

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.5 - phase: 0
(433 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
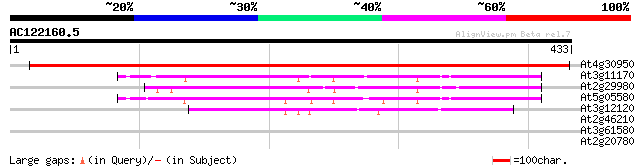
Score E
Sequences producing significant alignments: (bits) Value
At4g30950 chloroplast omega-6 fatty acid desaturase (fad6) 695 0.0
At3g11170 omega-3 fatty acid desaturase, chloroplast precursor 127 2e-29
At2g29980 omega-3 fatty acid desaturase 127 2e-29
At5g05580 temperature-sensitive omega-3 fatty acid desaturase, c... 125 6e-29
At3g12120 unknown protein 98 1e-20
At2g46210 putative fatty acid desaturase/cytochrome b5 fusion pr... 41 0.001
At3g61580 delta-8 sphingolipid desaturase (sld1) 40 0.003
At2g20780 putative sugar transporter 30 3.4
>At4g30950 chloroplast omega-6 fatty acid desaturase (fad6)
Length = 448
Score = 695 bits (1794), Expect = 0.0
Identities = 316/418 (75%), Positives = 366/418 (86%), Gaps = 1/418 (0%)
Query: 16 SAPYSPGLFNLKGDGLIHKGFRHQSQKHLTP-RNKVTVIRAVAIPVEPAPVESAEYRKQL 74
SA SPG++ +K L+ KG H+S++ + P + ++ I+AVA PV P +SAE R+QL
Sbjct: 29 SARVSPGVYAVKPIDLLLKGRTHRSRRCVAPVKRRIGCIKAVAAPVAPPSADSAEDREQL 88
Query: 75 AERYGFEQIGEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISATSYALGLLMISKAP 134
AE YGF QIGE LP+NVTLKD++ +LPK+VFEIDD+KA KSVLIS TSY LGL MI+K+P
Sbjct: 89 AESYGFRQIGEDLPENVTLKDIMDTLPKEVFEIDDLKALKSVLISVTSYTLGLFMIAKSP 148
Query: 135 WYLLPLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHD 194
WYLLPLAWAWTGTA+TGFFVIGHDCAHKSFS NKLVEDIVGTLAFLPL+YPYEPWRFKHD
Sbjct: 149 WYLLPLAWAWTGTAITGFFVIGHDCAHKSFSKNKLVEDIVGTLAFLPLVYPYEPWRFKHD 208
Query: 195 KHHAKTNMLKEDTAWQPVWQDEFESSPFLRKAIIYGYGPFRCWMSIAHWLVSHFDLKTFR 254
+HHAKTNML DTAWQPV +EFESSP +RKAII+GYGP R W+SIAHW+ HF+LK FR
Sbjct: 209 RHHAKTNMLVHDTAWQPVPPEEFESSPVMRKAIIFGYGPIRPWLSIAHWVNWHFNLKKFR 268
Query: 255 PNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYHFWMSTFTMVHHTAP 314
+E+ RVKISLACVFAF+A+GWPLI+YK GI+GW+KFWLMPWLGYHFWMSTFTMVHHTAP
Sbjct: 269 ASEVNRVKISLACVFAFMAVGWPLIVYKVGILGWVKFWLMPWLGYHFWMSTFTMVHHTAP 328
Query: 315 HIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIPSYNLRAAHKSI 374
HIPFK ++EWNAAQAQLNGTVHC YP WIEILCHDINVHIPHHISP+IPSYNLRAAH+SI
Sbjct: 329 HIPFKPADEWNAAQAQLNGTVHCDYPSWIEILCHDINVHIPHHISPRIPSYNLRAAHESI 388
Query: 375 QENWGKYLNEASWNWRLMKTIMTVCHVYDKQQNYVSFDEVAPKESRPIRFLKKVMPDY 432
QENWGKY N A+WNWRLMKTIMTVCHVYDK++NY+ FD +AP+ES+PI FLKK MP+Y
Sbjct: 389 QENWGKYTNLATWNWRLMKTIMTVCHVYDKEENYIPFDRLAPEESQPITFLKKAMPNY 446
>At3g11170 omega-3 fatty acid desaturase, chloroplast precursor
Length = 446
Score = 127 bits (318), Expect = 2e-29
Identities = 95/342 (27%), Positives = 151/342 (43%), Gaps = 24/342 (7%)
