

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.3 - phase: 0 /pseudo
(835 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
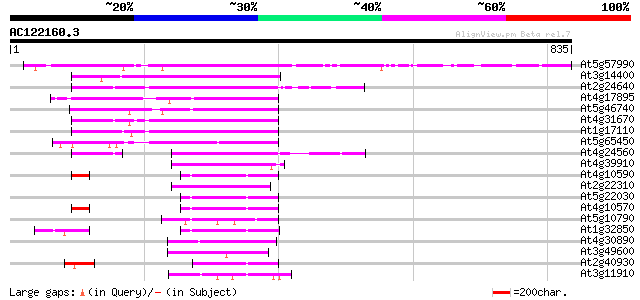
Score E
Sequences producing significant alignments: (bits) Value
At5g57990 ubiquitin-specific protease 23 (UBP23) 436 e-122
At3g14400 unknown protein 168 1e-41
At2g24640 putative ubiquitin carboxyl terminal hydrolase 166 7e-41
At4g17895 ubiquitin-specific protease 20 (UBP20) 163 3e-40
At5g46740 putative protein 160 3e-39
At4g31670 unknown protein 158 1e-38
At1g17110 ubiquitin-specific protease 15 (UBP15) 153 4e-37
At5g65450 putative protein 147 2e-35
At4g24560 putative protein 118 2e-26
At4g39910 ubiquitin-specific protease (AtUBP3) 87 3e-17
At4g10590 unknown protein 87 4e-17
At2g22310 putative ubiquitin carboxyl terminal hydrolase 86 1e-16
At5g22030 ubiquitin-specific protease 8 (UBP8) 85 2e-16
At4g10570 putative protein 85 2e-16
At5g10790 ubiquitin specific protease -like protein 83 6e-16
At1g32850 hypothetical protein 83 6e-16
At4g30890 ubiquitin-specific protease 24 (UBP24) 80 6e-15
At3g49600 putative protein 79 1e-14
At2g40930 ubiquitin-specific protease (UBP5) 78 2e-14
At3g11910 ubiquitin carboxyl-terminal hydrolase like protein 74 3e-13
>At5g57990 ubiquitin-specific protease 23 (UBP23)
Length = 859
Score = 436 bits (1121), Expect = e-122
Identities = 328/836 (39%), Positives = 450/836 (53%), Gaps = 89/836 (10%)
Query: 21 RKIEFIPVKKPFKGFSN---DFNIETLNPTTSEHRQLVSANIQSSQPQPSK--NSEFSEF 75
RKIEF P +KPF GFSN DF IETLNP +S R L S P K +S+ E
Sbjct: 38 RKIEFHPARKPFNGFSNGRSDFKIETLNPCSSNQRLL-------SAPSAKKPDSSDLLEH 90
Query: 76 GLDPELSLGITFRRIGAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFC 135
G +P+L+ ITFR+IGAGL NLGNTCFLNSVLQCLTYTEPLAA LQ+ H+ CH+AGFC
Sbjct: 91 GFEPDLTFSITFRKIGAGLQNLGNTCFLNSVLQCLTYTEPLAATLQTAAHQKYCHVAGFC 150
Query: 136 ALCAIQNHVSCALQATGRIVSPQDMVGNLRCI---LMSCFFNEQSRHITQFPKG*AGRCT 192
ALCAIQ HV A QA GRI++P+D+V NLRCI +C + ++ + +C+
Sbjct: 151 ALCAIQKHVRTARQANGRILAPKDLVSNLRCISRNFRNCRQEDAHEYMINLLEC-MHKCS 209
Query: 193 *VYGELT*VNA*MLPAFWSTE*ITWCFREKFGS*---DLWWSSSKSASNKFDPFLDLSLE 249
G +P+ S + FG + SNKFDPFLDLSL+
Sbjct: 210 LPSG---------VPSESSDAYRRSLVHKIFGGSLRSQVKCEQCSHCSNKFDPFLDLSLD 260
Query: 250 ILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYA 309
I KADSLQ+AL+ FTA ELLD G K Y C+RCKQKV+A KQLT+ KAPYVL +HLKRF A
Sbjct: 261 ISKADSLQRALSRFTAVELLDNGAKVYQCERCKQKVKAKKQLTVSKAPYVLTVHLKRFEA 320
Query: 310 HDPNIKIKKKVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNN 369
H KI +KV F SA+++KPFVSG +G++KY+LYGVLVH G S+HSGHY C+VRTS+
Sbjct: 321 HRSE-KIDRKVDFTSAIDMKPFVSGPHEGNLKYTLYGVLVHYGRSSHSGHYACFVRTSSG 379
Query: 370 MWYTLDDTRVSHVGEQEVLNQQAYMLFYVRDKKSITPRKPVNIAKEEISKTNVIGSRYPS 429
MWY+LDD RV V E+ V NQ+AYMLFYVRD+++ P+ V + K+E TN R
Sbjct: 380 MWYSLDDNRVVQVSEKTVFNQKAYMLFYVRDRQNAVPKNSVPVVKKESFATN----RASL 435
Query: 430 TPTNALKDYPNGHVENKFCGV-PLTAETQKNLLNADPSRISDALIQQKNSGILAKSLMHN 488
+ +KD NG K CG L A L + PS + A++ QK+ + AK +N
Sbjct: 436 IVASNIKDQVNGSTVIKECGFGALVANGLAPLKSCGPS--TPAVLTQKD--LNAKETQNN 491
Query: 489 ETPVSERTSKELTQMNSSDELPVAKSELECLSSLDHSGKDNVSGNQKCLVAP-AGDKPNL 547
+S +KE+ + + PV +L + L + QK + P A + +L
Sbjct: 492 --AISNVEAKEILE-TENGSAPVKTCDLAAPTVLVQKDLNTKEIFQKEVPLPQANGEGSL 548
Query: 548 FTEDT-----LLKEGVDSPLVEPIVSNPQTFIGKHASDRTSPSDEVSLSKYFSILFGQNW 602
