

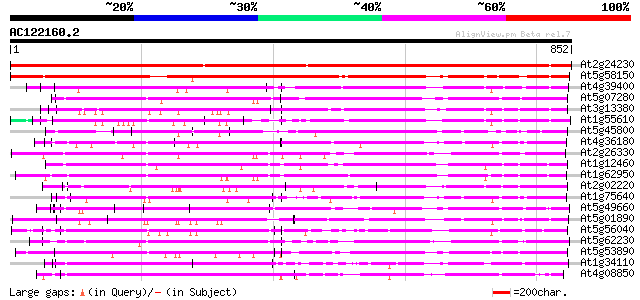
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122160.2 - phase: 0
(852 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g24230 putative receptor-like protein kinase 1181 0.0
At5g58150 receptor-like protein kinase 740 0.0
At4g39400 brassinosteroid insensitive 1 gene (BRI1) 369 e-102
At5g07280 receptor-like protein kinase-like protein 361 e-100
At3g13380 brassinosteroid receptor kinase, putative 347 2e-95
At1g55610 receptor kinase, putative 337 1e-92
At5g45800 receptor kinase-like protein 316 4e-86
At4g36180 receptor protein kinase like protein 310 3e-84
At2g26330 putative receptor-like protein kinase, ERECTA 303 3e-82
At1g12460 unknown protein 300 2e-81
At1g62950 protein kinase like protein 300 3e-81
At2g02220 putative receptor-like protein kinase 298 7e-81
At1g75640 receptor-like protein kinase, putative 298 9e-81
At5g49660 receptor protein kinase 297 1e-80
At5g01890 unknown protein 292 5e-79
At5g56040 receptor protein kinase-like protein 288 1e-77
At5g62230 unknown protein 287 2e-77
At5g53890 receptor protein kinase-like protein 286 3e-77
At1g34110 putative protein 281 8e-76
At4g08850 receptor protein kinase like protein 273 4e-73
>At2g24230 putative receptor-like protein kinase
Length = 853
Score = 1181 bits (3054), Expect = 0.0
Identities = 600/855 (70%), Positives = 701/855 (81%), Gaps = 8/855 (0%)
Query: 1 MGLGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVY 60
+GLG +G L L+ K Q+PNTD F+VSEF K+MGL SSS+ YNFS+ CSW+G++
Sbjct: 3 LGLGFWGYALFLSLFLKQSHCQEPNTDGFFVSEFYKQMGL-SSSQAYNFSAPFCSWQGLF 61
Query: 61 CDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLN 120
CDS EHV+ L SG+ L+G IPD TIGKL+KL SLDLSNNKI+ LPSDFWSL +LK+LN
Sbjct: 62 CDSKNEHVIMLIASGMSLSGQIPDNTIGKLSKLQSLDLSNNKISALPSDFWSLNTLKNLN 121
Query: 121 LSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPS 180
LS N ISGS ++N+GNFG LE D+S N+FS IPEA+ SLVSL+VLKLDHN F SIP
Sbjct: 122 LSFNKISGSFSSNVGNFGQLELLDISYNNFSGAIPEAVDSLVSLRVLKLDHNGFQMSIPR 181
Query: 181 GILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLN 240
G+L CQSLVSIDLSSNQL G+LP GFG AFPKL TL+LA N I+G ++F+ +KSI LN
Sbjct: 182 GLLGCQSLVSIDLSSNQLEGSLPDGFGSAFPKLETLSLAGNKIHGRDTDFADMKSISFLN 241
Query: 241 ISGNSFQGSIIEVFVLKLEALDLSRNQFQGHIS-QVKYNWSHLVYLDLSENQLSGEIFQN 299
ISGN F GS+ VF LE DLS+N+FQGHIS QV NW LVYLDLSEN+LSG + +N
Sbjct: 242 ISGNQFDGSVTGVFKETLEVADLSKNRFQGHISSQVDSNWFSLVYLDLSENELSG-VIKN 300
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
L LKHL+LA NRF+R FP+IEML GLEYLNLS T+L GHIP EIS L +L+ LD+
Sbjct: 301 LTLLKKLKHLNLAWNRFNRGMFPRIEMLSGLEYLNLSNTNLSGHIPREISKLSDLSTLDV 360
Query: 360 SMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIK 419
S NHL G IP+L K+L ID S NNL+G +P IL+ LP M+++NFS+NNLT C+ +
Sbjct: 361 SGNHLAGHIPILSIKNLVAIDVSRNNLTGEIPMSILEKLPWMERFNFSFNNLTFCSGKFS 420
Query: 420 PDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAG-ILFLAFGC 478
+ + SFFGS NSCPIAANP+ FK++R V G+KLAL +TLS + L G ++F+AFGC
Sbjct: 421 AETLNRSFFGSTNSCPIAANPALFKRKRSVTG-GLKLALAVTLSTMCLLIGALIFVAFGC 479
Query: 479 RRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADL 538
RRK K E K S +EEQ+ISGPFSFQTDSTTWVADVKQA +VPVVIFEKPLLNITF+DL
Sbjct: 480 RRKTKSGEAKDLSVKEEQSISGPFSFQTDSTTWVADVKQANAVPVVIFEKPLLNITFSDL 539
Query: 539 LSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKH 598
LSATSNFDR TLLA+GKFGPVYRGFLPG IHVAVKVLV GSTL+D+EAARELEFLGRIKH
Sbjct: 540 LSATSNFDRDTLLADGKFGPVYRGFLPGGIHVAVKVLVHGSTLSDQEAARELEFLGRIKH 599
Query: 599 PNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTW-EEADNGIQN 657
PNLVPLTGYC+AGDQRIAIY+YMENGNLQNLL+DLP GVQ+TDDW+TDTW EE DNG QN
Sbjct: 600 PNLVPLTGYCIAGDQRIAIYEYMENGNLQNLLHDLPFGVQTTDDWTTDTWEEETDNGTQN 659
Query: 658 VGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGL 717
+G+EG + TWRFRHKIALGTARALAFLHHGCSPPIIHR VKASSVYLD + EPRLSDFGL
Sbjct: 660 IGTEGPVATWRFRHKIALGTARALAFLHHGCSPPIIHRDVKASSVYLDQNWEPRLSDFGL 719
Query: 718 AKIFGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYT 777
AK+FG+GLD+EI GSPGY+PPEF QPE E PTPKSDVYCFGVVLFEL+TGKKP+ DDY
Sbjct: 720 AKVFGNGLDDEIIHGSPGYLPPEFLQPEHELPTPKSDVYCFGVVLFELMTGKKPIEDDYL 779
Query: 778 DDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTM 837
D+K+ T LVSWVR LVRKNQ S+AIDPKI +TGS+EQ+EEALK +GYLCTADLP KRP+M
Sbjct: 780 DEKD-TNLVSWVRSLVRKNQASKAIDPKIQETGSEEQMEEALK-IGYLCTADLPSKRPSM 837
Query: 838 QQIVGLLKDIEPTTS 852
QQ+VGLLKDIEP ++
Sbjct: 838 QQVVGLLKDIEPKSN 852
>At5g58150 receptor-like protein kinase
Length = 785
Score = 740 bits (1910), Expect = 0.0
Identities = 417/858 (48%), Positives = 548/858 (63%), Gaps = 84/858 (9%)
Query: 1 MGLGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVY 60
M L ++GS+L +F KHL S PNTD +++S F M L +S + + FSS +CSW GV
Sbjct: 1 MRLSLWGSLLFFSFFVKHLTSLDPNTDAYHLSSFFSAMRLPNSPQAHTFSS-LCSWPGVV 59
Query: 61 CDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLN 120
+ E+V+ ++ SG+ L+G IPD TIGK++KL +LDLS NKIT+LPSD WSL+ L+SLN
Sbjct: 60 VCDSSENVLHISASGLDLSGSIPDNTIGKMSKLQTLDLSGNKITSLPSDLWSLSLLESLN 119
Query: 121 LSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPS 180
LSSN IS L +NIGNF L DLS NS S +IP A+S+LV+L LKL +N F +P
Sbjct: 120 LSSNRISEPLPSNIGNFMSLHTLDLSFNSISGKIPAAISNLVNLTTLKLHNNDFQFGVPP 179
Query: 181 GILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLN 240
++ C+SL+SIDLSSN+L+ +LP GFG AFP L+ SLN
Sbjct: 180 ELVHCRSLLSIDLSSNRLNESLPVGFGSAFPLLK-----------------------SLN 216
Query: 241 ISGNSFQGSIIEVFVLKLEALDLSRNQFQGHISQV----KYNWSHLVYLDLSENQLSGEI 296
+S N FQGS+I V +E +DLS N+F GHI Q+ K+NWS L++LDLS+N G I
Sbjct: 217 LSRNLFQGSLIGVLHENVETVDLSENRFDGHILQLIPGHKHNWSSLIHLDLSDNSFVGHI 276
Query: 297 FQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNA 356
F L+++ L HL+LACNRF Q+FP+I L L YLNLS+T+L IP EIS L +L
Sbjct: 277 FNGLSSAHKLGHLNLACNRFRAQEFPEIGKLSALHYLNLSRTNLTNIIPREISRLSHLKV 336
Query: 357 LDLSMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCAS 416
LDLS N+L G +P+L K+++V+D S N L G +P +L+ L M+++NFS+NNLT C
Sbjct: 337 LDLSSNNLTGHVPMLSVKNIEVLDLSLNKLDGDIPRPLLEKLAMMQRFNFSFNNLTFCNP 396
Query: 417 EIKPDIMKTSFFGSVNSCPIAANPSFFK-KRRDVGHRGMKLALVLTLSLIFALAGILFL- 474
+ ++ SF N+CP AA P K K+ + + G+K+ L L +S+ F L G+L +
Sbjct: 397 NFSQETIQRSFINIRNNCPFAAKPIITKGKKVNKKNTGLKIGLGLAISMAFLLIGLLLIL 456
Query: 475 -AFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNI 533
A RRK++ W K E N Q DSTT D+KQAT +PVV+ +KPL+ +
Sbjct: 457 VALRVRRKSRTWATKLAINNTEPNSPD----QHDSTT---DIKQATQIPVVMIDKPLMKM 509
Query: 534 TFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFL 593
T ADL +AT NFDRGT+L EGK GP Y LPG A+KV+ G+TLTD E + E L
Sbjct: 510 TLADLKAATFNFDRGTMLWEGKSGPTYGAVLPGGFRAALKVIPSGTTLTDTEVSIAFERL 569
Query: 594 GRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADN 653
RI HPNL PL GYC+A +QRIAIY+ ++ NLQ+LL++ N
Sbjct: 570 ARINHPNLFPLCGYCIATEQRIAIYEDLDMVNLQSLLHN--------------------N 609
Query: 654 GIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLS 713
G + WR RHKIALGTARALAFLHHGC PP++H VKA+++ LD EPRL+
Sbjct: 610 GDDSA-------PWRLRHKIALGTARALAFLHHGCIPPMVHGEVKAATILLDSSQEPRLA 662
Query: 714 DFGLAKIFGSGLDEEI--ARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKP 771
DFGL K+ LDE+ + GY PPE Q SPT +SDVY FGVVL EL++GKKP
Sbjct: 663 DFGLVKL----LDEQFPGSESLDGYTPPE--QERNASPTLESDVYSFGVVLLELVSGKKP 716
Query: 772 VGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLP 831
GD LV+WVRGLVR+ Q RAIDP + +T +++I EA+K +GYLCTADLP
Sbjct: 717 EGD----------LVNWVRGLVRQGQGLRAIDPTMQETVPEDEIAEAVK-IGYLCTADLP 765
Query: 832 FKRPTMQQIVGLLKDIEP 849
+KRPTMQQ+VGLLKDI P
Sbjct: 766 WKRPTMQQVVGLLKDISP 783
>At4g39400 brassinosteroid insensitive 1 gene (BRI1)
Length = 1196
Score = 369 bits (946), Expect = e-102
Identities = 270/822 (32%), Positives = 412/822 (49%), Gaps = 95/822 (11%)
Query: 68 VVELNLSGIGLTGPI-PDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNH 125
++ L+LS +GPI P+ N L L L NN T +P + + L SL+LS N+
Sbjct: 392 LLTLDLSSNNFSGPILPNLCQNPKNTLQELYLQNNGFTGKIPPTLSNCSELVSLHLSFNY 451
Query: 126 ISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKC 185
+SG++ +++G+ L + L N EIP+ L + +L+ L LD N IPSG+ C
Sbjct: 452 LSGTIPSSLGSLSKLRDLKLWLNMLEGEIPQELMYVKTLETLILDFNDLTGEIPSGLSNC 511
Query: 186 QSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGN 244
+L I LS+N+L+G +P G L L L+ N+ G + + +S++ L+++ N
Sbjct: 512 TNLNWISLSNNRLTGEIPKWIG-RLENLAILKLSNNSFSGNIPAELGDCRSLIWLDLNTN 570
Query: 245 SFQGSIIEV-----------FVLKLEALDLSRN-------------QFQGHISQVKYNWS 280
F G+I F+ + + + +FQG S+ S
Sbjct: 571 LFNGTIPAAMFKQSGKIAANFIAGKRYVYIKNDGMKKECHGAGNLLEFQGIRSEQLNRLS 630
Query: 281 HLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSL 340
+++ G +N+ ++ L ++ N S +I + L LNL +
Sbjct: 631 TRNPCNITSRVYGGHTSPTFDNNGSMMFLDMSYNMLSGYIPKEIGSMPYLFILNLGHNDI 690
Query: 341 VGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNK--HLQVIDFSHNNLSGPVPSF-ILKS 397
G IPDE+ L LN LDLS N LDG+IP + L ID S+NNLSGP+P ++
Sbjct: 691 SGSIPDEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGPIPEMGQFET 750
Query: 398 LPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLA 457
P K NN LC + C + + +R G R LA
Sbjct: 751 FPPAK----FLNNPGLCGYPLP-------------RCDPSNADGYAHHQRSHGRRPASLA 793
Query: 458 LVLTLSLIFALA---GILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTW-VA 513
+ + L+F+ G++ + R++ + E + Y E SG + ++T W +
Sbjct: 794 GSVAMGLLFSFVCIFGLILVGREMRKRRRKKEAELEMYAEGHGNSGDRT--ANNTNWKLT 851
Query: 514 DVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVK 573