Query: 84 GEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISATSYALGLLMISKAP----WYLLP 139
G P P N L D+ ++PK + + WKS+ A+ + + A W + P
Sbjct: 91 GAPPPFN--LADIRAAIPKHCWVKNP---WKSLSYVVRDVAIVFALAAGAAYLNNWIVWP 145
Query: 140 LAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHAK 199
L W GT FV+GHDC H SFS + + +VG L ++ PY WR H HH
Sbjct: 146 LYWLAQGTMFWALFVLGHDCGHGSFSNDPKLNSVVGHLLHSSILVPYHGWRISHRTHHQN 205
Query: 200 TNMLKEDTAWQPVWQDEFESSP-------FLRKAIIYGYGPFRCWMSIAHWLVSHF--DL 250
++ D +W P+ + + + F ++ Y PF W SH+ D
Sbjct: 206 HGHVENDESWHPMSEKIYNTLDKPTRFFRFTLPLVMLAY-PFYLWARSPGKKGSHYHPDS 264
Query: 251 KTFRPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYHFWMSTFTMVH 310
F P E K V S AC A A+ + + G + +K + +P+ W+ T +H
Sbjct: 265 DLFLPKERKDVLTSTACWTAMAAL-LVCLNFTIGPIQMLKLYGIPYWINVMWLDFVTYLH 323
Query: 311 HTA--PHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIPSYNLR 368
H +P+ +EW+ + L T+ Y I + HDI H+ HH+ P+IP Y+L
Sbjct: 324 HHGHEDKLPWYRGKEWSYLRGGLT-TLDRDY-GLINNIHHDIGTHVIHHLFPQIPHYHLV 381
Query: 369 AAHKSIQENWGKYLNEASWNWRLMKTIMTVCHVYDKQQNYVS 410
A ++ + GKY E + L ++ + K+ +YVS
Sbjct: 382 EATEAAKPVLGKYYREPDKSGPLPLHLLEILAKSIKEDHYVS 423
>At2g29980 omega-3 fatty acid desaturase
Length = 386
Score = 127 bits (318), Expect = 2e-29
Identities = 88/332 (26%), Positives = 151/332 (44%), Gaps = 30/332 (9%)
Query: 105 FEIDDVKA------WKSVLISATSY---------ALGLLMISKAPWYLLPLAWAWTGTAV 149
F+I D++A W + + SY AL + + W+L PL WA GT
Sbjct: 34 FKIGDIRAAIPKHCWVKSPLRSMSYVVRDIIAVAALAIAAVYVDSWFLWPLYWAAQGTLF 93
Query: 150 TGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHAKTNMLKEDTAW 209
FV+GHDC H SFS L+ +VG + ++ PY WR H HH ++ D +W
Sbjct: 94 WAIFVLGHDCGHGSFSDIPLLNSVVGHILHSFILVPYHGWRISHRTHHQNHGHVENDESW 153
Query: 210 QPVWQDEFESSPFLRKAIIY-------GYGPFRCWMSIAHWLVSHFD--LKTFRPNEIKR 260
P+ + ++ P + + Y Y + C+ S SHF+ F P+E K
Sbjct: 154 VPLPERVYKKLPHSTRMLRYTVPLPMLAYPLYLCYRSPGK-EGSHFNPYSSLFAPSERKL 212
Query: 261 VKISLACVFAFIAIGWPLIIYKTGIMGWIKFWLMPWLGYHFWMSTFTMVHHTA--PHIPF 318
+ S C ++ + + + + G + +K + +P++ + W+ T +HH +P+
Sbjct: 213 IATSTTC-WSIMFVSLIALSFVFGPLAVLKVYGVPYIIFVMWLDAVTYLHHHGHDEKLPW 271
Query: 319 KNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIPSYNLRAAHKSIQENW 378
+EW+ + L T+ Y + I HDI H+ HH+ P+IP Y+L A K+ +
Sbjct: 272 YRGKEWSYLRGGLT-TIDRDYGIFNNI-HHDIGTHVIHHLFPQIPHYHLVDATKAAKHVL 329
Query: 379 GKYLNEASWNWRLMKTIMTVCHVYDKQQNYVS 410
G+Y E + + ++ K+ +YVS
Sbjct: 330 GRYYREPKTSGAIPIHLVESLVASIKKDHYVS 361
>At5g05580 temperature-sensitive omega-3 fatty acid desaturase,
chloroplast precursor (sp|P48622)
Length = 435
Score = 125 bits (313), Expect = 6e-29