ED+ +L E V SP ++ +N QT + P E S+ + S L N
Sbjct: 549 VKEDSKAACLILPEKV-SPHLDG-SANAQTLV---KLPTLGPKAENSVEEKNS-LNNLN- 601
Query: 603 CYFSFLDYLSVIHCQ-KNSPAEVDAVAAQDSVTNLSEHAGLVGTSNAVVPQGLVLKVSSK 661
+ L VI+ N P E + Q +L E A + + LK++S+
Sbjct: 602 ---EPANSLKVINVSVGNPPVEKAVLIDQTMGHHLEESATSIES----------LKLTSE 648
Query: 662 RSLKDSSLDQKQVKKSKKKFPKYKGSIMQLSLIRPSMRCLGPRKKNHKKNRR--CLLGLK 719
R ++ K+ +K K K K + +L+ LG RKK ++ R + +
Sbjct: 649 R---ETLTTPKKTRKPKTKTLKVEFKFFKLA--------LGLRKKKVQRRERLSTTVAGE 697
Query: 720 YHSKEKLKKNAISSDVGPSISRKEHLRSSLMKHDAEREIINRMDQNCAVLATATQVENMS 779
S+E L K + I+ + + SS D + + L+ A+Q
Sbjct: 698 IISEELLSKKRVKYQDTSLIAPSKMISSS----DGAVTSDQQQPVGSSDLSEASQNAKRK 753
Query: 780 RCSLQDDKRDQMQNTVVTQGLEETVVARWDDVELPSSQALESKTDQIARIGYVGDE 835
R S+ K ++T+G+ ETVVA+WD+ E+ +SQ SK++ + IGYV DE
Sbjct: 754 RESVLLQKE---AVNILTRGVPETVVAKWDE-EISASQKRGSKSEGASSIGYVADE 805
>At3g14400 unknown protein
Length = 661
Score = 168 bits (425), Expect = 1e-41
Identities = 115/317 (36%), Positives = 168/317 (52%), Gaps = 11/317 (3%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSC--HIAGF----CALCAIQNHVSC 146
GL NLGNTC+LNSVLQCLT+T PLA + + KH S C ++ G C C ++ ++
Sbjct: 25 GLRNLGNTCYLNSVLQCLTFTPPLANFCLTHKHSSHCDTYVDGERKRDCPFCIVEKRIAR 84
Query: 147 ALQATGRIVSPQDMVGNLRCILMSCFFNEQSRHITQFPKG*AGRCT*VYGELT*VNA*ML 206
+L +P + L+ I F + +F + C L +
Sbjct: 85 SLSVDLTTDAPNKISSCLK-IFAEHFKLGRQEDAHEFLRYVIDACHNTSLRLKKLRYNGN 143
Query: 207 PAFWSTE*ITWCFREKFGS*DLWWSSSKSASNKFDPFLDLSLEILKADSLQKALANFTAA 266
F + F S + S + SNK D +D+SLEIL++ S++++L F +
Sbjct: 144 EPFNGNSVVKEIFGGALQS-QVKCLSCGAESNKADEIMDISLEILQSSSVKESLQKFFQS 202
Query: 267 ELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSAL 326
E+LDG K Y C+ C++ V A KQ++I +AP +L I LKRF KI K + F L
Sbjct: 203 EILDGNNK-YRCESCEKLVTARKQMSILQAPNILVIQLKRFGGIFGG-KIDKAISFGEIL 260
Query: 327 NLKPFVS-GSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQ 385
L F+S S D +Y L+G++VHSGFS SGHYY YV+ S WY +D+ VS Q
Sbjct: 261 VLSNFMSKASKDPQPEYKLFGIIVHSGFSPESGHYYAYVKDSLGRWYCCNDSFVSLSTLQ 320
Query: 386 EVLNQQAYMLFYVRDKK 402
EVL+++AY+LF+ R +
Sbjct: 321 EVLSEKAYILFFSRSNQ 337
>At2g24640 putative ubiquitin carboxyl terminal hydrolase
Length = 672
Score = 166 bits (419), Expect = 7e-41
Identities = 137/442 (30%), Positives = 208/442 (46%), Gaps = 35/442 (7%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCALQATG 152
GL N GN+CF N VLQCL++T PL AYL HK C +C LC +NH+ A +
Sbjct: 175 GLTNCGNSCFANVVLQCLSWTRPLVAYLLERGHKRECRRNDWCFLCEFENHLDRANYSRF 234
Query: 153 RIVSPQDMVGNLRCILMSCFFNEQ--SRHITQFPKG*AGR-CT*VYGELT*VNA*MLPAF 209
SP +++ L I + + Q + + +F C +G V P
Sbjct: 235 PF-SPMNIISRLPNIGGNLGYGRQEDAHELMRFAIDMMQSVCLDEFGGEKVVP----PRA 289
Query: 210 WSTE*ITWCFREKFGS*DLWWSSSKSASNKFDPFLDLSLEIL-KADSLQKALANFTAAEL 268
T I + F S + ++ + S++++ +DL++EI A SL++ L FTA E
Sbjct: 290 QETTLIQYIFGGLLQS-QVQCTACSNVSDQYENMMDLTVEIHGDAVSLEECLDQFTAKEW 348
Query: 269 LDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSALNL 328
L G + Y C RC V+A K+L+I AP +L I LKRF K+ K++ F +L
Sbjct: 349 LQG-DNLYKCDRCDDYVKACKRLSIRCAPNILTIALKRFQGGRFG-KLNKRISFPETFDL 406
Query: 329 KPFVSGSSDGDVKYSLYGVLVHSGFSTHS--GHYYCYVRTSNNMWYTLDDTRVSHVGEQE 386
P++SG +G Y LY V+VH S GHY CYV+ WY +DD+ V V ++
Sbjct: 407 GPYMSGGGEGSDVYKLYAVIVHLDMLNASFFGHYICYVKDFRGNWYRIDDSEVEKVELED 466
Query: 387 VLNQQAYMLFYVRDKKSITPRKPVNIAKEEISKTNVIGSRYPSTPTNALKDYPNGHVENK 446
