173 LVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVG 232
L+ + +A AL L + +A I AGV L ++L EP + AL +
Sbjct: 484 LLNEGTQRGKKDAATALFNLCIYQGNKGKA-IRAGVIPTLTRLLTEPGSGMVDEALAILA 542
Query: 233 NIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGL 292
I+S I + +P L+ + ++ A L ++ +G+ + + + GL
Sbjct: 543 -ILSSHPEGKAIIGSSDAVPSLVEFIRTGSPRNRENAAAVLVHLCSGDPQHLVEAQKLGL 601
Query: 293 IAPLVNLLQNAEFDITKEVAQALTNVT 319
+ PL++L N ++ AQ L ++
Sbjct: 602 MGPLIDLAGNGTDRGKRKAAQLLERIS 628
Score = 34.7 bits (78), Expect = 0.12
Identities = 46/215 (21%), Positives = 88/215 (40%), Gaps = 4/215 (1%)
Query: 1 MVADIWSDDNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAA 60
++ + S +S+ E + LS+ + ++ +G +P VQ L KG + + AA
Sbjct: 398 LLVGLLSTPDSRIQEHSVTALLNLSICENNKGAIVSAGAIPGIVQVLKKGSM-EARENAA 456
Query: 61 WALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVL 120
L +++ EN + GA+P V LL+ + AA AL N+ +G
Sbjct: 457 ATLFSLSV-IDENKVTIGALGAIPPLVVLLNEGTQRGKKDAATALFNLC--IYQGNKGKA 513
Query: 121 SHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEV 180
+I L++L + G + A+ L+ A+ A+P+L + +
Sbjct: 514 IRAGVIPTLTRLLTEPGSGMVDEALAILAILSSHPEGKAIIGSSDAVPSLVEFIRTGSPR 573
Query: 181 VLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQI 215
A L +L G ++ + G+ G L+ +
Sbjct: 574 NRENAAAVLVHLCSGDPQHLVEAQKLGLMGPLIDL 608
>At3g60350 Arm repeat containing protein - like
Length = 928
Score = 54.7 bits (130), Expect = 1e-07
Identities = 64/218 (29%), Positives = 93/218 (42%), Gaps = 13/218 (5%)
Query: 6 WSDDNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTN 65
W LE L+ D +V ++G V V + Q +AA AL N
Sbjct: 523 WPHGCDGVLERAAGALANLAADDKCSMEVARAGGVHALVMLARNCKYEGAQEQAARALAN 582
Query: 66 IAA---GTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSH 122
+AA N V + GA+ V+L SP + V+ +AA AL N+A D + R+ + +
Sbjct: 583 LAAHGDSNGNNAAVGQEAGALEALVQLTQSPHEGVKQEAAGALWNLAFDD-KNRESIAAF 641
Query: 123 GALILLL----SQLNEQEGLY-RLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSK 177
G + L+ S N GL R+ A+W LS + +P L LV S+
Sbjct: 642 GGVEALVALAKSSSNASTGLQERVAGALWGLS---VSEANSIAIGHEGGIPPLIALVRSE 698
Query: 178 DEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQI 215
E V A AL LS + ++ V E GV LVQ+
Sbjct: 699 AEDVHETAAGALWNLSFNPGNALRIVEEGGVVA-LVQL 735
>At2g44900 hypothetical protein
Length = 930
Score = 52.8 bits (125), Expect = 4e-07
Identities = 62/210 (29%), Positives = 90/210 (42%), Gaps = 10/210 (4%)
Query: 6 WSDDNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTN 65
W + LE L+ D +V ++G V V + +Q +AA AL N
Sbjct: 532 WPNGCDGVLERAAGALANLAADDKCSMEVAKAGGVHALVMLARNCKYEGVQEQAARALAN 591
Query: 66 IAA-GTSENTKVVV--DHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSH 122
+AA G S N V + GA+ V+L SP + VR +AA AL N++ D + ++
Sbjct: 592 LAAHGDSNNNNAAVGQEAGALEALVQLTKSPHEGVRQEAAGALWNLSFDDKNRESISVAG 651
Query: 123 G--ALILLL-SQLNEQEGLY-RLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKD 178
G AL+ L S N GL R A+W LS + +P L L S+
Sbjct: 652 GVEALVALAQSCSNASTGLQERAAGALWGLS---VSEANSVAIGREGGVPPLIALARSEA 708
Query: 179 EVVLTEACWALSYLSDGTSDNIQAVIEAGV 208