VK+A S+ + FEKPL +TFADLL AT+ F +L+ G FG VY+ L VA+K
Sbjct: 852 GVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIK 911
Query: 574 VLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDL 633
L+ S D E E+E +G+IKH NLVPL GYC GD+R+ +Y++M+ G+L+++L+D
Sbjct: 912 KLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHD- 970
Query: 634 PLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPII 693
G+ W R KIA+G+AR LAFLHH CSP II
Sbjct: 971 ------------------------PKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHII 1006
Query: 694 HRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIA----RGSPGYVPPEFSQPEFESP 749
HR +K+S+V LD +LE R+SDFG+A++ S +D ++ G+PGYVPPE+ Q F
Sbjct: 1007 HRDMKSSNVLLDENLEARVSDFGMARLM-SAMDTHLSVSTLAGTPGYVPPEYYQ-SFRCS 1064
Query: 750 TPKSDVYCFGVVLFELLTGKKPV-GDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKIC- 807
T K DVY +GVVL ELLTGK+P D+ D+ LV WV+ K + S DP++
Sbjct: 1065 T-KGDVYSYGVVLLELLTGKRPTDSPDFGDN----NLVGWVKQHA-KLRISDVFDPELMK 1118
Query: 808 -DTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
D + ++ + LK V C D ++RPTM Q++ + K+I+
Sbjct: 1119 EDPALEIELLQHLK-VAVACLDDRAWRRPTMVQVMAMFKEIQ 1159
Score = 115 bits (287), Expect = 1e-25
Identities = 124/406 (30%), Positives = 191/406 (46%), Gaps = 33/406 (8%)
Query: 26 TDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDT 85
T F+ S F + S +Y + S+K V D N N + G
Sbjct: 12 TTLFFFSFFSLSFQASPSQSLYREIHQLISFKDVLPDKNLLPDWSSNKNPCTFDG----- 66
Query: 86 TIGKLNKLHSLDLSNNKI----TTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLE 141
+ +K+ S+DLS+ + + + S SLT L+SL LS++HI+GS++ L
Sbjct: 67 VTCRDDKVTSIDLSSKPLNVGFSAVSSSLLSLTGLESLFLSNSHINGSVSG-FKCSASLT 125
Query: 142 NFDLSKNSFSDEIPE--ALSSLVSLKVLKLDHNMFVRSIP---SGILKCQSLVSIDLSSN 196
+ DLS+NS S + +L S LK L + N P SG LK SL +DLS+N
Sbjct: 126 SLDLSRNSLSGPVTTLTSLGSCSGLKFLNVSSNTL--DFPGKVSGGLKLNSLEVLDLSAN 183
Query: 197 QLSGTLPHGF--GDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVF 254
+SG G+ D +L+ L ++ N I G V + SR ++ L++S N+F I +
Sbjct: 184 SISGANVVGWVLSDGCGELKHLAISGNKISGDV-DVSRCVNLEFLDVSSNNFSTGIPFLG 242
Query: 255 VLK-LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLAC 313
L+ LD+S N+ G S+ + L L++S NQ G I S L++LSLA
Sbjct: 243 DCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPPLPLKS--LQYLSLAE 300
Query: 314 NRFSRQKFPKIEMLLG----LEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP 369
N+F+ + P + L G L L+LS G +P L +L LS N+ G++P
Sbjct: 301 NKFTGE-IP--DFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLESLALSSNNFSGELP 357
Query: 370 ---LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
LLK + L+V+D S N SG +P + + + S NN +
Sbjct: 358 MDTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNNFS 403
Score = 112 bits (281), Expect = 7e-25
Identities = 107/358 (29%), Positives = 171/358 (46%), Gaps = 20/358 (5%)
Query: 47 YNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTL 106
++ + + C++ GV C +K ++L+ + + +++ L L SL LSN+ I
Sbjct: 55 WSSNKNPCTFDGVTCRDDKVTSIDLSSKPLNVGFSAVSSSLLSLTGLESLFLSNSHINGS 114
Query: 107 PSDFWSLTSLKSLNLSSNHISGSLT--NNIGNFGLLENFDLSKNS--FSDEIPEALSSLV 162
S F SL SL+LS N +SG +T ++G+ L+ ++S N+ F ++ L L
Sbjct: 115 VSGFKCSASLTSLDLSRNSLSGPVTTLTSLGSCSGLKFLNVSSNTLDFPGKVSGGL-KLN 173
Query: 163 SLKVLKLDHNMFV-RSIPSGILK--CQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLA 219
SL+VL L N ++ +L C L + +S N++SG + L L+++
Sbjct: 174 SLEVLDLSANSISGANVVGWVLSDGCGELKHLAISGNKISGDVD---VSRCVNLEFLDVS 230
Query: 220 ENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKY 277
NN G+ ++ L+ISGN G +L+ L++S NQF G I +
Sbjct: 231 SNNFSTGIPFLGDCSALQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPPLPL 290
Query: 278 NWSHLVYLDLSENQLSGEIFQNLNNSMN-LKHLSLACNRFSRQKFPKIEMLLGLEYLNLS 336
L YL L+EN+ +GEI L+ + + L L L+ N F P LE L LS
Sbjct: 291 --KSLQYLSLAENKFTGEIPDFLSGACDTLTGLDLSGNHFYGAVPPFFGSCSLLESLALS 348
Query: 337 KTSLVGHIP-DEISHLGNLNALDLSMNHLDGKIP-LLKN--KHLQVIDFSHNNLSGPV 390
+ G +P D + + L LDLS N G++P L N L +D S NN SGP+
Sbjct: 349 SNNFSGELPMDTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNNFSGPI 406
>At5g07280 receptor-like protein kinase-like protein
Length = 1192
Score = 361 bits (927), Expect = e-100
Identities = 270/829 (32%), Positives = 395/829 (47%), Gaps = 92/829 (11%)
Query: 70 ELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISG 128
EL L+ + G IP+ + KL L +LDL +N T +P W T+L S N + G
Sbjct: 405 ELLLTNNQINGSIPED-LWKL-PLMALDLDSNNFTGEIPKSLWKSTNLMEFTASYNRLEG 462
Query: 129 SLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSL 188
L IGN L+ LS N + EIP + L SL VL L+ NMF IP + C SL
Sbjct: 463 YLPAEIGNAASLKRLVLSDNQLTGEIPREIGKLTSLSVLNLNANMFQGKIPVELGDCTSL 522
Query: 189 VSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSN-------------FSRLKS 235
++DL SN L G +P A +L+ L L+ NN+ G + + S L+
Sbjct: 523 TTLDLGSNNLQGQIPDKI-TALAQLQCLVLSYNNLSGSIPSKPSAYFHQIEMPDLSFLQH 581
Query: 236 IVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLS 293
++S N G I E L L + LS N G I ++L LDLS N L+
Sbjct: 582 HGIFDLSYNRLSGPIPEELGECLVLVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALT 641
Query: 294 GEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGN 353
G I + + NS+ L+ L+LA N+ + +L L LNL+K L G +P + +L
Sbjct: 642 GSIPKEMGNSLKLQGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLDGPVPASLGNLKE 701
Query: 354 LNALDLSMNHLDGKIP-------------LLKNK-------------HLQVIDFSHNNLS 387
L +DLS N+L G++ + +NK L+ +D S N LS
Sbjct: 702 LTHMDLSFNNLSGELSSELSTMEKLVGLYIEQNKFTGEIPSELGNLTQLEYLDVSENLLS 761
Query: 388 GPVPSFILKSLPKMKKYNFSYNNLT--LCASEIKPDIMKTSFFGSVNSCPIAANPSFFKK 445
G +P+ I LP ++ N + NNL + + + D K G+ C
Sbjct: 762 GEIPTKIC-GLPNLEFLNLAKNNLRGEVPSDGVCQDPSKALLSGNKELCGRVVGS----- 815
Query: 446 RRDVGHRGMKLAL---VLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPF 502
D G KL + L L F + +F+ F RR VKQ E S
Sbjct: 816 --DCKIEGTKLRSAWGIAGLMLGFTIIVFVFV-FSLRRWAMTKRVKQRDDPERMEESRLK 872
Query: 503 SFQTDSTTWVADVK--QATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVY 560
F + +++ + + S+ + +FE+PLL + D++ AT +F + ++ +G FG VY
Sbjct: 873 GFVDQNLYFLSGSRSREPLSINIAMFEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVY 932
Query: 561 RGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDY 620
+ LPG VAVK L T + E E+E LG++KHPNLV L GYC ++++ +Y+Y
Sbjct: 933 KACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEY 992
Query: 621 MENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARA 680
M NG+L D W G+ V W R KIA+G AR
Sbjct: 993 MVNGSL-------------------DHWLRNQTGMLEV------LDWSKRLKIAVGAARG 1027
Query: 681 LAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGS--GLDEEIARGSPGYVP 738
LAFLHHG P IIHR +KAS++ LD D EP+++DFGLA++ + + G+ GY+P
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIAGTFGYIP 1087
Query: 739 PEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQT 798
PE+ Q T K DVY FGV+L EL+TGK+P G D+ + E LV W + + +
Sbjct: 1088 PEYGQS--ARATTKGDVYSFGVILLELVTGKEPTGPDF-KESEGGNLVGWAIQKINQGKA 1144
Query: 799 SRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
IDP + + + L ++ LC A+ P KRP M ++ LK+I
Sbjct: 1145 VDVIDPLLVSVAL-KNSQLRLLQIAMLCLAETPAKRPNMLDVLKALKEI 1192
Score = 152 bits (384), Expect = 8e-37
Identities = 118/353 (33%), Positives = 173/353 (48%), Gaps = 8/353 (2%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTL--PSDFWSLTSLKSLNL 121
N +H+ L+LSG LTG +P + +L +L LDLS+N + PS F SL +L SL++
Sbjct: 111 NLKHLQTLDLSGNSLTGLLP-RLLSELPQLLYLDLSDNHFSGSLPPSFFISLPALSSLDV 169
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSG 181
S+N +SG + IG L N + NSFS +IP + ++ LK F +P
Sbjct: 170 SNNSLSGEIPPEIGKLSNLSNLYMGLNSFSGQIPSEIGNISLLKNFAAPSCFFNGPLPKE 229
Query: 182 ILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLN 240
I K + L +DLS N L ++P FG+ L LNL + G + KS+ SL
Sbjct: 230 ISKLKHLAKLDLSYNPLKCSIPKSFGE-LHNLSILNLVSAELIGLIPPELGNCKSLKSLM 288
Query: 241 ISGNSFQGSI-IEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
+S NS G + +E+ + L RNQ G + W L L L+ N+ SGEI
Sbjct: 289 LSFNSLSGPLPLELSEIPLLTFSAERNQLSGSLPSWMGKWKVLDSLLLANNRFSGEIPHE 348
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
+ + LKHLSLA N S ++ LE ++LS L G I + +L L L
Sbjct: 349 IEDCPMLKHLSLASNLLSGSIPRELCGSGSLEAIDLSGNLLSGTIEEVFDGCSSLGELLL 408
Query: 360 SMNHLDGKIPL-LKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNL 411
+ N ++G IP L L +D NN +G +P + KS + ++ SYN L
Sbjct: 409 TNNQINGSIPEDLWKLPLMALDLDSNNFTGEIPKSLWKS-TNLMEFTASYNRL 460
>At3g13380 brassinosteroid receptor kinase, putative
Length = 1164
Score = 347 bits (890), Expect = 2e-95
Identities = 280/870 (32%), Positives = 417/870 (47%), Gaps = 134/870 (15%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWS-----LTSLKSLNLSSNH 125
L+LSG LTG +P + L SL+L NNK++ DF S L+ + +L L N+
Sbjct: 307 LDLSGNSLTGQLPQS-FTSCGSLQSLNLGNNKLS---GDFLSTVVSKLSRITNLYLPFNN 362
Query: 126 ISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLK---LDHNMFVRSIPSGI 182
ISGS+ ++ N L DLS N F+ E+P SL S VL+ + +N ++P +
Sbjct: 363 ISGSVPISLTNCSNLRVLDLSSNEFTGEVPSGFCSLQSSSVLEKLLIANNYLSGTVPVEL 422
Query: 183 LKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV--------------- 227
KC+SL +IDLS N L+G +P PKL L + NN+ GG+
Sbjct: 423 GKCKSLKTIDLSFNALTGLIPKEIW-TLPKLSDLVMWANNLTGGIPESICVDGGNLETLI 481
Query: 228 -----------SNFSRLKSIVSLNISGNSFQGSI-IEVFVL-KLEALDLSRNQFQGHISQ 274
+ S+ +++ +++S N G I + + L KL L L N G+I
Sbjct: 482 LNNNLLTGSLPESISKCTNMLWISLSSNLLTGEIPVGIGKLEKLAILQLGNNSLTGNIPS 541