VL+Q+AYML Y R + PR P N+ EE T T + +G VE+
Sbjct: 467 VLSQRAYMLLYSR----VQPR-PSNLRSEESQDEK-------KTDTLNTESNQDGSVESS 514
Query: 447 FCGVPLTAETQKNLLNADPSRISDALIQQKNSGILAKSLMHNETPVSERTSKELTQMNSS 506
GV + +L N S D ++++S + PVSE + ++++
Sbjct: 515 --GVGTNDTSVSSLCNGIISHSEDPEYEKESS-------LSASVPVSEEGKEVDVKVDTV 565
Query: 507 DELPVAKSELECLSSLDHSGKD 528
D ++E S DH ++
Sbjct: 566 DSESNRSIDMEHDSGTDHQEEE 587
>At4g17895 ubiquitin-specific protease 20 (UBP20)
Length = 695
Score = 163 bits (413), Expect = 3e-40
Identities = 117/354 (33%), Positives = 172/354 (48%), Gaps = 42/354 (11%)
Query: 61 SSQPQPSKNSEFSEFGLDPELSLGITFRRIGAGLWNLGNTCFLNSVLQCLTYTEPLAAYL 120
+S+ +P K S F F +PE G+ GAGLWNLGN+CFLNSV QC T+T PL L
Sbjct: 151 NSKNEPRK-SLFYGFRQEPEPVTGV-----GAGLWNLGNSCFLNSVFQCFTHTVPLIESL 204
Query: 121 QSGKHKSSCHIAG--FCALCAIQNHVSCALQATGRIVSPQDMVGNLRCILMSCFFNEQSR 178
S +++ CH FC + AI+ H+ AL+ ++P NL F Q
Sbjct: 205 LSFRYEVPCHCGNEFFCVIRAIRYHIEAALRPERCPIAPYFFFDNLNYFSPD-FQRYQQE 263
Query: 179 HITQFPKG*AGRCT*VYGELT*VNA*MLPAFWSTE*ITWCFREKFGS*DLWWSSSKSA-- 236
+F + + + T FR S D++ S
Sbjct: 264 DAHEFLQAFLEKLEICGSDRT------------------SFRGDITSQDVFSGRLISGLR 305
Query: 237 -------SNKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVK 289
S ++ + LSLEI D+L AL +FT E LD +Q C C +KV K
Sbjct: 306 CCNCDYVSETYEKSVGLSLEIEDVDTLGSALESFTRVEKLD---EQLTCDNCNEKVSKEK 362
Query: 290 QLTIHKAPYVLAIHLKRFYAHDPNI-KIKKKVRFDSALNLKPFVSGSSDGDV--KYSLYG 346
QL + K P V HLKRF + + KI K V+ ++L+P++ + +V KY LY
Sbjct: 363 QLLLDKLPLVATFHLKRFKNNGLYMEKIYKHVKIPLEIDLQPYMRNIQENEVSTKYHLYA 422
Query: 347 VLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFYVRD 400
++ H G+S GHY YVR++ +W+ DD++V+ + E VL+Q +Y+LFY R+
Sbjct: 423 LVEHFGYSVAYGHYSSYVRSAPKIWHHFDDSKVTRIDEDMVLSQDSYILFYARE 476
>At5g46740 putative protein
Length = 732
Score = 160 bits (405), Expect = 3e-39
Identities = 111/322 (34%), Positives = 160/322 (49%), Gaps = 24/322 (7%)
Query: 90 IGAGLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAG--FCALCAIQNHVSCA 147
+GAGL+N GNTCF+ SVLQC T+T PL L+S + + C+ FC + A+++H+ A
Sbjct: 161 VGAGLYNSGNTCFIASVLQCFTHTVPLIDSLRSFMYGNPCNCGNEKFCVMQALRDHIELA 220
Query: 148 LQATGRIVSPQDMVGNLRCILMSCFFNEQS---RHITQFPKG*AGRCT*VYGELT*VNA* 204
L+++G ++ NL N Q + F C +L V++
Sbjct: 221 LRSSGYGINIDRFRDNLTYFSSDFMINHQEDAHEFLQSFLDKLERCCLDPKNQLGSVSSQ 280
Query: 205 MLPAFWSTE*ITWCFREKFGS*---DLWWSSSKSASNKFDPFLDLSLEILKADSLQKALA 261
L + FG L + S SN F+P L SLEI ++L KAL
Sbjct: 281 DLNIVDNV----------FGGGLMSTLCCCNCNSVSNTFEPSLGWSLEIEDVNTLWKALE 330
Query: 262 NFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNI-KIKKKV 320
+FT E L E Q C CK+KV KQL K P V HLKRF + KI +
Sbjct: 331 SFTCVEKL---EDQLTCDNCKEKVTKEKQLRFDKLPPVATFHLKRFTNDGVTMEKIFDHI 387
Query: 321 RFDSALNLKPFVSGSSDGDV--KYSLYGVLVHSGFSTHSGHYYCYVRTSNNMWYTLDDTR 378
F L+L PF+S + D +V +Y LY + H G GHY YVR++ W+ DD++
Sbjct: 388 EFPLELDLSPFMSSNHDPEVSTRYHLYAFVEHIGIRATFGHYSSYVRSAPETWHNFDDSK 447
Query: 379 VSHVGEQEVLNQQAYMLFYVRD 400
V+ + E+ VL++ AY+LFY R+
Sbjct: 448 VTRISEERVLSRPAYILFYARE 469
>At4g31670 unknown protein
Length = 631
Score = 158 bits (399), Expect = 1e-38
Identities = 109/316 (34%), Positives = 157/316 (49%), Gaps = 20/316 (6%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCALQATG 152
GL N GN+CF N +LQCL++T PL AYL HK C +C LC Q HV A Q+
Sbjct: 169 GLMNCGNSCFANVILQCLSWTRPLVAYLLEKGHKRECMRNDWCFLCEFQTHVERASQSRF 228