Identities = 95/342 (27%), Positives = 154/342 (44%), Gaps = 24/342 (7%)
Query: 84 GEPLPDNVTLKDVITSLPKKVFEIDDVKAWKSVLISATSYALGLLMISKA--PWYLLPLA 141
G P P N L D+ ++PK + + + S ++ + GL ++ W L PL
Sbjct: 84 GAPPPFN--LADIRAAIPKHCW-VKNPWMSMSYVVRDVAIVFGLAAVAAYFNNWLLWPLY 140
Query: 142 WAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHAKTN 201
W GT FV+GHDC H SFS + + + G L ++ PY WR H HH
Sbjct: 141 WFAQGTMFWALFVLGHDCGHGSFSNDPRLNSVAGHLLHSSILVPYHGWRISHRTHHQNHG 200
Query: 202 MLKEDTAWQP----VWQDEFESSPFLRKAIIYGY--GPFRCWMSIAHWLVSHF--DLKTF 253
++ D +W P ++++ +++ R + + PF W SH+ D F
Sbjct: 201 HVENDESWHPLPESIYKNLEKTTQMFRFTLPFPMLAYPFYLWNRSPGKQGSHYHPDSDLF 260
Query: 254 RPNEIKRVKISLACVFAFIAIGWPLIIYKTGIMG---WIKFWLMPWLGYHFWMSTFTMVH 310
P E K V S AC A A L++ +MG +K + +P+ + W+ T +H
Sbjct: 261 LPKEKKDVLTSTACWTAMAA----LLVCLNFVMGPIQMLKLYGIPYWIFVMWLDFVTYLH 316
Query: 311 HTA--PHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIPSYNLR 368
H +P+ +EW+ + L T+ Y WI + HDI H+ HH+ P+IP Y+L
Sbjct: 317 HHGHEDKLPWYRGKEWSYLRGGLT-TLDRDY-GWINNIHHDIGTHVIHHLFPQIPHYHLV 374
Query: 369 AAHKSIQENWGKYLNEASWNWRLMKTIMTVCHVYDKQQNYVS 410
A ++ + GKY E + L ++ KQ ++VS
Sbjct: 375 EATEAAKPVLGKYYREPKNSGPLPLHLLGSLIKSMKQDHFVS 416
>At3g12120 unknown protein
Length = 383
Score = 97.8 bits (242), Expect = 1e-20
Identities = 71/267 (26%), Positives = 119/267 (43%), Gaps = 18/267 (6%)
Query: 139 PLAWAWTGTAVTGFFVIGHDCAHKSFSTNKLVEDIVGTLAFLPLIYPYEPWRFKHDKHHA 198
PL WA G +TG +VI H+C H +FS + ++D VG + L+ PY W++ H +HH+
Sbjct: 87 PLYWACQGCVLTGIWVIAHECGHHAFSDYQWLDDTVGLIFHSFLLVPYFSWKYSHRRHHS 146
Query: 199 KTNMLKEDTAWQP------VWQDEFESSP-----FLRKAIIYG---YGPFRCWMSIAHWL 244
T L+ D + P W ++ ++P L + G Y F
Sbjct: 147 NTGSLERDEVFVPKQKSAIKWYGKYLNNPLGRIMMLTVQFVLGWPLYLAFNVSGRPYDGF 206
Query: 245 VSHFDLKTFRPNEIKRVKISLACVFAFIAIGWPLIIYKT--GIMGWIKFWLMPWLGYHFW 302
HF N+ +R++I L+ +A+ + L Y G+ I + +P L + +
Sbjct: 207 ACHFFPNAPIYNDRERLQIYLSDA-GILAVCFGLYRYAAAQGMASMICLYGVPLLIVNAF 265
Query: 303 MSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKI 362
+ T + HT P +P +S EW+ + L TV Y ++ + + H+ HH+ +
Sbjct: 266 LVLITYLQHTHPSLPHYDSSEWDWLRGAL-ATVDRDYGILNKVFHNITDTHVAHHLFSTM 324
Query: 363 PSYNLRAAHKSIQENWGKYLNEASWNW 389
P YN A K+I+ G Y W
Sbjct: 325 PHYNAMEATKAIKPILGDYYQFDGTPW 351
>At2g46210 putative fatty acid desaturase/cytochrome b5 fusion
protein
Length = 449
Score = 41.2 bits (95), Expect = 0.001
Identities = 56/269 (20%), Positives = 110/269 (40%), Gaps = 49/269 (18%)
Query: 155 IGHDCAHKSFST----NKLVEDIVGT-LAFLPLIYPYEPWRFKHDKHHAKTNMLKEDTAW 209
+GHD H + ++ NKL++ + G L + + + W++ H+ HH N L D