Query: 275 VKYNWSHLVYLDLSENQLSGEIFQNLNNSMNL--------KHLSLACN------------ 314
N +L++LDL+ N L+G + L + L K + N
Sbjct: 542 ELGNCKNLIWLDLNSNNLTGNLPGELASQAGLVMPGSVSGKQFAFVRNEGGTDCRGAGGL 601
Query: 315 ------RFSR-------QKFPKIEMLLGLE-----------YLNLSKTSLVGHIPDEISH 350
R R PK + G+ YL+LS ++ G IP
Sbjct: 602 VEFEGIRAERLEHFPMVHSCPKTRIYSGMTMYMFSSNGSMIYLDLSYNAVSGSIPLGYGA 661
Query: 351 LGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSY 408
+G L L+L N L G IP K + V+D SHN+L G +P L L + + S
Sbjct: 662 MGYLQVLNLGHNLLTGTIPDSFGGLKAIGVLDLSHNDLQGFLPG-SLGGLSFLSDLDVSN 720
Query: 409 NNLT--LCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGH---RGMKLALVLTLS 463
NNLT + T + + C + P R H + +A ++
Sbjct: 721 NNLTGPIPFGGQLTTFPLTRYANNSGLCGVPLPPCSSGSRPTRSHAHPKKQSIATGMSAG 780
Query: 464 LIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTW-VADVKQATSVP 522
++F+ I+ L R K+ K+ RE+ S P S S++W ++ V + S+
Sbjct: 781 IVFSFMCIVMLIMALYRARKV--QKKEKQREKYIESLPTS---GSSSWKLSSVHEPLSIN 835
Query: 523 VVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLT 582
V FEKPL +TFA LL AT+ F +++ G FG VY+ L VA+K L+ +
Sbjct: 836 VATFEKPLRKLTFAHLLEATNGFSADSMIGSGGFGDVYKAKLADGSVVAIKKLIQVTGQG 895
Query: 583 DEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDD 642
D E E+E +G+IKH NLVPL GYC G++R+ +Y+YM+ G+L+ +L+
Sbjct: 896 DREFMAEMETIGKIKHRNLVPLLGYCKIGEERLLVYEYMKYGSLETVLH----------- 944
Query: 643 WSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSV 702
E+ G G+ W R KIA+G AR LAFLHH C P IIHR +K+S+V
Sbjct: 945 ------EKTKKG-------GIFLDWSARKKIAIGAARGLAFLHHSCIPHIIHRDMKSSNV 991
Query: 703 YLDYDLEPRLSDFGLAKIFGSGLDEEIA----RGSPGYVPPEFSQPEFESPTPKSDVYCF 758
LD D R+SDFG+A++ S LD ++ G+PGYVPPE+ Q F T K DVY +
Sbjct: 992 LLDQDFVARVSDFGMARLV-SALDTHLSVSTLAGTPGYVPPEYYQ-SFRC-TAKGDVYSY 1048
Query: 759 GVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPK-ICDTGSDEQIEE 817
GV+L ELL+GKKP+ D + E LV W + L R+ + + +DP+ + D D ++
Sbjct: 1049 GVILLELLSGKKPI--DPEEFGEDNNLVGWAKQLYREKRGAEILDPELVTDKSGDVELLH 1106
Query: 818 ALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
LK + C D PFKRPTM Q++ + K++
Sbjct: 1107 YLK-IASQCLDDRPFKRPTMIQVMTMFKEL 1135
Score = 124 bits (312), Expect = 2e-28
Identities = 112/358 (31%), Positives = 175/358 (48%), Gaps = 25/358 (6%)
Query: 60 YCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-----TLPSDFWSLT 114
Y S ++V +N S L G + + ++ ++DLSNN+ + T +DF
Sbjct: 144 YVFSTCLNLVSVNFSHNKLAGKLKSSPSASNKRITTVDLSNNRFSDEIPETFIADF--PN 201
Query: 115 SLKSLNLSSNHISGSLTNNIGNFGLLEN---FDLSKNSFS-DEIPEALSSLVSLKVLKLD 170
SLK L+LS N+++G + +FGL EN F LS+NS S D P +LS+ L+ L L
Sbjct: 202 SLKHLDLSGNNVTGDFSRL--SFGLCENLTVFSLSQNSISGDRFPVSLSNCKLLETLNLS 259
Query: 171 HNMFVRSIPSGIL--KCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV- 227
N + IP Q+L + L+ N SG +P L L+L+ N++ G +
Sbjct: 260 RNSLIGKIPGDDYWGNFQNLRQLSLAHNLYSGEIPPELSLLCRTLEVLDLSGNSLTGQLP 319
Query: 228 SNFSRLKSIVSLNISGNSFQGSIIEVFVLKLEA---LDLSRNQFQGHISQVKYNWSHLVY 284
+F+ S+ SLN+ N G + V KL L L N G + N S+L
Sbjct: 320 QSFTSCGSLQSLNLGNNKLSGDFLSTVVSKLSRITNLYLPFNNISGSVPISLTNCSNLRV 379
Query: 285 LDLSENQLSGEI---FQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLV 341
LDLS N+ +GE+ F +L +S L+ L +A N S ++ L+ ++LS +L
Sbjct: 380 LDLSSNEFTGEVPSGFCSLQSSSVLEKLLIANNYLSGTVPVELGKCKSLKTIDLSFNALT 439
Query: 342 GHIPDEISHLGNLNALDLSMNHLDGKIP---LLKNKHLQVIDFSHNNLSGPVPSFILK 396
G IP EI L L+ L + N+L G IP + +L+ + ++N L+G +P I K
Sbjct: 440 GLIPKEIWTLPKLSDLVMWANNLTGGIPESICVDGGNLETLILNNNLLTGSLPESISK 497
Score = 124 bits (310), Expect = 3e-28
Identities = 122/415 (29%), Positives = 183/415 (43%), Gaps = 58/415 (13%)
Query: 47 YNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTL 106
Y C+W+GV C S+ V+ L+L GLTG + + L+ L SL L N ++
Sbjct: 58 YGSGRDPCTWRGVSCSSD-GRVIGLDLRNGGLTGTLNLNNLTALSNLRSLYLQGNNFSSG 116
Query: 107 PSDFWSLTSLKSLNLSSNHISGS-------------LTNNIGNFGL-------------- 139
S S SL+ L+LSSN ++ S ++ N + L
Sbjct: 117 DSSSSSGCSLEVLDLSSNSLTDSSIVDYVFSTCLNLVSVNFSHNKLAGKLKSSPSASNKR 176
Query: 140 LENFDLSKNSFSDEIPEALSSLV--SLKVLKLDHNMFVRSIPS-GILKCQSLVSIDLSSN 196
+ DLS N FSDEIPE + SLK L L N C++L LS N
Sbjct: 177 ITTVDLSNNRFSDEIPETFIADFPNSLKHLDLSGNNVTGDFSRLSFGLCENLTVFSLSQN 236
Query: 197 QLSGTLPHGFGDAFP-------KLRTLNLAENNIYG---GVSNFSRLKSIVSLNISGNSF 246
+S GD FP L TLNL+ N++ G G + +++ L+++ N +
Sbjct: 237 SIS-------GDRFPVSLSNCKLLETLNLSRNSLIGKIPGDDYWGNFQNLRQLSLAHNLY 289
Query: 247 QGSI---IEVFVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNS 303
G I + + LE LDLS N G + Q + L L+L N+LSG+ + +
Sbjct: 290 SGEIPPELSLLCRTLEVLDLSGNSLTGQLPQSFTSCGSLQSLNLGNNKLSGDFLSTVVSK 349
Query: 304 MN-LKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALD---L 359
++ + +L L N S + L L+LS G +P L + + L+ +
Sbjct: 350 LSRITNLYLPFNNISGSVPISLTNCSNLRVLDLSSNEFTGEVPSGFCSLQSSSVLEKLLI 409
Query: 360 SMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
+ N+L G +P L K K L+ ID S N L+G +P I +LPK+ NNLT
Sbjct: 410 ANNYLSGTVPVELGKCKSLKTIDLSFNALTGLIPKEIW-TLPKLSDLVMWANNLT 463
Score = 117 bits (293), Expect = 3e-26
Identities = 108/357 (30%), Positives = 165/357 (45%), Gaps = 17/357 (4%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT--TLPSDFWSLTSLKSLNLSSNHISG 128
L+LSG +TG + G L LS N I+ P + L++LNLS N + G
Sbjct: 206 LDLSGNNVTGDFSRLSFGLCENLTVFSLSQNSISGDRFPVSLSNCKLLETLNLSRNSLIG 265
Query: 129 SLTNNI--GNFGLLENFDLSKNSFSDEIPEALSSLV-SLKVLKLDHNMFVRSIPSGILKC 185
+ + GNF L L+ N +S EIP LS L +L+VL L N +P C
Sbjct: 266 KIPGDDYWGNFQNLRQLSLAHNLYSGEIPPELSLLCRTLEVLDLSGNSLTGQLPQSFTSC 325
Query: 186 QSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGN 244
SL S++L +N+LSG ++ L L NNI G V + + ++ L++S N
Sbjct: 326 GSLQSLNLGNNKLSGDFLSTVVSKLSRITNLYLPFNNISGSVPISLTNCSNLRVLDLSSN 385
Query: 245 SFQGSIIEVFV-----LKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
F G + F LE L ++ N G + L +DLS N L+G I +
Sbjct: 386 EFTGEVPSGFCSLQSSSVLEKLLIANNYLSGTVPVELGKCKSLKTIDLSFNALTGLIPKE 445
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLG--LEYLNLSKTSLVGHIPDEISHLGNLNAL 357
+ L L + N + P+ + G LE L L+ L G +P+ IS N+ +
Sbjct: 446 IWTLPKLSDLVMWANNLT-GGIPESICVDGGNLETLILNNNLLTGSLPESISKCTNMLWI 504
Query: 358 DLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
LS N L G+IP + K + L ++ +N+L+G +PS L + + + + NNLT
Sbjct: 505 SLSSNLLTGEIPVGIGKLEKLAILQLGNNSLTGNIPS-ELGNCKNLIWLDLNSNNLT 560
>At1g55610 receptor kinase, putative
Length = 1166
Score = 337 bits (865), Expect = 1e-92
Identities = 262/847 (30%), Positives = 411/847 (47%), Gaps = 112/847 (13%)
Query: 35 LKKMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLH 94
L+ + L+S+ N S CS + + + ++ ++ L+G +P +GK L
Sbjct: 377 LRVLDLSSNGFTGNVPSGFCSLQ------SSPVLEKILIANNYLSGTVP-MELGKCKSLK 429
Query: 95 SLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIG-NFGLLENFDLSKNSFSD 152
++DLS N++T +P + W L +L L + +N+++G++ + G LE L+ N +
Sbjct: 430 TIDLSFNELTGPIPKEIWMLPNLSDLVMWANNLTGTIPEGVCVKGGNLETLILNNNLLTG 489
Query: 153 EIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPK 212
IPE++S ++ + L N IPSGI L + L +N LSG +P G+
Sbjct: 490 SIPESISRCTNMIWISLSSNRLTGKIPSGIGNLSKLAILQLGNNSLSGNVPRQLGNC-KS 548
Query: 213 LRTLNLAENNIYGGVSN--FSRLKSIVSLNISGNSFQ-------------GSIIEVFVLK 257
L L+L NN+ G + S+ ++ ++SG F G ++E ++
Sbjct: 549 LIWLDLNSNNLTGDLPGELASQAGLVMPGSVSGKQFAFVRNEGGTDCRGAGGLVEFEGIR 608
Query: 258 LEALDL-----SRNQFQGHISQVKYNWS---HLVYLDLSENQLSGEIFQNLNNSMNLKHL 309
E L+ S + + Y +S ++Y D+S N +SG I N L+ L
Sbjct: 609 AERLERLPMVHSCPATRIYSGMTMYTFSANGSMIYFDISYNAVSGFIPPGYGNMGYLQVL 668
Query: 310 SLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP 369
+L NR + L + L+LS +L G++P + L L+ LD+S N+L G IP
Sbjct: 669 NLGHNRITGTIPDSFGGLKAIGVLDLSHNNLQGYLPGSLGSLSFLSDLDVSNNNLTGPIP 728
Query: 370 LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFG 429
F + PV + NN LC ++P G
Sbjct: 729 -----------FGGQLTTFPVSRYA--------------NNSGLCGVPLRP-------CG 756
Query: 430 SVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQ 489
S PI + K++ V A++ ++ F +L +A RK + E K+
Sbjct: 757 SAPRRPITSR--IHAKKQTVA-----TAVIAGIAFSFMCFVMLVMALYRVRKVQKKEQKR 809
Query: 490 GSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGT 549
Y E SG S++ S V + S+ V FEKPL +TFA LL AT+ F T
Sbjct: 810 EKYIESLPTSGSCSWKLSS------VPEPLSINVATFEKPLRKLTFAHLLEATNGFSAET 863
Query: 550 LLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCV 609
++ G FG VY+ L VA+K L+ + D E E+E +G+IKH NLVPL GYC
Sbjct: 864 MVGSGGFGEVYKAQLRDGSVVAIKKLIRITGQGDREFMAEMETIGKIKHRNLVPLLGYCK 923
Query: 610 AGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRF 669
G++R+ +Y+YM+ G+L+ +L++ ++ G+ W
Sbjct: 924 VGEERLLVYEYMKWGSLETVLHE-----------------------KSSKKGGIYLNWAA 960
Query: 670 RHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEI 729
R KIA+G AR LAFLHH C P IIHR +K+S+V LD D E R+SDFG+A++ S LD +
Sbjct: 961 RKKIAIGAARGLAFLHHSCIPHIIHRDMKSSNVLLDEDFEARVSDFGMARLV-SALDTHL 1019
Query: 730 A----RGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTL 785
+ G+PGYVPPE+ Q F T K DVY +GV+L ELL+GKKP+ D + E L
Sbjct: 1020 SVSTLAGTPGYVPPEYYQ-SFRC-TAKGDVYSYGVILLELLSGKKPI--DPGEFGEDNNL 1075
Query: 786 VSWVRGLVRKNQTSRAIDPK-ICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLL 844
V W + L R+ + + +DP+ + D D ++ LK + C D PFKRPTM Q++ +
Sbjct: 1076 VGWAKQLYREKRGAEILDPELVTDKSGDVELFHYLK-IASQCLDDRPFKRPTMIQLMAMF 1134
Query: 845 KDIEPTT 851
K+++ T
Sbjct: 1135 KEMKADT 1141
Score = 128 bits (321), Expect = 2e-29
Identities = 131/456 (28%), Positives = 202/456 (43%), Gaps = 92/456 (20%)