Query: 153 RIVSPQDMVGNLRCILMSCFFNEQS------RHITQFPKG*AGRCT*VYGELT*VNA*ML 206
SP +++ L I + + Q R+ + C +G V
Sbjct: 229 PF-SPMNIISRLTNIGGTLGYGRQEDAHEFMRYAIDMMQS---VCLDEFGGEKIVP---- 280
Query: 207 PAFWSTE*ITWCFREKFGS*DLWWSSSKSASNKFDPFLDLSLEIL-KADSLQKALANFTA 265
P T I + F S + + S++++ +DL +E+ A SL++ L FTA
Sbjct: 281 PRSQETTLIQYIFGGLLQS-QVQCTVCNHVSDQYENMMDLIVEMHGDAGSLEECLDQFTA 339
Query: 266 AELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSA 325
E L G + Y C RC V+A K+LTI +AP +L I LKR+ K+ K++ F
Sbjct: 340 EEWLHG-DNMYKCDRCSDYVKACKRLTIRRAPNILTIALKRYQGGRYG-KLNKRISFPET 397
Query: 326 LNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHS--GHYYCYVRTSNNMWYTLDDTRVSHVG 383
L+L P++S DG Y LY V+VH S GHY CY++ WY +DD+ + V
Sbjct: 398 LDLNPYMSEGGDGSDVYKLYAVIVHLDMLNASFFGHYICYIKDFCGNWYRIDDSEIESVE 457
Query: 384 EQEVLNQQAYMLFYVR 399
++VL+Q+AYML Y R
Sbjct: 458 LEDVLSQRAYMLLYSR 473
>At1g17110 ubiquitin-specific protease 15 (UBP15)
Length = 924
Score = 153 bits (386), Expect = 4e-37
Identities = 109/315 (34%), Positives = 161/315 (50%), Gaps = 18/315 (5%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCALQATG 152
GL N GN+C+ N+VLQ LT T+PL AYL H SC +C +C ++ HV L+ +G
Sbjct: 439 GLVNCGNSCYANAVLQSLTCTKPLVAYLLRRSHSRSCSGKDWCLMCELEQHVMM-LRESG 497
Query: 153 RIVSPQDMVGNLRCILMSCFFNEQSRH-----ITQFPKG*AGRCT*VYGELT*VNA*MLP 207
+S ++ ++R I +C + S+ + C G T V+ P
Sbjct: 498 GPLSASRILSHMRSI--NCQIGDGSQEDAHEFLRLLVASMQSICLERLGGETKVD----P 551
Query: 208 AFWSTE*ITWCFREKFGS*DLWWSSSKSASNKFDPFLDLSLEILK-ADSLQKALANFTAA 266
T + F + S + S +++ +DL+LEI +SLQ AL FT
Sbjct: 552 RLQETTLVQHMFGGRLRS-KVKCLRCDHESERYENIMDLTLEIYGWVESLQDALTQFTRP 610
Query: 267 ELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSAL 326
E LDG E Y C RC V+A K+L+IH+AP +L I LKRF KI K + F L
Sbjct: 611 EDLDG-ENMYRCSRCAGYVRARKELSIHEAPNILTIVLKRFQEGRYG-KINKCISFPEML 668
Query: 327 NLKPFVSGSSDGDVKYSLYGVLVHSGF--STHSGHYYCYVRTSNNMWYTLDDTRVSHVGE 384
++ PF++ + D Y LY V+VH ++ SGHY YV+ WY +DD+ + V
Sbjct: 669 DMIPFMTRTGDVPPLYMLYAVIVHLDTLNASFSGHYISYVKDLRGNWYRIDDSEIHPVPM 728
Query: 385 QEVLNQQAYMLFYVR 399
+V+++ AYMLFY+R
Sbjct: 729 TQVMSEGAYMLFYMR 743
>At5g65450 putative protein
Length = 731
Score = 147 bits (372), Expect = 2e-35
Identities = 123/366 (33%), Positives = 179/366 (48%), Gaps = 55/366 (15%)
Query: 64 PQPSKNSEFS----EFGLDPELSLGITFRRIGA---GLWNLGNTCFLNSVLQCLTYTEPL 116
PQ SK S+ S E EL + + R+ GL NLGN+C+ N+VLQCL +T PL
Sbjct: 294 PQSSKKSQPSSSIDEMSFSYELFVKLYCDRVELQPFGLVNLGNSCYANAVLQCLAFTRPL 353
Query: 117 AAYLQSGKHKSSCHIAGFCALCAIQNHVSCA---------------LQATGRIVSP---Q 158
+YL G H +C +C +C ++ + A LQ G+ + P +
Sbjct: 354 ISYLIRGLHSKTCRKKSWCFVCEFEHLILKARGGESPLSPIKILSKLQKIGKHLGPGKEE 413
Query: 159 DMVGNLRCILMSCFFNEQSRHITQFPKG*AGRCT*VYGELT*VNA*MLPAFWSTE*ITWC 218
D LRC + + QS + + P AG P T +
Sbjct: 414 DAHEFLRCAVDTM----QSVFLKEAPA--AG-----------------PFAEETTLVGLT 450
Query: 219 FREKFGS*DLWWSSSKSASNKFDPFLDLSLEIL-KADSLQKALANFTAAELLDGGEKQYH 277
F S + + S + + +DL++EI SL++ALA FTA E+LDG E +Y
Sbjct: 451 FGGYLHS-KIKCMACLHKSERPELMMDLTVEIDGDIGSLEEALAQFTAYEVLDG-ENRYF 508
Query: 278 CQRCKQKVQAVKQLTIHKAPYVLAIHLKRFYAHDPNIKIKKKVRFDSALNLKPFVSGSSD 337
C RCK +A K+L I + P +L + LKRF + D K+ K + F L++ P++S +
Sbjct: 509 CGRCKSYQKAKKKLMILEGPNILTVVLKRFQS-DNFGKLSKPIHFPELLDISPYMSDPNH 567
Query: 338 GDVK-YSLYGVLVH-SGFST-HSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYM 394