E V A AL L+ + ++ V E GV
Sbjct: 709 EDVHETAAGALWNLAFNPGNALRIVEEGGV 738
>At1g23030 unknown protein
Length = 612
Score = 51.6 bits (122), Expect = 9e-07
Identities = 52/241 (21%), Positives = 112/241 (45%), Gaps = 5/241 (2%)
Query: 63 LTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSH 122
+ +++ +++N ++ + GA+P+ V LL+S + A + N++ ++L++
Sbjct: 355 IRSLSKRSTDNRILIAEAGAIPVLVNLLTSEDVATQENAITCVLNLS-IYENNKELIMFA 413
Query: 123 GALILLLSQLNEQEGLYRLR-NAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVV 181
GA+ ++ L + G R NA TL + + A+PAL L+ +
Sbjct: 414 GAVTSIVQVL--RAGTMEARENAAATLFSLSLADENKIIIGGSGAIPALVDLLENGTPRG 471
Query: 182 LTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQ 241
+A AL L + +AV AG+ LV++L + + + T+ ++++ +
Sbjct: 472 KKDAATALFNLCIYHGNKGRAV-RAGIVTALVKMLSDSTRHRMVDEALTILSVLANNQDA 530
Query: 242 TQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQ 301
AIV LP L+ +L + ++ A L ++ + E++ + G + PL++L +
Sbjct: 531 KSAIVKANTLPALIGILQTDQTRNRENAAAILLSLCKRDTEKLITIGRLGAVVPLMDLSK 590
Query: 302 N 302
N
Sbjct: 591 N 591
Score = 38.9 bits (89), Expect = 0.006
Identities = 55/269 (20%), Positives = 111/269 (40%), Gaps = 4/269 (1%)
Query: 74 TKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLN 133
TK D + V+ LSS + R A + +++ S R L+ GA+ +L++ L
Sbjct: 324 TKNSGDMSVIRALVQRLSSRSTEDRRNAVSEIRSLSKRSTDNRILIAEAGAIPVLVNLLT 383
Query: 134 EQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACWALSYLS 193
E + NA+ + N + L A+ ++ ++ + A L LS
Sbjct: 384 -SEDVATQENAITCVLNLSIYENNKELIMFAGAVTSIVQVLRAGTMEARENAAATLFSLS 442
Query: 194 DGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPC 253
+ I + +G LV +L + A + N+ + +A V G++
Sbjct: 443 LADENKI-IIGGSGAIPALVDLLENGTPRGKKDAATALFNLCIYHGNKGRA-VRAGIVTA 500
Query: 254 LLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQ 313
L+ +L+ + + T+ ++ A N++ A+++A + L+ +LQ + + A
Sbjct: 501 LVKMLSDSTRHRMVDEALTILSVLANNQDAKSAIVKANTLPALIGILQTDQTRNRENAAA 560
Query: 314 ALTNVTSGGTHEHIKYLVSQGCIKPLCDL 342
L ++ T E + + G + PL DL
Sbjct: 561 ILLSLCKRDT-EKLITIGRLGAVVPLMDL 588
>At5g58680 unknown protein
Length = 357
Score = 48.9 bits (115), Expect = 6e-06
Identities = 51/231 (22%), Positives = 100/231 (43%), Gaps = 19/231 (8%)
Query: 170 LKCLVFSKDEVVLT---------------EACWALSYLSDGTSDNIQAVIEAGVCGRLVQ 214
L C V + D+V+ +A + LS +N + +AG LV
Sbjct: 51 LSCAVDNSDDVIRNLITHLESSSSIEEQKQAAMEIRLLSKNKPENRIKLAKAGAIKPLVS 110
Query: 215 ILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLS 274
++ L + V N+ D+ + + IV+ G + L++ L + K+ A L
Sbjct: 111 LISSSDLQLQEYGVTAVLNLSLCDENK-EMIVSSGAVKPLVNALRLGTPTTKENAACALL 169
Query: 275 NITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKYLVSQG 334
++ +I + +G I LVNLL+N F K+ + AL ++ S T+E+ V G
Sbjct: 170 RLSQVEENKI-TIGRSGAIPLLVNLLENGGFRAKKDASTALYSLCS--TNENKTRAVESG 226
Query: 335 CIKPLCDLLVCPDPRIVTVCLEGLENFLKVGEAEKSISNTGDVNLYAQTIK 385
+KPL +L++ + +V + + E++ ++ G V + + ++
Sbjct: 227 IMKPLVELMIDFESDMVDKSAFVMNLLMSAPESKPAVVEEGGVPVLVEIVE 277
Score = 38.9 bits (89), Expect = 0.006
Identities = 65/310 (20%), Positives = 113/310 (35%), Gaps = 49/310 (15%)