Query: 47 YNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTL 106
Y CSW+GV C S+ +V L+L GLTG + + L L +L L N ++
Sbjct: 59 YESGRGSCSWRGVSC-SDDGRIVGLDLRNSGLTGTLNLVNLTALPNLQNLYLQGNYFSSG 117
Query: 107 PSDFWSLTSLKSLNLSSNHISG-----------------SLTNN--IGNFGL-------L 140
S L+ L+LSSN IS +++NN +G G L
Sbjct: 118 GDSSGSDCYLQVLDLSSNSISDYSMVDYVFSKCSNLVSVNISNNKLVGKLGFAPSSLQSL 177
Query: 141 ENFDLSKNSFSDEIPEALSS--LVSLKVLKLDH--------------------------N 172
DLS N SD+IPE+ S SLK L L H N
Sbjct: 178 TTVDLSYNILSDKIPESFISDFPASLKYLDLTHNNLSGDFSDLSFGICGNLTFFSLSQNN 237
Query: 173 MFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHG-FGDAFPKLRTLNLAENNIYGGVSNFS 231
+ P + C+ L ++++S N L+G +P+G + +F L+ L+LA N + G +
Sbjct: 238 LSGDKFPITLPNCKFLETLNISRNNLAGKIPNGEYWGSFQNLKQLSLAHNRLSGEIPPEL 297
Query: 232 RL--KSIVSLNISGNSFQGSIIEVFV--LKLEALDLSRNQFQG-HISQVKYNWSHLVYLD 286
L K++V L++SGN+F G + F + L+ L+L N G ++ V + + YL
Sbjct: 298 SLLCKTLVILDLSGNTFSGELPSQFTACVWLQNLNLGNNYLSGDFLNTVVSKITGITYLY 357
Query: 287 LSENQLSGEIFQNLNNSMNLKHLSLACNRF---------SRQKFPKIEMLL--------- 328
++ N +SG + +L N NL+ L L+ N F S Q P +E +L
Sbjct: 358 VAYNNISGSVPISLTNCSNLRVLDLSSNGFTGNVPSGFCSLQSSPVLEKILIANNYLSGT 417
Query: 329 ---------GLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP---LLKNKHL 376
L+ ++LS L G IP EI L NL+ L + N+L G IP +K +L
Sbjct: 418 VPMELGKCKSLKTIDLSFNELTGPIPKEIWMLPNLSDLVMWANNLTGTIPEGVCVKGGNL 477
Query: 377 QVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
+ + ++N L+G +P I + M + S N LT
Sbjct: 478 ETLILNNNLLTGSIPESISR-CTNMIWISLSSNRLT 512
Score = 111 bits (277), Expect = 2e-24
Identities = 87/298 (29%), Positives = 151/298 (50%), Gaps = 10/298 (3%)
Query: 66 EHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSN 124
+++ +L+L+ L+G IP L LDLS N + LPS F + L++LNL +N
Sbjct: 277 QNLKQLSLAHNRLSGEIPPELSLLCKTLVILDLSGNTFSGELPSQFTACVWLQNLNLGNN 336
Query: 125 HISGSLTNNI-GNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGIL 183
++SG N + + ++ N+ S +P +L++ +L+VL L N F ++PSG
Sbjct: 337 YLSGDFLNTVVSKITGITYLYVAYNNISGSVPISLTNCSNLRVLDLSSNGFTGNVPSGFC 396
Query: 184 KCQS---LVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSL 239
QS L I +++N LSGT+P G L+T++L+ N + G + L ++ L
Sbjct: 397 SLQSSPVLEKILIANNYLSGTVPMELGKC-KSLKTIDLSFNELTGPIPKEIWMLPNLSDL 455
Query: 240 NISGNSFQGSIIEVFVLK---LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEI 296
+ N+ G+I E +K LE L L+ N G I + ++++++ LS N+L+G+I
Sbjct: 456 VMWANNLTGTIPEGVCVKGGNLETLILNNNLLTGSIPESISRCTNMIWISLSSNRLTGKI 515
Query: 297 FQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNL 354
+ N L L L N S ++ L +L+L+ +L G +P E++ L
Sbjct: 516 PSGIGNLSKLAILQLGNNSLSGNVPRQLGNCKSLIWLDLNSNNLTGDLPGELASQAGL 573
Score = 70.5 bits (171), Expect = 4e-12
Identities = 85/342 (24%), Positives = 139/342 (39%), Gaps = 80/342 (23%)
Query: 1 MGLGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVY 60
M LG S+ + F L P E ++ L + + +++ VC G
Sbjct: 420 MELGKCKSLKTIDLSFNELTGPIPK--EIWMLPNLSDLVMWANNLTGTIPEGVCVKGG-- 475
Query: 61 CDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSL 119
++ L L+ LTG IP++ I + + + LS+N++T +PS +L+ L L
Sbjct: 476 ------NLETLILNNNLLTGSIPES-ISRCTNMIWISLSSNRLTGKIPSGIGNLSKLAIL 528
Query: 120 NLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSL--------------- 164
L +N +SG++ +GN L DL+ N+ + ++P L+S L
Sbjct: 529 QLGNNSLSGNVPRQLGNCKSLIWLDLNSNNLTGDLPGELASQAGLVMPGSVSGKQFAFVR 588
Query: 165 -------------------KVLKLDHNMFVRSIP-----SGILKCQ-----SLVSIDLSS 195
+ +L+ V S P SG+ S++ D+S
Sbjct: 589 NEGGTDCRGAGGLVEFEGIRAERLERLPMVHSCPATRIYSGMTMYTFSANGSMIYFDISY 648
Query: 196 NQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF 254
N +SG +P G+G+ L+ LNL N I G + +F LK+I
Sbjct: 649 NAVSGFIPPGYGN-MGYLQVLNLGHNRITGTIPDSFGGLKAI------------------ 689
Query: 255 VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEI 296
LDLS N QG++ + S L LD+S N L+G I
Sbjct: 690 ----GVLDLSHNNLQGYLPGSLGSLSFLSDLDVSNNNLTGPI 727
>At5g45800 receptor kinase-like protein
Length = 666
Score = 316 bits (809), Expect = 4e-86
Identities = 230/684 (33%), Positives = 357/684 (51%), Gaps = 96/684 (14%)
Query: 185 CQSLVS-IDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFS-RLKSIVSLNIS 242
C ++++ + L S +L+GT+ +LR L+L+ N++ G + + + +VS+N+S
Sbjct: 53 CSAVITHVVLPSRKLNGTVSWNPIRNLTRLRVLDLSNNSLDGSLPTWLWSMPGLVSVNLS 112
Query: 243 GNSFQGSIIEVFV-----LKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIF 297
N F GSI + V ++ L+LS N+F+ ++ + ++L LDLS N L G +
Sbjct: 113 RNRFGGSIRVIPVNGSVLSAVKELNLSFNRFKHAVNFTGF--TNLTTLDLSHNSL-GVLP 169
Query: 298 QNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNAL 357
L + L+HL ++ + + P I L L+YL+LS+ S+ G P + +L +L L
Sbjct: 170 LGLGSLSGLRHLDISRCKINGSVKP-ISGLKSLDYLDLSENSMNGSFPVDFPNLNHLQFL 228
Query: 358 DLSMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASE 417
+LS N G + K + F H +F +N+
Sbjct: 229 NLSANRFSGSVGFDKYRKFGKSAFLHGG-------------------DFVFND------- 262
Query: 418 IKPDIMKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLS--------LIFALA 469
K + ++ P +P ++R HR LV+ LS +IFA A
Sbjct: 263 -----SKIPYHHRIHRLP-HRHPPPVRQRNVKTHRTNHTPLVIGLSSSLGALIIVIFAAA 316
Query: 470 GILFLA-FGCRRKNKMWEVKQGSYRE-EQNISGPFSFQTDS-TTWVADVKQATSVPVVIF 526
IL R W + + + + SGPF F T+S ++WVAD+K+ T+ PVV+
Sbjct: 317 IILIRRRMKSARTKSRWAISNPTPLDFKMEKSGPFEFGTESGSSWVADIKEPTAAPVVMA 376
Query: 527 EKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEA 586
KPL+N+TF DL+ ATS+F +++++G GP+YR LPG++HVA+KVL + +A
Sbjct: 377 SKPLMNLTFKDLIVATSHFGTESVISDGTCGPLYRAVLPGDLHVAIKVLERIRDVDQNDA 436
Query: 587 ARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTD 646
E L R+KHPNL+ L+GYC+AG +++ +Y++M NG+L L++LP G + +DWS D
Sbjct: 437 VTAFEALTRLKHPNLLTLSGYCIAGKEKLILYEFMANGDLHRWLHELPAGETNVEDWSAD 496
Query: 647 TWEEADNGIQNVG-SEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLD 705
TWE +VG S T W RH+IA+G AR LA+LHH H + A+++ L
Sbjct: 497 TWE------SHVGDSSPEKTNWLIRHRIAIGVARGLAYLHH---VGTTHGHLVATNILLT 547
Query: 706 YDLEPRLSDFGLAKIFGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFEL 765
LEPR+SDFG+ I +G D + EF DVY FGV+LFEL
Sbjct: 548 ETLEPRISDFGINNIARTGDD------------TNKNNVEF-------DVYSFGVILFEL 588
Query: 766 LTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALK--EVG 823
LTGK+ G D V VR LV++ + A+D ++ + E + E ++ +G
Sbjct: 589 LTGKQ--GSDEN--------VKSVRRLVKERRGEEALDSRL-RLAAGESVNEMVESLRIG 637
Query: 824 YLCTADLPFKRPTMQQIVGLLKDI 847
Y CTA+ P KRPTMQQ++GLLKDI
Sbjct: 638 YFCTAETPVKRPTMQQVLGLLKDI 661
Score = 84.0 bits (206), Expect = 3e-16
Identities = 76/226 (33%), Positives = 104/226 (45%), Gaps = 31/226 (13%)
Query: 55 SWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSL 113
SW C + HVV L L G + I L +L LDLSNN + +LP+ WS+
Sbjct: 47 SWFSSNCSAVITHVV---LPSRKLNGTVSWNPIRNLTRLRVLDLSNNSLDGSLPTWLWSM 103
Query: 114 TSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNM 173
L S+NLS N GS+ IP S L ++K L L N
Sbjct: 104 PGLVSVNLSRNRFGGSIR---------------------VIPVNGSVLSAVKELNLSFNR 142
Query: 174 FVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRL 233
F ++ +L ++DLS N L G LP G G + LR L+++ I G V S L
Sbjct: 143 FKHAV--NFTGFTNLTTLDLSHNSL-GVLPLGLG-SLSGLRHLDISRCKINGSVKPISGL 198
Query: 234 KSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKY 277
KS+ L++S NS GS F + L+ L+LS N+F G + KY
Sbjct: 199 KSLDYLDLSENSMNGSFPVDFPNLNHLQFLNLSANRFSGSVGFDKY 244
Score = 68.9 bits (167), Expect = 1e-11
Identities = 57/187 (30%), Positives = 95/187 (50%), Gaps = 14/187 (7%)
Query: 158 LSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGT---LPHGFGDAFPKLR 214
+ +L L+VL L +N S+P+ + LVS++LS N+ G+ +P G ++
Sbjct: 76 IRNLTRLRVLDLSNNSLDGSLPTWLWSMPGLVSVNLSRNRFGGSIRVIPVN-GSVLSAVK 134
Query: 215 TLNLAENNIYGGVSNFSRLKSIVSLNISGNSFQGSIIEVFVLK-LEALDLSRNQFQGHIS 273
LNL+ N V NF+ ++ +L++S NS + + L L LD+SR + G +
Sbjct: 135 ELNLSFNRFKHAV-NFTGFTNLTTLDLSHNSLGVLPLGLGSLSGLRHLDISRCKINGSVK 193
Query: 274 QVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFS-------RQKFPKIEM 326
+ L YLDLSEN ++G + N +L+ L+L+ NRFS +KF K
Sbjct: 194 PIS-GLKSLDYLDLSENSMNGSFPVDFPNLNHLQFLNLSANRFSGSVGFDKYRKFGKSAF 252
Query: 327 LLGLEYL 333
L G +++
Sbjct: 253 LHGGDFV 259
>At4g36180 receptor protein kinase like protein
Length = 1136
Score = 310 bits (793), Expect = 3e-84
Identities = 250/803 (31%), Positives = 385/803 (47%), Gaps = 62/803 (7%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N + + EL L+ LTG IP I + L LD N + +P + +LK L+L
Sbjct: 354 NLKRLEELKLANNSLTGEIP-VEIKQCGSLDVLDFEGNSLKGQIPEFLGYMKALKVLSLG 412
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI 182
N SG + +++ N LE +L +N+ + P L +L SL L L N F ++P I
Sbjct: 413 RNSFSGYVPSSMVNLQQLERLNLGENNLNGSFPVELMALTSLSELDLSGNRFSGAVPVSI 472
Query: 183 LKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNI 241
+L ++LS N SG +P G+ F KL L+L++ N+ G V S L ++ + +
Sbjct: 473 SNLSNLSFLNLSGNGFSGEIPASVGNLF-KLTALDLSKQNMSGEVPVELSGLPNVQVIAL 531
Query: 242 SGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
GN+F G + E F ++ L ++LS N F G I Q LV L LS+N +SG I
Sbjct: 532 QGNNFSGVVPEGFSSLVSLRYVNLSSNSFSGEIPQTFGFLRLLVSLSLSDNHISGSIPPE 591
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
+ N L+ L L NR + L L+ L+L + +L G IP EIS +LN+L L
Sbjct: 592 IGNCSALEVLELRSNRLMGHIPADLSRLPRLKVLDLGQNNLSGEIPPEISQSSSLNSLSL 651
Query: 360 SMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT-LCAS 416
NHL G IP +L +D S NNL+G +P+ + + +N S NNL +
Sbjct: 652 DHNHLSGVIPGSFSGLSNLTKMDLSVNNLTGEIPASLALISSNLVYFNVSSNNLKGEIPA 711