GD YSLY V+VH ST SGHY CY++T + W+ +DD+ V V + VL + AYM
Sbjct: 568 GDHPVYSLYAVVVHLDAMSTLFSGHYVCYIKTLDGDWFKIDDSNVFPVQLETVLLEGAYM 627
Query: 395 LFYVRD 400
L Y RD
Sbjct: 628 LLYARD 633
>At4g24560 putative protein
Length = 1008
Score = 118 bits (295), Expect = 2e-26
Identities = 96/293 (32%), Positives = 136/293 (45%), Gaps = 36/293 (12%)
Query: 241 DPFLDLSLEIL-KADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYV 299
+ +DL++EI +L AL FT E+LDG E +Y C CK +A K+L I + P V
Sbjct: 688 EKMMDLTVEIDGDISTLDDALRRFTRTEILDG-ENKYRCGSCKSYERAKKKLKITEPPNV 746
Query: 300 LAIHLKRFYAHDPNIKIKKKVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSGF--STHS 357
L I LKRF A K+ K +RF L+L P+VSG S+ Y LYGV+VH + S
Sbjct: 747 LTIALKRFQAGKFG-KLNKLIRFPETLDLAPYVSGGSEKSHDYKLYGVIVHLDVMNAAFS 805
Query: 358 GHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFYVRDKKSITPRKPVNIAKEEI 417
GHY CY+R + N WY DD+ V + +L + AYMLFY R + TP + K E
Sbjct: 806 GHYVCYIR-NQNKWYKADDSTVVTSDVERILTKGAYMLFYAR--CTPTPPRLAVCTKTEA 862
Query: 418 SKTNVIGSRYPSTPTNALKDYPNGHVENKFCGVPLTAETQKNLLNADPSRISDALIQQKN 477
S NK VPL +K+ ++ S S L
Sbjct: 863 S--------------------------NKKSRVPLPKANEKSTISRSVSTSSPELSSNTP 896
Query: 478 SGILAKSLMHNETPVSERTSKELTQMNSSDELPVAKSEL-ECLSSLDHSGKDN 529
G + ++ + +R K L + ++SD + S EC S D + D+
Sbjct: 897 GGGRSGNIQSFYSSF-QRLQKILEEDSASDSSSLFDSNSDECSCSTDSTSMDD 948
Score = 47.8 bits (112), Expect = 3e-05
Identities = 26/75 (34%), Positives = 36/75 (47%), Gaps = 1/75 (1%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCALQATG 152
GL N+GN+CF N V QCL +T PL Y H +C C C + V A +
Sbjct: 543 GLINVGNSCFANVVFQCLMFTPPLTTYFLQQFHSRACTKKEQCFTCGFEKLVVKAKEEKS 602
Query: 153 RIVSPQDMVGNLRCI 167
+ SP ++ L+ I
Sbjct: 603 PL-SPNGLLSQLQNI 616
>At4g39910 ubiquitin-specific protease (AtUBP3)
Length = 371
Score = 87.4 bits (215), Expect = 3e-17
Identities = 62/181 (34%), Positives = 87/181 (47%), Gaps = 19/181 (10%)
Query: 241 DPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVL 300
+ FLDLSL+I + S+ L NF++ E L +K + C +C +A K++ I K P++L
Sbjct: 197 ETFLDLSLDIEQNSSITSCLKNFSSTETLHAEDK-FFCDKCCSLQEAQKRMKIKKPPHIL 255
Query: 301 AIHLKRF-YAHDPNIKIKKKVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGH 359
IHLKRF Y K R L LK + DV+YSL+ V+VH G + GH
Sbjct: 256 VIHLKRFKYIEQLGRYKKLSYRVVFPLELKLSNTVEPYADVEYSLFAVVVHVGSGPNHGH 315
Query: 360 YYCYVRTSNNMWYTLDDTRVSHVGEQEVL------------NQQAYMLFYVRDKKSITPR 407
Y V+ S+N W DD V + E V Y+LFY +S+ P
Sbjct: 316 YVSLVK-SHNHWLFFDDENVEMIEESAVQTFFGSSQEYSSNTDHGYILFY----ESLGPT 370
Query: 408 K 408
K
Sbjct: 371 K 371
Score = 38.9 bits (89), Expect = 0.012
Identities = 24/77 (31%), Positives = 37/77 (47%), Gaps = 5/77 (6%)
Query: 93 GLWNLGNTCFLNSVLQ----CLTYTEPLAAYLQSGKHKSSCHIAGFCALCAIQNHVSCAL 148
G N GNTC+ NSVLQ C+ + E L Y S K + L + + +S
Sbjct: 24 GFENFGNTCYCNSVLQALYFCVPFREQLLEYYTSNKSVADAEENLMTCLADLFSQISSQK 83
Query: 149 QATGRIVSPQDMVGNLR 165
+ TG +++P+ V L+
Sbjct: 84 KKTG-VIAPKRFVQRLK 99
>At4g10590 unknown protein
Length = 910
Score = 87.0 bits (214), Expect = 4e-17
Identities = 59/148 (39%), Positives = 74/148 (49%), Gaps = 5/148 (3%)
Query: 255 SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRF-YAHDPN 313
SL L F A E L G + + C CK+ QA K+L + K P +L HLKRF Y+
Sbjct: 748 SLFSCLEAFLAEEPL-GPDDMWFCPSCKEHRQANKKLDLWKLPDILVFHLKRFTYSRYLK 806
Query: 314 IKIKKKVRFD-SALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRT-SNNMW 371