Query: 7 SDDNSQQLEATTEFRKRLSVDKPPID-DVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTN 65
S +Q +A E R LS +KP + ++G + V + D QLQ A+ N
Sbjct: 72 SSSIEEQKQAAMEIRL-LSKNKPENRIKLAKAGAIKPLVSLISSSDL-QLQEYGVTAVLN 129
Query: 66 IAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGAL 125
++ EN +++V GAV V L + AA AL ++ + +
Sbjct: 130 LSL-CDENKEMIVSSGAVKPLVNALRLGTPTTKENAACALLRLS--QVEENKITIGRSGA 186
Query: 126 ILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEA 185
I LL L E G ++A L + C
Sbjct: 187 IPLLVNLLENGGFRAKKDASTALYSLC--------------------------------- 213
Query: 186 CWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAI 245
T++N +E+G+ LV+++++ S + V N++ A+
Sbjct: 214 ---------STNENKTRAVESGIMKPLVELMIDFE-SDMVDKSAFVMNLLMSAPESKPAV 263
Query: 246 VNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEF 305
V G +P L+ ++ + K+ + L + + V G + PLV L Q +
Sbjct: 264 VEEGGVPVLVEIVEAGTQRQKEISVSILLQLCEESVVYRTMVAREGAVPPLVALSQGSAS 323
Query: 306 DITKEVAQAL 315
K A+AL
Sbjct: 324 RGAKVKAEAL 333
>At4g31890 unknown protein
Length = 518
Score = 47.4 bits (111), Expect = 2e-05
Identities = 39/153 (25%), Positives = 63/153 (40%), Gaps = 5/153 (3%)
Query: 9 DNSQQLEATTEFRKRLSVDKPPIDDVIQSGVVPRFVQFLDKGDFPQLQLEAAWALTNIAA 68
D +++ A +E R D + G +P V +D Q+ + +AL N+
Sbjct: 156 DCRKKITAASEVRLLAKEDSEARVTLAMLGAIPPLVSMIDDSRIVDAQIASLYALLNLGI 215
Query: 69 GTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDS--PRGRDLVLSHGALI 126
G N +V GAV +KL+ SP + A + N G S + ++ S GA+I
Sbjct: 216 GNDANKAAIVKAGAVHKMLKLIESPNTPDQEIAEAVVANFLGLSALDSNKPIIGSSGAII 275
Query: 127 LL---LSQLNEQEGLYRLRNAVWTLSNFCRGKP 156
L L L+E +A+ L N +P
Sbjct: 276 FLVKTLQNLDETSSSQAREDALRALYNLSIYQP 308
>At2g23140 hypothetical protein
Length = 924
Score = 46.2 bits (108), Expect = 4e-05
Identities = 51/217 (23%), Positives = 93/217 (42%), Gaps = 11/217 (5%)
Query: 211 RLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIV-NHGLLPCLLSLLTHPKKSIKKEA 269
+LV+ L SL A + +++ +M + ++ N G + L+ LL + ++ A
Sbjct: 624 KLVEELKSSSLDTQRQATAEL-RLLAKHNMDNRIVIGNSGAIVLLVELLYSTDSATQENA 682
Query: 270 CRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNVTSGGTHEHIKY 329
L N++ + + +A+ +AG I PL+++L+N + + A L +++ E K
Sbjct: 683 VTALLNLSINDNNK-KAIADAGAIEPLIHVLENGSSEAKENSAATLFSLS---VIEENKI 738
Query: 330 LVSQ-GCIKPLCDLLVCPDPRIVTVCLEGLENFLKVGEAEKSISNTGDVNLYAQTIKDVE 388
+ Q G I PL DLL PR L N E + I +G V +
Sbjct: 739 KIGQSGAIGPLVDLLGNGTPRGKKDAATALFNLSIHQENKAMIVQSGAVRYLIDLMDPAA 798
Query: 389 GL-EKG---LEKFLKVGEAEKRFGNTGDVNLYAQMIE 421
G+ +K L + E G G + L +++E
Sbjct: 799 GMVDKAVAVLANLATIPEGRNAIGQEGGIPLLVEVVE 835
Score = 32.0 bits (71), Expect = 0.75
Identities = 55/272 (20%), Positives = 104/272 (38%), Gaps = 45/272 (16%)
Query: 41 PRFVQFLDKGDFPQLQLEAAWALTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCDDVRGQ 100
P+F+ +G F + E + A + V V+ L S D + Q
Sbjct: 580 PKFMDRRTRGQFWRRPSERLGSRIVSAPSNETRRDLSEVETQVKKLVEELKSSSLDTQRQ 639
Query: 101 AAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPAL 160
A L +A + R ++ + GA++LL+