Query: 417 EIKPDIMKTS-FFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLA 475
+ I TS F G+ C N + + K+ L++ ++ I A LF
Sbjct: 712 SLGSRINNTSEFSGNTELCGKPLNRRCESSTAEGKKKKRKMILMIVMAAIGAFLLSLFCC 771
Query: 476 FGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLL---- 531
F K W K ++Q+ +G T+ + V+ +TS +P L
Sbjct: 772 FYVYTLLK-WRKK----LKQQSTTGEKKRSPGRTSAGSRVRSSTSRSSTENGEPKLVMFN 826
Query: 532 -NITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAAREL 590
IT A+ + AT FD +L+ ++G +++ + ++++ L GS L + +E
Sbjct: 827 NKITLAETIEATRQFDEENVLSRTRYGLLFKANYNDGMVLSIRRLPNGSLLNENLFKKEA 886
Query: 591 EFLGRIKHPNLVPLTGYCVA-GDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWE 649
E LG++KH N+ L GY D R+ +YDYM NGNL LL +
Sbjct: 887 EVLGKVKHRNITVLRGYYAGPPDLRLLVYDYMPNGNLSTLL------------------Q 928
Query: 650 EADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLE 709
EA + +G + W RH IALG AR L FLH ++H +K +V D D E
Sbjct: 929 EASH------QDGHVLNWPMRHLIALGIARGLGFLHQS---NMVHGDIKPQNVLFDADFE 979
Query: 710 PRLSDFGLAKIFGSGLDEEIAR----GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFEL 765
+SDFGL ++ G+ GYV PE + T +SD+Y FG+VL E+
Sbjct: 980 AHISDFGLDRLTIRSPSRSAVTANTIGTLGYVSPEATLS--GEITRESDIYSFGIVLLEI 1037
Query: 766 LTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSD-EQIEEALK--EV 822
LTGK+PV +T D++ +V WV+ +++ Q + ++P + + + + EE L +V
Sbjct: 1038 LTGKRPV--MFTQDED---IVKWVKKQLQRGQVTELLEPGLLELDPESSEWEEFLLGIKV 1092
Query: 823 GYLCTADLPFKRPTMQQIVGLLK 845
G LCTA P RPTM +V +L+
Sbjct: 1093 GLLCTATDPLDRPTMSDVVFMLE 1115
Score = 163 bits (413), Expect = 3e-40
Identities = 138/390 (35%), Positives = 195/390 (49%), Gaps = 39/390 (10%)
Query: 54 CSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWS 112
C W+GV C +++ V E+ L + L+G I D G L L L L +N T+P+
Sbjct: 58 CDWRGVGCTNHR--VTEIRLPRLQLSGRISDRISG-LRMLRKLSLRSNSFNGTIPTSLAY 114
Query: 113 LTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHN 172
T L S+ L N +SG L + N LE F+++ N S EIP L S SL+ L + N
Sbjct: 115 CTRLLSVFLQYNSLSGKLPPAMRNLTSLEVFNVAGNRLSGEIPVGLPS--SLQFLDISSN 172
Query: 173 MFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFS 231
F IPSG+ L ++LS NQL+G +P G+ L+ L L N + G + S S
Sbjct: 173 TFSGQIPSGLANLTQLQLLNLSYNQLTGEIPASLGN-LQSLQYLWLDFNLLQGTLPSAIS 231
Query: 232 RLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQG------------HISQVKY 277
S+V L+ S N G I + + KLE L LS N F G I Q+ +
Sbjct: 232 NCSSLVHLSASENEIGGVIPAAYGALPKLEVLSLSNNNFSGTVPFSLFCNTSLTIVQLGF 291
Query: 278 N-WSHLV-------------YLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPK 323
N +S +V LDL EN++SG L N ++LK+L ++ N FS + P
Sbjct: 292 NAFSDIVRPETTANCRTGLQVLDLQENRISGRFPLWLTNILSLKNLDVSGNLFSGEIPPD 351
Query: 324 IEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDF 381
I L LE L L+ SL G IP EI G+L+ LD N L G+IP L K L+V+
Sbjct: 352 IGNLKRLEELKLANNSLTGEIPVEIKQCGSLDVLDFEGNSLKGQIPEFLGYMKALKVLSL 411
Query: 382 SHNNLSGPVPSFILKSLPKMKKYNFSYNNL 411
N+ SG VPS ++ +L ++++ N NNL
Sbjct: 412 GRNSFSGYVPSSMV-NLQQLERLNLGENNL 440
Score = 151 bits (381), Expect = 2e-36
Identities = 131/407 (32%), Positives = 188/407 (46%), Gaps = 35/407 (8%)
Query: 38 MGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLD 97
+GL SS + + SS+ S + +N + LNLS LTG IP ++G L L L
Sbjct: 158 VGLPSSLQFLDISSNTFSGQIPSGLANLTQLQLLNLSYNQLTGEIP-ASLGNLQSLQYLW 216
Query: 98 LSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPE 156
L N + TLPS + +SL L+ S N I G + G LE LS N+FS +P
Sbjct: 217 LDFNLLQGTLPSAISNCSSLVHLSASENEIGGVIPAAYGALPKLEVLSLSNNNFSGTVPF 276
Query: 157 ALSSLVSLKVLKLDHNMFVRSI-PSGILKCQ-------------------------SLVS 190
+L SL +++L N F + P C+ SL +
Sbjct: 277 SLFCNTSLTIVQLGFNAFSDIVRPETTANCRTGLQVLDLQENRISGRFPLWLTNILSLKN 336
Query: 191 IDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGNSFQGS 249
+D+S N SG +P G+ +L L LA N++ G + + S+ L+ GNS +G
Sbjct: 337 LDVSGNLFSGEIPPDIGN-LKRLEELKLANNSLTGEIPVEIKQCGSLDVLDFEGNSLKGQ 395
Query: 250 IIEV--FVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLK 307
I E ++ L+ L L RN F G++ N L L+L EN L+G L +L
Sbjct: 396 IPEFLGYMKALKVLSLGRNSFSGYVPSSMVNLQQLERLNLGENNLNGSFPVELMALTSLS 455
Query: 308 HLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGK 367
L L+ NRFS I L L +LNLS G IP + +L L ALDLS ++ G+
Sbjct: 456 ELDLSGNRFSGAVPVSISNLSNLSFLNLSGNGFSGEIPASVGNLFKLTALDLSKQNMSGE 515
Query: 368 IP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
+P L ++QVI NN SG VP SL ++ N S N+ +
Sbjct: 516 VPVELSGLPNVQVIALQGNNFSGVVPEG-FSSLVSLRYVNLSSNSFS 561
Score = 96.3 bits (238), Expect = 6e-20
Identities = 87/264 (32%), Positives = 115/264 (42%), Gaps = 54/264 (20%)
Query: 38 MGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLD 97
M LTS S++ + S + S SN ++ LNLSG G +G IP ++G L KL +LD
Sbjct: 449 MALTSLSEL-DLSGNRFSGAVPVSISNLSNLSFLNLSGNGFSGEIP-ASVGNLFKLTALD 506
Query: 98 LS-------------------------NNKITTLPSDFWSLTSLKSLNLSS--------- 123
LS NN +P F SL SL+ +NLSS
Sbjct: 507 LSKQNMSGEVPVELSGLPNVQVIALQGNNFSGVVPEGFSSLVSLRYVNLSSNSFSGEIPQ 566
Query: 124 ---------------NHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLK 168
NHISGS+ IGN LE +L N IP LS L LKVL
Sbjct: 567 TFGFLRLLVSLSLSDNHISGSIPPEIGNCSALEVLELRSNRLMGHIPADLSRLPRLKVLD 626
Query: 169 LDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS 228
L N IP I + SL S+ L N LSG +P F L ++L+ NN+ G +
Sbjct: 627 LGQNNLSGEIPPEISQSSSLNSLSLDHNHLSGVIPGSF-SGLSNLTKMDLSVNNLTGEIP 685
Query: 229 NFSRL--KSIVSLNISGNSFQGSI 250
L ++V N+S N+ +G I
Sbjct: 686 ASLALISSNLVYFNVSSNNLKGEI 709
>At2g26330 putative receptor-like protein kinase, ERECTA
Length = 976
Score = 303 bits (775), Expect = 3e-82
Identities = 270/974 (27%), Positives = 417/974 (42%), Gaps = 192/974 (19%)
Query: 3 LGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNF----SSSVCSWKG 58
+ +F +++L FLF ++E +KK ++ +Y++ SS C W+G
Sbjct: 1 MALFRDIVLLGFLFCLSLVATVTSEEGATLLEIKKSFKDVNNVLYDWTTSPSSDYCVWRG 60
Query: 59 VYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLK 117
V C++ +VV LNLS + L G I IG L L S+DL N+++ +P + +SL+
Sbjct: 61 VSCENVTFNVVALNLSDLNLDGEI-SPAIGDLKSLLSIDLRGNRLSGQIPDEIGDCSSLQ 119
Query: 118 SLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLD------- 170
+L+LS N +SG + +I LE L N IP LS + +LK+L L
Sbjct: 120 NLDLSFNELSGDIPFSISKLKQLEQLILKNNQLIGPIPSTLSQIPNLKILDLAQNKLSGE 179
Query: 171 -----------------------------------------HNMFVRSIPSGILKCQSLV 189
+N SIP I C +
Sbjct: 180 IPRLIYWNEVLQYLGLRGNNLVGNISPDLCQLTGLWYFDVRNNSLTGSIPETIGNCTAFQ 239
Query: 190 SIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQG 248
+DLS NQL+G +P G F ++ TL+L N + G + S ++++ L++SGN G
Sbjct: 240 VLDLSYNQLTGEIPFDIG--FLQVATLSLQGNQLSGKIPSVIGLMQALAVLDLSGNLLSG 297
Query: 249 SIIEVF--------------------------VLKLEALDLSRNQFQGHISQVKYNWSHL 282
SI + + KL L+L+ N GHI + L
Sbjct: 298 SIPPILGNLTFTEKLYLHSNKLTGSIPPELGNMSKLHYLELNDNHLTGHIPPELGKLTDL 357
Query: 283 VYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVG 342
L+++ N L G I +L++ NL L++ N+FS + L + YLNLS ++ G
Sbjct: 358 FDLNVANNDLEGPIPDHLSSCTNLNSLNVHGNKFSGTIPRAFQKLESMTYLNLSSNNIKG 417
Query: 343 HIPDEISHLGNLNALDLSMNHLDGKIP--------LLKN------------------KHL 376
IP E+S +GNL+ LDLS N ++G IP LLK + +
Sbjct: 418 PIPVELSRIGNLDTLDLSNNKINGIIPSSLGDLEHLLKMNLSRNHITGVVPGDFGNLRSI 477
Query: 377 QVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT----------------LCASEIKP 420
ID S+N++SGP+P L L + NNLT + + +
Sbjct: 478 MEIDLSNNDISGPIPEE-LNQLQNIILLRLENNNLTGNVGSLANCLSLTVLNVSHNNLVG 536
Query: 421 DIMKTSFFGSVNSCPIAANP----SFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAF 476
DI K + F + NP S+ R +++++ L A+ G++ L
Sbjct: 537 DIPKNNNFSRFSPDSFIGNPGLCGSWLNSPCHDSRRTVRVSISRAAILGIAIGGLVILLM 596
Query: 477 ----GCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLN 532
CR N + + ++ P ++ T +VI +
Sbjct: 597 VLIAACRPHNPPPFL-------DGSLDKPVTYSTPK--------------LVILHMNMAL 635
Query: 533 ITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEF 592
+ D++ T N ++ G VY+ L VA+K L + + ++ ELE
Sbjct: 636 HVYEDIMRMTENLSEKYIIGHGASSTVYKCVLKNCKPVAIKRLYSHNPQSMKQFETELEM 695
Query: 593 LGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEAD 652
L IKH NLV L Y ++ + YDY+ENG+L +LL+ + T DW T
Sbjct: 696 LSSIKHRNLVSLQAYSLSHLGSLLFYDYLENGSLWDLLHGPT--KKKTLDWDT------- 746
Query: 653 NGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRL 712
R KIA G A+ LA+LHH CSP IIHR VK+S++ LD DLE RL
Sbjct: 747 -----------------RLKIAYGAAQGLAYLHHDCSPRIIHRDVKSSNILLDKDLEARL 789
Query: 713 SDFGLAK--IFGSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK 770
+DFG+AK G+ GY+ PE+++ T KSDVY +G+VL ELLT +K
Sbjct: 790 TDFGIAKSLCVSKSHTSTYVMGTIGYIDPEYART--SRLTEKSDVYSYGIVLLELLTRRK 847
Query: 771 PVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADL 830
V D+ + L + N+ DP I T D + + + ++ LCT
Sbjct: 848 AVDDE-------SNLHHLIMSKTGNNEVMEMADPDITSTCKDLGVVKKVFQLALLCTKRQ 900
Query: 831 PFKRPTMQQIVGLL 844
P RPTM Q+ +L
Sbjct: 901 PNDRPTMHQVTRVL 914
>At1g12460 unknown protein
Length = 882
Score = 300 bits (768), Expect = 2e-81
Identities = 257/870 (29%), Positives = 402/870 (45%), Gaps = 127/870 (14%)
Query: 55 SWKGVYCDSNK--EHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFW 111
S+ G+ C+ + +V N S G P + L + L+L N+ T LP D++
Sbjct: 57 SFNGITCNPQGFVDKIVLWNTSLAGTLAP----GLSNLKFIRVLNLFGNRFTGNLPLDYF 112
Query: 112 SLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVS-LKVLKLD 170
L +L ++N+SSN +SG + I L DLSKN F+ EIP +L K + L
Sbjct: 113 KLQTLWTINVSSNALSGPIPEFISELSSLRFLDLSKNGFTGEIPVSLFKFCDKTKFVSLA 172