KI V F L+L +V +D Y LY V H G GHY Y + +N W
Sbjct: 807 NKIDTFVNFPVHDLDLSKYVKNKNDQSYLYELYAVSNHYG-GLGGGHYTAYAKLIDDNEW 865
Query: 372 YTLDDTRVSHVGEQEVLNQQAYMLFYVR 399
Y DD+ VS V E E+ N AY+LFY R
Sbjct: 866 YHFDDSHVSSVNESEIKNSAAYVLFYRR 893
Score = 46.2 bits (108), Expect = 8e-05
Identities = 19/28 (67%), Positives = 22/28 (77%)
Query: 92 AGLWNLGNTCFLNSVLQCLTYTEPLAAY 119
AGL NLGNTCF+NS LQCL +T P+ Y
Sbjct: 303 AGLSNLGNTCFMNSALQCLAHTPPIVEY 330
>At2g22310 putative ubiquitin carboxyl terminal hydrolase
Length = 365
Score = 85.5 bits (210), Expect = 1e-16
Identities = 54/148 (36%), Positives = 79/148 (52%), Gaps = 3/148 (2%)
Query: 241 DPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVL 300
+ FLDLSL+I + S+ L NF++ E L +K + C +C +A K++ I K P++L
Sbjct: 192 ETFLDLSLDIEQNSSITSCLKNFSSTETLHAEDK-FFCDKCCSLQEAQKRMKIKKPPHIL 250
Query: 301 AIHLKRFYAHDPNIKIKK-KVRFDSALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGH 359
IHLKRF + + KK R L LK + D++YSL+ V+VH G + GH
Sbjct: 251 VIHLKRFKYMEQLGRYKKLSYRVVFPLELKLSNTVDEYVDIEYSLFAVVVHVGSGPNHGH 310
Query: 360 YYCYVRTSNNMWYTLDDTRVSHVGEQEV 387
Y V+ S+N W DD V + E V
Sbjct: 311 YVSLVK-SHNHWLFFDDESVEIIEESAV 337
Score = 34.7 bits (78), Expect = 0.23
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 3/75 (4%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYL--QSGKHKSSCHIAGFCALCAIQNHVSCALQA 150
G N GNTC+ NSVLQ L + P L +K+ L + + +S +
Sbjct: 24 GFENFGNTCYCNSVLQALYFCAPFREQLLEHYANNKADAEENLLTCLADLFSQISSQKKK 83
Query: 151 TGRIVSPQDMVGNLR 165
TG +++P+ V L+
Sbjct: 84 TG-VIAPKRFVQRLK 97
>At5g22030 ubiquitin-specific protease 8 (UBP8)
Length = 901
Score = 84.7 bits (208), Expect = 2e-16
Identities = 52/148 (35%), Positives = 78/148 (52%), Gaps = 5/148 (3%)
Query: 255 SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRF-YAHDPN 313
SL K L F E L G + ++C CK+ QA+K+L + + P +L IHLKRF Y+
Sbjct: 753 SLFKCLEAFLTEEPL-GPDDMWYCPGCKEHRQAIKKLDLWRLPEILVIHLKRFSYSRFMK 811
Query: 314 IKIKKKVRFD-SALNLKPFVS-GSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNMW 371
K++ V F L+L ++S + +Y LY + H G S GHY YV + W
Sbjct: 812 NKLEAYVDFPLDNLDLSSYISYKNGQTTYRYMLYAISNHYG-SMGGGHYTAYVHHGGDRW 870
Query: 372 YTLDDTRVSHVGEQEVLNQQAYMLFYVR 399
Y DD+ V + ++++ AY+LFY R
Sbjct: 871 YDFDDSHVHQISQEKIKTSAAYVLFYKR 898
Score = 40.4 bits (93), Expect = 0.004
Identities = 20/38 (52%), Positives = 25/38 (65%), Gaps = 6/38 (15%)
Query: 82 SLGITFRRIGAGLWNLGNTCFLNSVLQCLTYTEPLAAY 119
+LG+T GL NLGNTCF+NS LQCL +T L +
Sbjct: 305 TLGLT------GLQNLGNTCFMNSSLQCLAHTPKLVDF 336
>At4g10570 putative protein
Length = 928
Score = 84.7 bits (208), Expect = 2e-16
Identities = 58/148 (39%), Positives = 73/148 (49%), Gaps = 5/148 (3%)
Query: 255 SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRF-YAHDPN 313
SL L F A E L G + + C CK+ QA K+L + K P +L HLKRF Y+
Sbjct: 754 SLFSCLEAFLAEEPL-GPDDMWFCPSCKEHRQANKKLDLWKLPDILVFHLKRFTYSRYLK 812
Query: 314 IKIKKKVRFD-SALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRT-SNNMW 371
KI V F L+L +V + Y LY V H G GHY Y + +N W
Sbjct: 813 NKIDTFVNFPVHDLDLSKYVKNKNGQSYLYELYAVSNHYG-GLGGGHYTAYAKLIDDNKW 871
Query: 372 YTLDDTRVSHVGEQEVLNQQAYMLFYVR 399
Y DD+ VS V E E+ N AY+LFY R
Sbjct: 872 YHFDDSHVSSVNESEIRNSAAYVLFYRR 899
Score = 46.2 bits (108), Expect = 8e-05
Identities = 19/28 (67%), Positives = 22/28 (77%)
Query: 92 AGLWNLGNTCFLNSVLQCLTYTEPLAAY 119