Sbjct: 640 ATAELRLLAKHNMDNRIVIGNSGAIVLLVE------------------------------ 669
Query: 161 EQVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPS 220
L++S D A AL LS +N +A+ +AG L+ +L S
Sbjct: 670 ------------LLYSTDSATQENAVTALLNLSI-NDNNKKAIADAGAIEPLIHVLENGS 716
Query: 221 LSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGN 280
A + T+ ++ ++ + + I G + L+ LL + KK+A L N++ +
Sbjct: 717 SEAKENSAATLFSLSVIEENKIK-IGQSGAIGPLVDLLGNGTPRGKKDAATALFNLSI-H 774
Query: 281 REQIQAVIEAGLIAPLVNLLQNAEFDITKEVA 312
+E ++++G + L++L+ A + K VA
Sbjct: 775 QENKAMIVQSGAVRYLIDLMDPAAGMVDKAVA 806
>At1g71020 unknown protein
Length = 628
Score = 46.2 bits (108), Expect = 4e-05
Identities = 51/240 (21%), Positives = 110/240 (45%), Gaps = 8/240 (3%)
Query: 63 LTNIAAGTSENTKVVVDHGAVPMFVKLLSSPCD-DVRGQAAWALGNIAGDSPRGRDLVLS 121
+ +++ +++N ++ + GA+P+ VKLL+S D + + A + N++ ++L++
Sbjct: 365 IRSLSKRSTDNRILIAEAGAIPVLVKLLTSDGDTETQENAVTCILNLS-IYEHNKELIML 423
Query: 122 HGAL--ILLLSQLNEQEGLYRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDE 179
GA+ I+L+ + E NA TL + + A+ AL L+
Sbjct: 424 AGAVTSIVLVLRAGSMEA---RENAAATLFSLSLADENKIIIGASGAIMALVDLLQYGSV 480
Query: 180 VVLTEACWALSYLSDGTSDNIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDD 239
+A AL L + +AV AG+ LV++L + S + T+ ++++ +
Sbjct: 481 RGKKDAATALFNLCIYQGNKGRAV-RAGIVKPLVKMLTDSSSERMADEALTILSVLASNQ 539
Query: 240 MQTQAIVNHGLLPCLLSLLTHPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNL 299
+ AI+ +P L+ L + ++ A L + + E++ ++ G + PL+ L
Sbjct: 540 VAKTAILRANAIPPLIDCLQKDQPRNRENAAAILLCLCKRDTEKLISIGRLGAVVPLMEL 599
Score = 42.0 bits (97), Expect = 7e-04
Identities = 57/264 (21%), Positives = 104/264 (38%), Gaps = 3/264 (1%)
Query: 79 DHGAVPMFVKLLSSPCDDVRGQAAWALGNIAGDSPRGRDLVLSHGALILLLSQLNEQEGL 138
D A+ V LSS + R A + +++ S R L+ GA+ +L+ L
Sbjct: 339 DMSAIRALVCKLSSQSIEDRRTAVSEIRSLSKRSTDNRILIAEAGAIPVLVKLLTSDGDT 398
Query: 139 YRLRNAVWTLSNFCRGKPQPALEQVRSALPALKCLVFSKDEVVLTEACWALSYLSDGTSD 198
NAV + N + L + A+ ++ LV + E A + +
Sbjct: 399 ETQENAVTCILNLSIYEHNKELIMLAGAVTSI-VLVLRAGSMEARENAAATLFSLSLADE 457
Query: 199 NIQAVIEAGVCGRLVQILLEPSLSALFPALRTVGNIVSGDDMQTQAIVNHGLLPCLLSLL 258
N + +G LV +L S+ A + N+ + +A V G++ L+ +L
Sbjct: 458 NKIIIGASGAIMALVDLLQYGSVRGKKDAATALFNLCIYQGNKGRA-VRAGIVKPLVKML 516
Query: 259 THPKKSIKKEACRTLSNITAGNREQIQAVIEAGLIAPLVNLLQNAEFDITKEVAQALTNV 318
T + T+ ++ A N+ A++ A I PL++ LQ + + A L +
Sbjct: 517 TDSSSERMADEALTILSVLASNQVAKTAILRANAIPPLIDCLQKDQPRNRENAAAILLCL 576
Query: 319 TSGGTHEHIKYLVSQGCIKPLCDL 342
T + I + G + PL +L
Sbjct: 577 CKRDTEKLIS-IGRLGAVVPLMEL 599
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,307,106
Number of Sequences: 26719
Number of extensions: 429603
Number of successful extensions: 1483
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 1160
Number of HSP's gapped (non-prelim): 173
length of query: 466
length of database: 11,318,596
effective HSP length: 103
effective length of query: 363
effective length of database: 8,566,539
effective search space: 3109653657
effective search space used: 3109653657
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC122160.6