Query: 171 HNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAEN--------- 221
HN SIP+ I+ C +LV D S N L G LP D P L +++ N
Sbjct: 173 HNNIFGSIPASIVNCNNLVGFDFSYNNLKGVLPPRICD-IPVLEYISVRNNLLSGDVSEE 231
Query: 222 --------------NIYGGVSNFSRL--KSIVSLNISGNSFQGSIIEVFVLK--LEALDL 263
N++ G++ F+ L K+I N+S N F G I E+ LE LD
Sbjct: 232 IQKCQRLILVDLGSNLFHGLAPFAVLTFKNITYFNVSWNRFGGEIGEIVDCSESLEFLDA 291
Query: 264 SRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPK 323
S N+ G I L LDL N+L+G I ++ +L + L N
Sbjct: 292 SSNELTGRIPTGVMGCKSLKLLDLESNKLNGSIPGSIGKMESLSVIRLGNNSIDGVIPRD 351
Query: 324 IEMLLGLEYLNLSKTSLVGHIPDEISH------------------------LGNLNALDL 359
I L L+ LNL +L+G +P++IS+ L N+ LDL
Sbjct: 352 IGSLEFLQVLNLHNLNLIGEVPEDISNCRVLLELDVSGNDLEGKISKKLLNLTNIKILDL 411
Query: 360 SMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASE 417
N L+G IP L +Q +D S N+LSGP+PS L SL + +N SYNNL S
Sbjct: 412 HRNRLNGSIPPELGNLSKVQFLDLSQNSLSGPIPSS-LGSLNTLTHFNVSYNNL----SG 466
Query: 418 IKPDIMKTSFFGSV---NSCPIAANP--------SFFKKRRDVGHRGMKLALVLTLSLIF 466
+ P + FGS N+ + +P K R+ + + +V+ + +
Sbjct: 467 VIPPVPMIQAFGSSAFSNNPFLCGDPLVTPCNSRGAAAKSRNSDALSISVIIVIIAAAVI 526
Query: 467 ALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPV--- 523
+ LA R + + R+++ I T TT +A ++ V +
Sbjct: 527 LFGVCIVLALNLRARKR---------RKDEEIL------TVETTPLASSIDSSGVIIGKL 571
Query: 524 VIFEKPLLNITFADLLSATSNF-DRGTLLAEGKFGPVYRGFLPGNIHVAVKVL-VVGSTL 581
V+F K L + + D + T D+ ++ G G VYR G + +AVK L +G
Sbjct: 572 VLFSKNLPS-KYEDWEAGTKALLDKENIIGMGSIGSVYRASFEGGVSIAVKKLETLGRIR 630
Query: 582 TDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTD 641
EE +E+ LG ++HPNL GY + ++ + +++ NG+L + L+ S+
Sbjct: 631 NQEEFEQEIGRLGGLQHPNLSSFQGYYFSSTMQLILSEFVPNGSLYDNLHLRIFPGTSSS 690
Query: 642 DWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASS 701
+TD W R +IALGTA+AL+FLH+ C P I+H VK+++
Sbjct: 691 YGNTDL------------------NWHRRFQIALGTAKALSFLHNDCKPAILHLNVKSTN 732
Query: 702 VYLDYDLEPRLSDFGLAKIF----GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYC 757
+ LD E +LSD+GL K GL ++ + GY+ PE +Q + + K DVY
Sbjct: 733 ILLDERYEAKLSDYGLEKFLPVMDSFGLTKKF-HNAVGYIAPELAQQSLRA-SEKCDVYS 790
Query: 758 FGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEE 817
+GVVL EL+TG+KPV + + + L +VR L+ S D ++ + +E I+
Sbjct: 791 YGVVLLELVTGRKPV--ESPSENQVLILRDYVRDLLETGSASDCFDRRLREFEENELIQ- 847
Query: 818 ALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
+ ++G LCT++ P KRP+M ++V +L+ I
Sbjct: 848 -VMKLGLLCTSENPLKRPSMAEVVQVLESI 876
>At1g62950 protein kinase like protein
Length = 890
Score = 300 bits (767), Expect = 3e-81
Identities = 274/911 (30%), Positives = 426/911 (46%), Gaps = 109/911 (11%)
Query: 9 VLVLTFLFKHL--ASQQPNTDEFYVS-EFLKKMGLTSSSKVYNFSSS------VC-SWKG 58
+ V+ F+F H+ S + +D E L + + YN +S +C S+ G
Sbjct: 7 IWVIMFIFVHIIITSSRSFSDSIITEREILLQFKDNINDDPYNSLASWVSNADLCNSFNG 66
Query: 59 VYCDSNKEHVVE-LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSL 116
V C N+E VE + L L G + G L L L L N+IT LP D+ L +L
Sbjct: 67 VSC--NQEGFVEKIVLWNTSLAGTLTPALSG-LTSLRVLTLFGNRITGNLPLDYLKLQTL 123
Query: 117 KSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLV-SLKVLKLDHNMFV 175
+N+SSN +SG + IG+ L DLSKN+F EIP +L K + L HN
Sbjct: 124 WKINVSSNALSGLVPEFIGDLPNLRFLDLSKNAFFGEIPNSLFKFCYKTKFVSLSHNNLS 183
Query: 176 RSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLK 234
SIP I+ C +L+ D S N ++G LP P L +++ N + G V S+ K
Sbjct: 184 GSIPESIVNCNNLIGFDFSYNGITGLLPRICD--IPVLEFVSVRRNLLSGDVFEEISKCK 241
Query: 235 SIVSLNISGNSFQG-SIIEVFVLK-LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQL 292
+ ++I NSF G + EV K L ++S N+F+G I ++ L +LD S N+L
Sbjct: 242 RLSHVDIGSNSFDGVASFEVIGFKNLTYFNVSGNRFRGEIGEIVDCSESLEFLDASSNEL 301
Query: 293 SGEIFQNLNNSMNLKHLSLACNRFSRQ------KFPKIEML--------------LG--- 329
+G + + +LK L L NR + K K+ ++ LG
Sbjct: 302 TGNVPSGITGCKSLKLLDLESNRLNGSVPVGMGKMEKLSVIRLGDNFIDGKLPLELGNLE 361
Query: 330 -LEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP-------------LLKNK- 374
L+ LNL +LVG IP+++S+ L LD+S N L+G+IP L +N+
Sbjct: 362 YLQVLNLHNLNLVGEIPEDLSNCRLLLELDVSGNGLEGEIPKNLLNLTNLEILDLHRNRI 421
Query: 375 ------------HLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDI 422
+Q +D S N LSGP+PS L++L ++ +N SYNNL S I P I
Sbjct: 422 SGNIPPNLGSLSRIQFLDLSENLLSGPIPSS-LENLKRLTHFNVSYNNL----SGIIPKI 476
Query: 423 MKTSFFGSVNSCPIAANPSFFK-KRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCR-R 480
+ N+ + +P G R K + T +I +A L C
Sbjct: 477 QASGASSFSNNPFLCGDPLETPCNALRTGSRSRKTKALSTSVIIVIIAAAAILVGICLVL 536
Query: 481 KNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLS 540
+ K+ REE+ ++ + T ++T + T +V+F K L + + D +
Sbjct: 537 VLNLRARKRRKKREEEIVTFDTTTPTQASTESGN-GGVTFGKLVLFSKSLPS-KYEDWEA 594
Query: 541 ATSNF-DRGTLLAEGKFGPVYRGFLPGNIHVAVKVL-VVGSTLTDEEAARELEFLGRIKH 598
T D+ ++ G G VYR G + +AVK L +G EE +E+ LG + H
Sbjct: 595 GTKALLDKDNIIGIGSIGAVYRASFEGGVSIAVKKLETLGRIRNQEEFEQEIGRLGSLSH 654
Query: 599 PNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNV 658
PNL GY + ++ + +++ NG+L + L+ S+ S E
Sbjct: 655 PNLASFQGYYFSSTMQLILSEFVTNGSLYDNLHPRVSHRTSSSSSSHGNTE--------- 705
Query: 659 GSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLA 718
W R +IA+GTA+AL+FLH+ C P I+H VK++++ LD E +LSD+GL
Sbjct: 706 ------LNWHRRFQIAVGTAKALSFLHNDCKPAILHLNVKSTNILLDERYEAKLSDYGLE 759
Query: 719 KIF----GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGD 774
K SGL + + GY+ PE +Q S K DVY +GVVL EL+TG+KPV
Sbjct: 760 KFLPVLNSSGLTK--FHNAVGYIAPELAQSLRVS--DKCDVYSYGVVLLELVTGRKPV-- 813
Query: 775 DYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKR 834
+ + E L VR L+ S D ++ +E I+ + ++G +CT + P KR
Sbjct: 814 ESPSENEVVILRDHVRNLLETGSASDCFDRRLRGFEENELIQ--VMKLGLICTTENPLKR 871
Query: 835 PTMQQIVGLLK 845
P++ ++V +L+
Sbjct: 872 PSIAEVVQVLE 882
>At2g02220 putative receptor-like protein kinase
Length = 1008
Score = 298 bits (764), Expect = 7e-81
Identities = 254/865 (29%), Positives = 390/865 (44%), Gaps = 154/865 (17%)
Query: 88 GKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLS 146
GK L L L N +T +P D + L L L + N +SGSL+ I N L D+S
Sbjct: 193 GKCVLLEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVRLDVS 252
Query: 147 KNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI---------------------LKC 185
N FS EIP+ L LK N F+ IP + L C
Sbjct: 253 WNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPSLNLLNLRNNSLSGRLMLNC 312
Query: 186 QSLV---SIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNI 241
+++ S+DL +N+ +G LP D +L+ +NLA N +G V +F +S+ ++
Sbjct: 313 TAMIALNSLDLGTNRFNGRLPENLPDC-KRLKNVNLARNTFHGQVPESFKNFESLSYFSL 371
Query: 242 SGNS-------------------------FQGSII-----------EVFVL--------- 256
S +S F G + +V V+
Sbjct: 372 SNSSLANISSALGILQHCKNLTTLVLTLNFHGEALPDDSSLHFEKLKVLVVANCRLTGSM 431
Query: 257 --------KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKH 308
+L+ LDLS N+ G I ++ L YLDLS N +GEI ++L +L
Sbjct: 432 PRWLSSSNELQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIPKSLTKLESLTS 491
Query: 309 LSLACNRFSRQKFP----KIEMLLGLEY---------LNLSKTSLVGHIPDEISHLGNLN 355
+++ N S FP + E L+Y + L +L G I +E +L L+
Sbjct: 492 RNISVNEPSPD-FPFFMKRNESARALQYNQIFGFPPTIELGHNNLSGPIWEEFGNLKKLH 550
Query: 356 ALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT- 412
DL N L G IP L L+ +D S+N LSG +P L+ L + K++ +YNNL+
Sbjct: 551 VFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIP-VSLQQLSFLSKFSVAYNNLSG 609
Query: 413 LCASEIKPDIMKTSFFGSVNSCPIAANP-------SFFKKRRDVGHRGMKLALVLTLSLI 465
+ S + S F S + C P + K+ R + +A+ + +
Sbjct: 610 VIPSGGQFQTFPNSSFESNHLCGEHRFPCSEGTESALIKRSRRSRGGDIGMAIGIAFGSV 669
Query: 466 FALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVI 525
F L + + RR++ G E S + + ++ + S VV+
Sbjct: 670 FLLTLLSLIVLRARRRS-------GEVDPEIEESESMNRK--------ELGEIGSKLVVL 714
Query: 526 FEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEE 585
F+ +++ DLL +T++FD+ ++ G FG VY+ LP VA+K L + E
Sbjct: 715 FQSNDKELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIERE 774
Query: 586 AARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWST 645
E+E L R +HPNLV L G+C + R+ IY YMENG+L L++
Sbjct: 775 FEAEVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHE------------- 821
Query: 646 DTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLD 705
+N G L W+ R +IA G A+ L +LH GC P I+HR +K+S++ LD
Sbjct: 822 ----------RNDGP--ALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLD 869
Query: 706 YDLEPRLSDFGLAKIFGSGLDEEIAR---GSPGYVPPEFSQPEFESPTPKSDVYCFGVVL 762
+ L+DFGLA++ S + ++ G+ GY+PPE+ Q T K DVY FGVVL
Sbjct: 870 ENFNSHLADFGLARLM-SPYETHVSTDLVGTLGYIPPEYGQASV--ATYKGDVYSFGVVL 926
Query: 763 FELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEV 822
ELLT K+PV D K L+SWV + +++ S DP I +D+++ L E+
Sbjct: 927 LELLTDKRPV--DMCKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMFRVL-EI 983
Query: 823 GYLCTADLPFKRPTMQQIVGLLKDI 847
LC ++ P +RPT QQ+V L D+
Sbjct: 984 ACLCLSENPKQRPTTQQLVSWLDDV 1008
Score = 149 bits (376), Expect = 6e-36
Identities = 158/578 (27%), Positives = 262/578 (44%), Gaps = 90/578 (15%)
Query: 50 SSSVCSWKGVYCDSNKE-HVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKI-TTLP 107
S+ C+W G+ C+SN V+ L L L+G + + ++GKL+++ L+LS N I ++P
Sbjct: 59 STDCCNWTGITCNSNNTGRVIRLELGNKKLSGKLSE-SLGKLDEIRVLNLSRNFIKDSIP 117
Query: 108 SDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEAL-SSLVSLKV 166
++L +L++L+LSSN +SG + +I N L++FDLS N F+ +P + + ++V