AGL NLGNTCF+NS LQCL +T P+ Y
Sbjct: 295 AGLSNLGNTCFMNSALQCLAHTPPIVEY 322
>At5g10790 ubiquitin specific protease -like protein
Length = 557
Score = 83.2 bits (204), Expect = 6e-16
Identities = 58/224 (25%), Positives = 100/224 (43%), Gaps = 53/224 (23%)
Query: 227 DLWWSSSKSASNKFDPFLDLSLEILKADSLQKA-----------------------LANF 263
D+ ++ S S +DPF+D+SL + + A L F
Sbjct: 311 DVTCTTCGSTSTTYDPFIDISLTLDSMNGFSPADCRKNRYSGGPSVNAIMPTLSGCLDFF 370
Query: 264 TAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRF---YAHDPNIKIKKKV 320
T +E L G +++ +CQ C +K ++ KQ++I + P +L +H+KRF + KI +
Sbjct: 371 TRSEKL-GPDQKLNCQSCGEKRESSKQMSIRRLPLLLCLHVKRFEHSLTRKTSRKIDSYL 429
Query: 321 RFDSALNLKPFV------------------------SGSSDGDVKYSLYGVLVHSGFSTH 356
++ LN+ P++ S SS ++ ++ V+ H G
Sbjct: 430 QYPFRLNMSPYLSSSIIGKRFGNRIFAFDGEGEYDSSSSSSPSAEFEIFAVVTHKGM-LE 488
Query: 357 SGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFYVRD 400
SGHY Y+R +WY DD ++ V E+ V + YMLFY ++
Sbjct: 489 SGHYVTYLRL-KGLWYRCDDAWINEVEEEVVRGCECYMLFYAQE 531
Score = 41.6 bits (96), Expect = 0.002
Identities = 18/33 (54%), Positives = 24/33 (72%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQSGKH 125
GL NLG+TCF+N+VLQ L + PL + SG+H
Sbjct: 178 GLNNLGSTCFMNAVLQALVHAPPLRNFWLSGQH 210
>At1g32850 hypothetical protein
Length = 887
Score = 83.2 bits (204), Expect = 6e-16
Identities = 57/150 (38%), Positives = 73/150 (48%), Gaps = 5/150 (3%)
Query: 255 SLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRF-YAHDPN 313
SL L F A E L G + ++C CK+ QA K+L + K P +L HLKRF Y+
Sbjct: 729 SLFSCLEAFLAEEPL-GPDDMWYCPGCKEHRQANKKLDLWKLPDILVFHLKRFTYSRYFK 787
Query: 314 IKIKKKVRFD-SALNLKPFVSGSSDGDVKYSLYGVLVHSGFSTHSGHYYCYVRTSNNM-W 371
KI V F L+L +V Y LY + H G GHY Y + + W
Sbjct: 788 NKIDTLVNFHIHDLDLSKYVKNEDGQSYLYELYAISNHYG-GLGGGHYTAYAKLMDETKW 846
Query: 372 YTLDDTRVSHVGEQEVLNQQAYMLFYVRDK 401
Y DD+RVS V E E+ AY+LFY R K
Sbjct: 847 YNFDDSRVSAVNESEIKTSAAYVLFYQRVK 876
Score = 46.2 bits (108), Expect = 8e-05
Identities = 31/89 (34%), Positives = 42/89 (46%), Gaps = 7/89 (7%)
Query: 37 NDFNIETLNPTTSEHRQLVSANIQSSQPQPSKNSEFSEFGLDP------ELSLGITFRRI 90
N+ + L P S+ +V S S +FS FG + + G +R
Sbjct: 236 NELAMVPLEPMRSDAMDIVRGGGTLSNGH-SNGFKFSFFGRNTFKDDVSSRTFGKGEKRG 294
Query: 91 GAGLWNLGNTCFLNSVLQCLTYTEPLAAY 119
GL NLGNTCF+NS LQCL +T P+ Y
Sbjct: 295 LGGLQNLGNTCFMNSTLQCLAHTPPIVEY 323
>At4g30890 ubiquitin-specific protease 24 (UBP24)
Length = 551
Score = 79.7 bits (195), Expect = 6e-15
Identities = 54/166 (32%), Positives = 81/166 (48%), Gaps = 6/166 (3%)
Query: 236 ASNKFDPFLDLSLEILK--ADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTI 293
AS P+L L L+I ++ AL F+A E L+G + V A K + I
Sbjct: 385 ASATVQPYLLLHLDIHPDGVQGIEDALHLFSAQEDLEGYRASVTGKT--GVVSASKSIKI 442
Query: 294 HKAPYVLAIHLKRF-YAHDPNIKIKKKVRFDSALNL-KPFVSGSSDGDVKYSLYGVLVHS 351
K ++ +HL RF Y + K++K V+F LNL + + S+ ++Y L + H
Sbjct: 443 QKLSKIMILHLMRFSYGSQGSTKLRKGVKFPLELNLNRSHLVSLSNESLRYELVATITHH 502
Query: 352 GFSTHSGHYYCYVRTSNNMWYTLDDTRVSHVGEQEVLNQQAYMLFY 397
G+ GHY R N W DD V+ +G + VL+ QAY+LFY
Sbjct: 503 GWDPSKGHYTTDARRKNGQWLRFDDASVTPIGTKLVLHDQAYVLFY 548
Score = 32.3 bits (72), Expect = 1.1
Identities = 15/29 (51%), Positives = 16/29 (54%)
Query: 93 GLWNLGNTCFLNSVLQCLTYTEPLAAYLQ 121
GL N GN CFLN+ LQ L P LQ
Sbjct: 198 GLINAGNLCFLNATLQALLSCSPFVQLLQ 226
>At3g49600 putative protein
Length = 1672