Sbjct: 158 VGHDSGHYTVTSTKPCNKLIQLLSGNCLTGISIAW----WKWTHNAHHIACNSLDHDPDL 213
Query: 210 Q--PVWQDEFESSPFLRKAIIYGYGPFRCWMSIAHWLVSH---------------FDLKT 252
Q P++ S+ F YG + +A +L+S+ ++T
Sbjct: 214 QHIPIFA---VSTKFFNSMTSRFYGRKLTFDPLARFLISYQHWTFYPVMCVGRINLFIQT 270
Query: 253 FRPNEIKRVKISLACVFAFIAIGW---PLIIYKTGIMGWIKFWLMPWLGY--------HF 301
F KR A A I + W PL++ + + W + ++ ++ + F
Sbjct: 271 FLLLFSKRHVPDRALNIAGILVFWTWFPLLV--SFLPNWQERFIFVFVSFAVTAIQHVQF 328
Query: 302 WMSTFTMVHHTAPHIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPK 361
++ F +T P N +W + Q GT+ + +++ + + HH+ P+
Sbjct: 329 CLNHFAADVYTGP----PNGNDW--FEKQTAGTLDISCRSFMDWFFGGLQFQLEHHLFPR 382
Query: 362 IPSYNLRAAHKSIQENWGKY-LNEASWNW 389
+P +LR ++E K+ L S +W
Sbjct: 383 LPRCHLRTVSPVVKELCKKHNLPYRSLSW 411
>At3g61580 delta-8 sphingolipid desaturase (sld1)
Length = 449
Score = 39.7 bits (91), Expect = 0.003
Identities = 61/261 (23%), Positives = 100/261 (37%), Gaps = 33/261 (12%)
Query: 155 IGHDCAHKSFSTNKLVEDIVGTLAFLPLI-YPYEPWRFKHDKHHAKTNMLKEDTAWQ--P 211
IGHD H +NK L+ L W++ H+ HH N L D Q P
Sbjct: 158 IGHDSGHYVIMSNKSYNRFAQLLSGNCLTGISIAWWKWTHNAHHLACNSLDYDPDLQHIP 217
Query: 212 VWQDEFESSPFLRKAIIYGYGPFRCWMSIAHWLVS--HFD-------------LKTFRPN 256
V+ S+ F Y + +A +LVS HF ++TF
Sbjct: 218 VFA---VSTKFFSSLTSRFYDRKLTFDPVARFLVSYQHFTYYPVMCFGRINLFIQTFLLL 274
Query: 257 EIKRVKISLACVFAFIAIGW---PLIIYKTGIMGWIKFWLMPWLGYHFWMST---FTMVH 310
KR A FA I + W PL++ + + W + + + + FT+ H
Sbjct: 275 FSKREVPDRALNFAGILVFWTWFPLLV--SCLPNWPERFFFVFTSFTVTALQHIQFTLNH 332
Query: 311 HTAP-HIPFKNSEEWNAAQAQLNGTVHCAYPQWIEILCHDINVHIPHHISPKIPSYNLRA 369
A ++ +W QA GT+ + +++ + + HH+ P++P +LR
Sbjct: 333 FAADVYVGPPTGSDWFEKQAA--GTIDISCRSYMDWFFGGLQFQLEHHLFPRLPRCHLRK 390
Query: 370 AHKSIQENWGKY-LNEASWNW 389
+QE K+ L S +W
Sbjct: 391 VSPVVQELCKKHNLPYRSMSW 411
>At2g20780 putative sugar transporter
Length = 547
Score = 29.6 bits (65), Expect = 3.4
Identities = 12/31 (38%), Positives = 20/31 (63%), Gaps = 1/31 (3%)
Query: 210 QPVWQDEFESSPFLRKAIIYGYGPFRCWMSI 240
+PVW++ SP +RK +I G+G +C+ I
Sbjct: 305 RPVWRELLSPSPVVRKMLIVGFG-IQCFQQI 334
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,468,301
Number of Sequences: 26719
Number of extensions: 458614
Number of successful extensions: 967
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 944
Number of HSP's gapped (non-prelim): 8
length of query: 433
length of database: 11,318,596
effective HSP length: 102
effective length of query: 331
effective length of database: 8,593,258
effective search space: 2844368398
effective search space used: 2844368398
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122160.5