Sbjct: 118 LSIFNLKNLQTLDLSSNDLSGGIPTSI-NLPALQSFDLSSNKFNGSLPSHICHNSTQIRV 176
Query: 167 LKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAF--PKLRTLNLAENNIY 224
+KL N F + SG KC L + L N L+G +P D F +L L + EN +
Sbjct: 177 VKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPE---DLFHLKRLNLLGIQENRLS 233
Query: 225 GGVSNFSR-LKSIVSLNISGNSFQGSIIEVF----VLK---------------------- 257
G +S R L S+V L++S N F G I +VF LK
Sbjct: 234 GSLSREIRNLSSLVRLDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPS 293
Query: 258 LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFS 317
L L+L N G + L LDL N+ +G + +NL + LK+++LA N F
Sbjct: 294 LNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKRLKNVNLARNTFH 353
Query: 318 RQKFPKIEMLLGLEYLNLSKTSLVG--------------------------HIPDEIS-H 350
Q + L Y +LS +SL +PD+ S H
Sbjct: 354 GQVPESFKNFESLSYFSLSNSSLANISSALGILQHCKNLTTLVLTLNFHGEALPDDSSLH 413
Query: 351 LGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI--LKSLPKMKKYNF 406
L L ++ L G +P L + LQ++D S N L+G +PS+I K+L + N
Sbjct: 414 FEKLKVLVVANCRLTGSMPRWLSSSNELQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNN 473
Query: 407 SYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSF-FKKRRDVGHRGMKLALVL----T 461
S+ EI + K S N +P F F +R+ R ++ + T
Sbjct: 474 SF------TGEIPKSLTKLESLTSRNISVNEPSPDFPFFMKRNESARALQYNQIFGFPPT 527
Query: 462 LSLIF-ALAGILFLAFGCRRKNKMWEVKQGSYREE--QNISGPFSFQTDSTTWVADVKQA 518
+ L L+G ++ FG +K ++++K + ++SG S + +++ + +
Sbjct: 528 IELGHNNLSGPIWEEFGNLKKLHVFDLKWNALSGSIPSSLSGMTSLEALD---LSNNRLS 584
Query: 519 TSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKF 556
S+PV L ++F S N G + + G+F
Sbjct: 585 GSIPV-----SLQQLSFLSKFSVAYNNLSGVIPSGGQF 617
Score = 110 bits (276), Expect = 3e-24
Identities = 102/343 (29%), Positives = 158/343 (45%), Gaps = 12/343 (3%)
Query: 81 PIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLL 140
P PD G +N S D N T S+ + + L L + +SG L+ ++G +
Sbjct: 48 PKPD---GWINSSSSTDCCNWTGITCNSN--NTGRVIRLELGNKKLSGKLSESLGKLDEI 102
Query: 141 ENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSG 200
+LS+N D IP ++ +L +L+ L L N IP+ I +L S DLSSN+ +G
Sbjct: 103 RVLNLSRNFIKDSIPLSIFNLKNLQTLDLSSNDLSGGIPTSI-NLPALQSFDLSSNKFNG 161
Query: 201 TLPHGFGDAFPKLRTLNLAENNIYGG-VSNFSRLKSIVSLNISGNSFQGSIIE-VFVLK- 257
+LP ++R + LA N G S F + + L + N G+I E +F LK
Sbjct: 162 SLPSHICHNSTQIRVVKLAVNYFAGNFTSGFGKCVLLEHLCLGMNDLTGNIPEDLFHLKR 221
Query: 258 LEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFS 317
L L + N+ G +S+ N S LV LD+S N SGEI + LK N F
Sbjct: 222 LNLLGIQENRLSGSLSREIRNLSSLVRLDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFI 281
Query: 318 RQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKH 375
+ L LNL SL G + + + LN+LDL N +G++P L K
Sbjct: 282 GGIPKSLANSPSLNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKR 341
Query: 376 LQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEI 418
L+ ++ + N G VP K+ + ++ S ++L +S +
Sbjct: 342 LKNVNLARNTFHGQVPE-SFKNFESLSYFSLSNSSLANISSAL 383
>At1g75640 receptor-like protein kinase, putative
Length = 1140
Score = 298 bits (763), Expect = 9e-81
Identities = 257/796 (32%), Positives = 380/796 (47%), Gaps = 93/796 (11%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGS 129
++ G +G IP + +L L ++ L N + +PSD SL L++LNL+ NH++G+
Sbjct: 391 VDFEGNKFSGQIPGF-LSQLRSLTTISLGRNGFSGRIPSDLLSLYGLETLNLNENHLTGA 449
Query: 130 LTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLV 189
+ + I L +LS N FS E+P + L SL VL + IP I L
Sbjct: 450 IPSEITKLANLTILNLSFNRFSGEVPSNVGDLKSLSVLNISGCGLTGRIPVSISGLMKLQ 509
Query: 190 SIDLSSNQLSGTLP-HGFGDAFPKLRTLNLAENNIYGGV--SNFSRLKSIVSLNISGNSF 246
+D+S ++SG LP FG P L+ + L NN+ GGV FS L S+ LN+S N F
Sbjct: 510 VLDISKQRISGQLPVELFG--LPDLQVVALG-NNLLGGVVPEGFSSLVSLKYLNLSSNLF 566
Query: 247 QGSIIEV--FVLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSM 304
G I + F+ L+ L LS N+ G I N S L L+L N L G I ++
Sbjct: 567 SGHIPKNYGFLKSLQVLSLSHNRISGTIPPEIGNCSSLEVLELGSNSLKGHIPVYVSKLS 626
Query: 305 NLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHL 364
LK L L+ N + +I LE L L+ SL G IP+ +S L NL ALDLS N L
Sbjct: 627 LLKKLDLSHNSLTGSIPDQISKDSSLESLLLNSNSLSGRIPESLSRLTNLTALDLSSNRL 686
Query: 365 DGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDI 422
+ IP L + + L + S N+L G +P + + N LC + +
Sbjct: 687 NSTIPSSLSRLRFLNYFNLSRNSLEGEIPEALAARFTNPTVF---VKNPGLCGKPLGIE- 742
Query: 423 MKTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCRRKN 482
CP +RR R KL L++TL++ AL L L C
Sbjct: 743 -----------CPNV-------RRR----RRRKLILLVTLAVAGAL---LLLLCCCGYVF 777
Query: 483 KMW----EVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADL 538
+W +++ G R+++ S + T D +V+F IT A+
Sbjct: 778 SLWKWRNKLRLGLSRDKKGTPSRTSRASSGGTRGEDNNGGPK--LVMFNN---KITLAET 832
Query: 539 LSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKH 598
L AT FD +L+ G++G V++ + ++V+ L+ G+++TD + E LGR+KH
Sbjct: 833 LEATRQFDEENVLSRGRYGLVFKATFRDGMVLSVRRLMDGASITDATFRNQAEALGRVKH 892
Query: 599 PNLVPLTG-YCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQN 657
N+ L G YC D R+ +YDYM NGNL LL +EA +
Sbjct: 893 KNITVLRGYYCGPPDLRLLVYDYMPNGNLATLL------------------QEASH---- 930
Query: 658 VGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGL 717
+G + W RH IALG AR L+FLH S IIH +K +V D D E LS+FGL
Sbjct: 931 --QDGHVLNWPMRHLIALGIARGLSFLH---SLSIIHGDLKPQNVLFDADFEAHLSEFGL 985
Query: 718 AKIFGSGLDEEIAR-----GSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPV 772
++ EE + GS GY+ PE + +SDVY FG+VL E+LTGKK V
Sbjct: 986 DRLTALTPAEEPSTSSTPVGSLGYIAPEAGLT--GETSKESDVYSFGIVLLEILTGKKAV 1043
Query: 773 GDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSD-EQIEEALK--EVGYLCTAD 829
+T+D++ +V WV+ ++K Q ++P + + + + EE L +VG LCT
Sbjct: 1044 --MFTEDED---IVKWVKRQLQKGQIVELLEPGLLELDPESSEWEEFLLGIKVGLLCTGG 1098
Query: 830 LPFKRPTMQQIVGLLK 845
RP+M +V +L+
Sbjct: 1099 DVVDRPSMADVVFMLE 1114
Score = 141 bits (355), Expect = 2e-33
Identities = 119/385 (30%), Positives = 181/385 (46%), Gaps = 39/385 (10%)
Query: 64 NKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLS 122
N ++ LN + LTG + D T+ K L +DLS+N I+ +P++F + +SL+ +NLS
Sbjct: 138 NLRNLQVLNAAHNSLTGNLSDVTVSK--SLRYVDLSSNAISGKIPANFSADSSLQLINLS 195
Query: 123 SNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGI 182
NH SG + +G LE L N IP AL++ SL + N IP +
Sbjct: 196 FNHFSGEIPATLGQLQDLEYLWLDSNQLQGTIPSALANCSSLIHFSVTGNHLTGLIPVTL 255
Query: 183 LKCQSLVSIDLSSNQLSGTLP-------HGFGDAF-----------------------PK 212
+SL I LS N +GT+P G+ + P
Sbjct: 256 GTIRSLQVISLSENSFTGTVPVSLLCGYSGYNSSMRIIQLGVNNFTGIAKPSNAACVNPN 315
Query: 213 LRTLNLAENNIYGGVSNF-SRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQ 269
L L++ EN I G + + L S+V L+ISGN F G + ++ L+ L ++ N
Sbjct: 316 LEILDIHENRINGDFPAWLTDLTSLVVLDISGNGFSGGVTAKVGNLMALQELRVANNSLV 375
Query: 270 GHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLG 329
G I N L +D N+ SG+I L+ +L +SL N FS + + L G
Sbjct: 376 GEIPTSIRNCKSLRVVDFEGNKFSGQIPGFLSQLRSLTTISLGRNGFSGRIPSDLLSLYG 435
Query: 330 LEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLS 387
LE LNL++ L G IP EI+ L NL L+LS N G++P + K L V++ S L+
Sbjct: 436 LETLNLNENHLTGAIPSEITKLANLTILNLSFNRFSGEVPSNVGDLKSLSVLNISGCGLT 495
Query: 388 GPVPSFILKSLPKMKKYNFSYNNLT 412
G +P I L K++ + S ++
Sbjct: 496 GRIPVSI-SGLMKLQVLDISKQRIS 519
Score = 98.6 bits (244), Expect = 1e-20
Identities = 100/374 (26%), Positives = 160/374 (42%), Gaps = 72/374 (19%)
Query: 93 LHSLDLSNNKITTLPSDFWSLTS----LKSLNLSSNHISGSLTNNIGNFGLLENFDLSKN 148
L +L+ N + P D+ ++ ++ L L H++G L+ +G L L N
Sbjct: 43 LGALESWNQSSPSAPCDWHGVSCFSGRVRELRLPRLHLTGHLSPRLGELTQLRKLSLHTN 102
Query: 149 SFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGD 208
+ +P +LS V L+ L L +N F P IL ++L ++ + N L+G L
Sbjct: 103 DINGAVPSSLSRCVFLRALYLHYNSFSGDFPPEILNLRNLQVLNAAHNSLTGNLSD--VT 160
Query: 209 AFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSR 265
LR ++L+ N I G + +NFS S+ +N+S N F G I + LE L L
Sbjct: 161 VSKSLRYVDLSSNAISGKIPANFSADSSLQLINLSFNHFSGEIPATLGQLQDLEYLWLDS 220
Query: 266 NQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIE 325
NQ QG I N S L++ ++ N L+G I L +L+ +SL+ N F+ +
Sbjct: 221 NQLQGTIPSALANCSSLIHFSVTGNHLTGLIPVTLGTIRSLQVISLSENSFTGT--VPVS 278
Query: 326 MLLG---------------------------------LEYLNLSKTSLVGHIPDEISHLG 352
+L G LE L++ + + G P ++ L
Sbjct: 279 LLCGYSGYNSSMRIIQLGVNNFTGIAKPSNAACVNPNLEILDIHENRINGDFPAWLTDLT 338
Query: 353 NLNALDLS------------------------MNHLDGKIPL-LKN-KHLQVIDFSHNNL 386
+L LD+S N L G+IP ++N K L+V+DF N
Sbjct: 339 SLVVLDISGNGFSGGVTAKVGNLMALQELRVANNSLVGEIPTSIRNCKSLRVVDFEGNKF 398
Query: 387 SGPVPSFI--LKSL 398
SG +P F+ L+SL
Sbjct: 399 SGQIPGFLSQLRSL 412
>At5g49660 receptor protein kinase
Length = 966
Score = 297 bits (761), Expect = 1e-80
Identities = 246/798 (30%), Positives = 382/798 (47%), Gaps = 92/798 (11%)
Query: 78 LTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSN-HISGSLTNNIG 135
L G IP +IG L L L+LS N ++ +P + +L++L+ L L N H++GS+ IG
Sbjct: 207 LHGNIP-RSIGNLTSLVDLELSGNFLSGEIPKEIGNLSNLRQLELYYNYHLTGSIPEEIG 265
Query: 136 NFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSS 195
N L + D+S + + IP+++ SL +L+VL+L +N IP + ++L + L
Sbjct: 266 NLKNLTDIDISVSRLTGSIPDSICSLPNLRVLQLYNNSLTGEIPKSLGNSKTLKILSLYD 325
Query: 196 NQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF 254
N L+G LP G + P + L+++EN + G + ++ + ++ + N F GSI E +
Sbjct: 326 NYLTGELPPNLGSSSPMI-ALDVSENRLSGPLPAHVCKSGKLLYFLVLQNRFTGSIPETY 384
Query: 255 --VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLA 312