Score = 78.6 bits (192), Expect = 1e-14
Identities = 47/155 (30%), Positives = 78/155 (50%), Gaps = 7/155 (4%)
Query: 236 ASNKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHK 295
AS+K + F L L + SL +L ++ + E L+G + QY C C +V A + + +
Sbjct: 839 ASSKMEDFYALELNVKGLKSLDASLNDYLSLEQLNG-DNQYFCGSCNARVDATRCIKLRT 897
Query: 296 APYVLAIHLKRFYAHDPNIKIKKKVR----FDSALNLKPFVSGSSDGDVKYSLYGVLVHS 351
P V+ LKR P KKK+ F L++ ++ SS + Y L VL+H
Sbjct: 898 LPPVITFQLKRCIFL-PKTTAKKKITSSFSFPQVLDMGSRLAESSQNKLTYDLSAVLIHK 956
Query: 352 GFSTHSGHYYCYVR-TSNNMWYTLDDTRVSHVGEQ 385
G + +SGHY +++ +W+ DD VS +G++
Sbjct: 957 GSAVNSGHYVAHIKDEKTGLWWEFDDEHVSELGKR 991
Score = 35.0 bits (79), Expect = 0.17
Identities = 23/58 (39%), Positives = 27/58 (45%), Gaps = 14/58 (24%)
Query: 53 QLVSANIQSSQPQPSKNSEFSEFGLDPELSLGITFRRIGAGLWNLGNTCFLNSVLQCL 110
Q S IQS P P+ + SE AGL NLG TC+ NS+LQCL
Sbjct: 686 QKTSEIIQSLGPDPTLDRRDSE--------------STPAGLTNLGATCYANSILQCL 729
>At2g40930 ubiquitin-specific protease (UBP5)
Length = 924
Score = 77.8 bits (190), Expect = 2e-14
Identities = 45/131 (34%), Positives = 74/131 (56%), Gaps = 5/131 (3%)
Query: 273 EKQYHCQRCKQKVQAVKQLTIHKAPYVLAIHLKRF-YAHDPNIKIKKKVRFD-SALNLKP 330
++ + C +C ++ QA K+L + + P VL IHLKRF Y+ K++ V F L+L
Sbjct: 786 DEMWFCPQCNERRQASKKLDLWRLPEVLVIHLKRFSYSRSMKHKLETFVNFPIHDLDLTK 845
Query: 331 FVSGSSDGDVK-YSLYGVLVHSGFSTHSGHYYCYVRT-SNNMWYTLDDTRVSHVGEQEVL 388
+V+ + + Y LY + H G SGHY +++ ++ WY DD+ +SH+ E +V
Sbjct: 846 YVANKNLSQPQLYELYALTNHYG-GMGSGHYTAHIKLLDDSRWYNFDDSHISHINEDDVK 904
Query: 389 NQQAYMLFYVR 399
+ AY+LFY R
Sbjct: 905 SGAAYVLFYRR 915
Score = 48.5 bits (114), Expect = 2e-05
Identities = 24/48 (50%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Query: 82 SLGITFRRIGAGLW---NLGNTCFLNSVLQCLTYTEPLAAYLQSGKHK 126
S G+T R AGL NLGNTCF+NS +QCL +T A+Y Q H+
Sbjct: 304 STGVTTRGSTAGLTGLLNLGNTCFMNSAIQCLVHTPEFASYFQEDYHQ 351
>At3g11910 ubiquitin carboxyl-terminal hydrolase like protein
Length = 1089
Score = 74.3 bits (181), Expect = 3e-13
Identities = 65/230 (28%), Positives = 98/230 (42%), Gaps = 50/230 (21%)
Query: 237 SNKFDPFLDLSLEILKADSLQKALANFTAAELLDGGEKQYHCQRCKQKVQAVKQLTIHKA 296
S + + F DL L++ + + + E L+G K YH + + A K +
Sbjct: 326 STRKESFYDLQLDVKGCKDVYASFDKYVEVERLEGDNK-YHAEGHDLQ-DAKKGVLFIDF 383
Query: 297 PYVLAIHLKRF---YAHDPNIKIKKKVRFDSALNLKP----FVSGSSDGDVK--YSLYGV 347
P VL + LKRF + D +KI + F L+L ++S +D V+ Y+L+ V
Sbjct: 384 PPVLQLQLKRFEYDFMRDTMVKINDRYEFPLQLDLDREDGRYLSPDADKSVRNLYTLHSV 443
Query: 348 LVHSGFSTHSGHYYCYVR-TSNNMWYTLDDTRVSHVGEQEVLNQQ--------------- 391
LVHSG H GHYY ++R T ++ WY DD RV+ + L +Q
Sbjct: 444 LVHSG-GVHGGHYYAFIRPTLSDQWYKFDDERVTKEDVKRALEEQYGGEEELPQNNPGFN 502
Query: 392 -----------AYMLFYVR-----------DKKSITPRKPVNIAKEEISK 419
AYML Y+R D+K I V + KE+ K
Sbjct: 503 NPPFKFTKYSNAYMLVYIRESDKDKIICNVDEKDIAEHLRVRLKKEQEEK 552
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,405,331
Number of Sequences: 26719
Number of extensions: 797510
Number of successful extensions: 2385
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 2261
Number of HSP's gapped (non-prelim): 90
length of query: 835
length of database: 11,318,596
effective HSP length: 108
effective length of query: 727
effective length of database: 8,432,944
effective search space: 6130750288
effective search space used: 6130750288
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC122160.3