L ++ N+ G I Q + H+ +DL+ N LSG I + N+ NL L +
Sbjct: 385 GSCKTLIRFRVASNRLVGTIPQGVMSLPHVSIIDLAYNSLSGPIPNAIGNAWNLSELFMQ 444
Query: 313 CNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--L 370
NR S ++ L L+LS L G IP E+ L LN L L NHLD IP L
Sbjct: 445 SNRISGVIPHELSHSTNLVKLDLSNNQLSGPIPSEVGRLRKLNLLVLQGNHLDSSIPDSL 504
Query: 371 LKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT--LCASEIKPDIMKTSFF 428
K L V+D S N L+G +P + + LP NFS N L+ + S I+ +++ SF
Sbjct: 505 SNLKSLNVLDLSSNLLTGRIPENLSELLPT--SINFSSNRLSGPIPVSLIRGGLVE-SFS 561
Query: 429 GSVNSC--PIAANPSFFKKRRDVGHRGMKLALV--LTLSLIFALAGILFLAFGCRRKNKM 484
+ N C P A + H KL+ + + +S+ + G++ R
Sbjct: 562 DNPNLCIPPTAGSSDLKFPMCQEPHGKKKLSSIWAILVSVFILVLGVIMFYLRQRMSKNR 621
Query: 485 WEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSN 544
++Q +E S FS+ K I+F S
Sbjct: 622 AVIEQ----DETLASSFFSYDV---------------------KSFHRISFDQREILESL 656
Query: 545 FDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLT---------DEEAARELEFLGR 595
D+ ++ G G VYR L VAVK L S ++E E+E LG
Sbjct: 657 VDKN-IVGHGGSGTVYRVELKSGEVVAVKKLWSQSNKDSASEDKMHLNKELKTEVETLGS 715
Query: 596 IKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGI 655
I+H N+V L Y + D + +Y+YM NGNL W+ G
Sbjct: 716 IRHKNIVKLFSYFSSLDCSLLVYEYMPNGNL---------------------WDALHKGF 754
Query: 656 QNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDF 715
++ WR RH+IA+G A+ LA+LHH SPPIIHR +K++++ LD + +P+++DF
Sbjct: 755 VHL-------EWRTRHQIAVGVAQGLAYLHHDLSPPIIHRDIKSTNILLDVNYQPKVADF 807
Query: 716 GLAKIF---GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPV 772
G+AK+ G + G+ GY+ PE++ T K DVY FGVVL EL+TGKKPV
Sbjct: 808 GIAKVLQARGKDSTTTVMAGTYGYLAPEYAYS--SKATIKCDVYSFGVVLMELITGKKPV 865
Query: 773 GDDYTDDKEATTLVSWVRGLV-RKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLP 831
+ ++K +V+WV + K +D ++ ++ + I AL+ V CT+ P
Sbjct: 866 DSCFGENK---NIVNWVSTKIDTKEGLIETLDKRLSESSKADMI-NALR-VAIRCTSRTP 920
Query: 832 FKRPTMQQIVGLLKDIEP 849
RPTM ++V LL D P
Sbjct: 921 TIRPTMNEVVQLLIDATP 938
Score = 133 bits (334), Expect = 5e-31
Identities = 114/387 (29%), Positives = 182/387 (46%), Gaps = 36/387 (9%)
Query: 42 SSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNN 101
S+ VY+ ++ C++ GV CD + V +L+LSG+ L+G PD L L LS+N
Sbjct: 48 STWNVYDVGTNYCNFTGVRCDG-QGLVTDLDLSGLSLSGIFPDGVCSYFPNLRVLRLSHN 106
Query: 102 KIT-------TLPS-------------------DFWSLTSLKSLNLSSNHISGSLTNNIG 135
+ T+P+ DF + SL+ +++S NH +GS +I
Sbjct: 107 HLNKSSSFLNTIPNCSLLRDLNMSSVYLKGTLPDFSQMKSLRVIDMSWNHFTGSFPLSIF 166
Query: 136 NFGLLENFDLSKNSFSD--EIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDL 193
N LE + ++N D +P+++S L L + L M +IP I SLV ++L
Sbjct: 167 NLTDLEYLNFNENPELDLWTLPDSVSKLTKLTHMLLMTCMLHGNIPRSIGNLTSLVDLEL 226
Query: 194 SSNQLSGTLPHGFGDAFPKLRTLNLAEN-NIYGGV-SNFSRLKSIVSLNISGNSFQGSII 251
S N LSG +P G+ LR L L N ++ G + LK++ ++IS + GSI
Sbjct: 227 SGNFLSGEIPKEIGN-LSNLRQLELYYNYHLTGSIPEEIGNLKNLTDIDISVSRLTGSIP 285
Query: 252 EVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHL 309
+ + L L L N G I + N L L L +N L+GE+ NL +S + L
Sbjct: 286 DSICSLPNLRVLQLYNNSLTGEIPKSLGNSKTLKILSLYDNYLTGELPPNLGSSSPMIAL 345
Query: 310 SLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP 369
++ NR S + L Y + + G IP+ L ++ N L G IP
Sbjct: 346 DVSENRLSGPLPAHVCKSGKLLYFLVLQNRFTGSIPETYGSCKTLIRFRVASNRLVGTIP 405
Query: 370 --LLKNKHLQVIDFSHNNLSGPVPSFI 394
++ H+ +ID ++N+LSGP+P+ I
Sbjct: 406 QGVMSLPHVSIIDLAYNSLSGPIPNAI 432
Score = 80.9 bits (198), Expect = 3e-15
Identities = 61/207 (29%), Positives = 102/207 (48%), Gaps = 3/207 (1%)
Query: 68 VVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKSLNLSSNHIS 127
++ L++S L+GP+P L+ L L N ++P + S +L ++SN +
Sbjct: 342 MIALDVSENRLSGPLPAHVCKSGKLLYFLVLQNRFTGSIPETYGSCKTLIRFRVASNRLV 401
Query: 128 GSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQS 187
G++ + + + DL+ NS S IP A+ + +L L + N IP + +
Sbjct: 402 GTIPQGVMSLPHVSIIDLAYNSLSGPIPNAIGNAWNLSELFMQSNRISGVIPHELSHSTN 461
Query: 188 LVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSF 246
LV +DLS+NQLSG +P G KL L L N++ + + S LKS+ L++S N
Sbjct: 462 LVKLDLSNNQLSGPIPSEVG-RLRKLNLLVLQGNHLDSSIPDSLSNLKSLNVLDLSSNLL 520
Query: 247 QGSIIE-VFVLKLEALDLSRNQFQGHI 272
G I E + L +++ S N+ G I
Sbjct: 521 TGRIPENLSELLPTSINFSSNRLSGPI 547
Score = 75.9 bits (185), Expect = 9e-14
Identities = 49/138 (35%), Positives = 76/138 (54%), Gaps = 3/138 (2%)
Query: 67 HVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNH 125
HV ++L+ L+GPIP+ IG L L + +N+I+ +P + T+L L+LS+N
Sbjct: 413 HVSIIDLAYNSLSGPIPNA-IGNAWNLSELFMQSNRISGVIPHELSHSTNLVKLDLSNNQ 471
Query: 126 ISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKC 185
+SG + + +G L L N IP++LS+L SL VL L N+ IP + +
Sbjct: 472 LSGPIPSEVGRLRKLNLLVLQGNHLDSSIPDSLSNLKSLNVLDLSSNLLTGRIPENLSEL 531
Query: 186 QSLVSIDLSSNQLSGTLP 203
SI+ SSN+LSG +P
Sbjct: 532 LP-TSINFSSNRLSGPIP 548
Score = 54.3 bits (129), Expect = 3e-07
Identities = 35/97 (36%), Positives = 58/97 (59%), Gaps = 3/97 (3%)
Query: 63 SNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKI-TTLPSDFWSLTSLKSLNL 121
S+ ++V+L+LS L+GPIP + +G+L KL+ L L N + +++P +L SL L+L
Sbjct: 457 SHSTNLVKLDLSNNQLSGPIP-SEVGRLRKLNLLVLQGNHLDSSIPDSLSNLKSLNVLDL 515
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEAL 158
SSN ++G + N+ L + + S N S IP +L
Sbjct: 516 SSNLLTGRIPENLSEL-LPTSINFSSNRLSGPIPVSL 551
>At5g01890 unknown protein
Length = 967
Score = 292 bits (748), Expect = 5e-79
Identities = 254/892 (28%), Positives = 396/892 (43%), Gaps = 166/892 (18%)
Query: 71 LNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-------------------------T 105
++ SG L+G IPD + L S+ L+NNK+T
Sbjct: 121 VDFSGNNLSGRIPDGFFEQCGSLRSVSLANNKLTGSIPVSLSYCSTLTHLNLSSNQLSGR 180
Query: 106 LPSDFWSLTSLKSL------------------------NLSSNHISGSLTNNIGNFGLLE 141
LP D W L SLKSL NLS N SG + ++IG L+
Sbjct: 181 LPRDIWFLKSLKSLDFSHNFLQGDIPDGLGGLYDLRHINLSRNWFSGDVPSDIGRCSSLK 240
Query: 142 NFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGT 201
+ DLS+N FS +P+++ SL S ++L N + IP I +L +DLS+N +GT
Sbjct: 241 SLDLSENYFSGNLPDSMKSLGSCSSIRLRGNSLIGEIPDWIGDIATLEILDLSANNFTGT 300
Query: 202 LPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIE-------- 252
+P G+ L+ LNL+ N + G + S +++S+++S NSF G +++
Sbjct: 301 VPFSLGN-LEFLKDLNLSANMLAGELPQTLSNCSNLISIDVSKNSFTGDVLKWMFTGNSE 359
Query: 253 ----------------------VFVLKLEALDLSRNQFQGHI--------SQVKYNWSH- 281
F+ L LDLS N F G + S ++ N S
Sbjct: 360 SSSLSRFSLHKRSGNDTIMPIVGFLQGLRVLDLSSNGFTGELPSNIWILTSLLQLNMSTN 419
Query: 282 ---------------LVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEM 326
LDLS N L+G + + +++LK L L NR S Q KI
Sbjct: 420 SLFGSIPTGIGGLKVAEILDLSSNLLNGTLPSEIGGAVSLKQLHLHRNRLSGQIPAKISN 479
Query: 327 LLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHN 384
L +NLS+ L G IP I L NL +DLS N+L G +P + K HL + SHN
Sbjct: 480 CSALNTINLSENELSGAIPGSIGSLSNLEYIDLSRNNLSGSLPKEIEKLSHLLTFNISHN 539
Query: 385 NLSGPVPS-FILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFF 443
N++G +P+ ++P + N +LC S + + SV+ PI NP+
Sbjct: 540 NITGELPAGGFFNTIP----LSAVTGNPSLCGSVVNRSCL------SVHPKPIVLNPNSS 589
Query: 444 KKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFS 503
G VL++S + A+ +A G + V S + + +
Sbjct: 590 NPTNGPALTGQIRKSVLSISALIAIGAAAVIAIGVVAVT-LLNVHARSSVSRHDAAAALA 648
Query: 504 FQTDSTTWVADVKQATSVPVVIF--EKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYR 561
T + K +V+F E + + T AD L ++ + L G FG VY+
Sbjct: 649 LSVGETFSCSPSKDQEFGKLVMFSGEVDVFDTTGADAL-----LNKDSELGRGGFGVVYK 703
Query: 562 GFLPGNIHVAVKVLVV-GSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDY 620
L VAVK L V G + EE RE+ LG+++H N+V + GY ++ I+++
Sbjct: 704 TSLQDGRPVAVKKLTVSGLIKSQEEFEREMRKLGKLRHKNVVEIKGYYWTQSLQLLIHEF 763
Query: 621 MENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARA 680
+ G+L L+ G E + TWR R I LG AR
Sbjct: 764 VSGGSLYRHLH---------------------------GDESVCLTWRQRFSIILGIARG 796
Query: 681 LAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLDEEIARG----SPGY 736
LAFLH S I H +KA++V +D E ++SDFGLA++ S LD + G + GY
Sbjct: 797 LAFLH---SSNITHYNMKATNVLIDAAGEAKVSDFGLARLLASALDRCVLSGKVQSALGY 853
Query: 737 VPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKN 796
PEF+ + T + DVY FG+++ E++TGK+PV +Y +D + L VR + +
Sbjct: 854 TAPEFACRTVKI-TDRCDVYGFGILVLEVVTGKRPV--EYAED-DVVVLCETVREGLEEG 909
Query: 797 QTSRAIDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIE 848
+ +DP++ E+ +K +G +C + +P RP M+++V +L+ I+
Sbjct: 910 RVEECVDPRLRGNFPAEEAIPVIK-LGLVCGSQVPSNRPEMEEVVKILELIQ 960
Score = 133 bits (335), Expect = 4e-31
Identities = 145/515 (28%), Positives = 216/515 (41%), Gaps = 115/515 (22%)
Query: 5 VFGSVLVLTFLFKHLASQQ--PNTDEFYVSEFLKKMGLTSS-SKVYNFSSS---VCSWKG 58
+F + L FLF + S + P ++ + + K GL SK+ +++S C+W G
Sbjct: 1 MFNGAVSLLFLFLAVVSARADPTFNDDVLGLIVFKAGLDDPLSKLSSWNSEDYDPCNWVG 60
Query: 59 VYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLTSLKS 118
CD V EL L L+G I + +L LH+L LSNN
Sbjct: 61 CTCDPATNRVSELRLDAFSLSGHIGRGLL-RLQFLHTLVLSNN----------------- 102
Query: 119 LNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEA-LSSLVSLKVLKLDHNMFVRS 177
+++G+L + G L+ D S N+ S IP+ SL+ + L +N S
Sbjct: 103 ------NLTGTLNPEFPHLGSLQVVDFSGNNLSGRIPDGFFEQCGSLRSVSLANNKLTGS 156
Query: 178 IPSGILKCQSLVSIDLSSNQLSGTLP---------------HGF--GD------AFPKLR 214
IP + C +L ++LSSNQLSG LP H F GD LR
Sbjct: 157 IPVSLSYCSTLTHLNLSSNQLSGRLPRDIWFLKSLKSLDFSHNFLQGDIPDGLGGLYDLR 216
Query: 215 TLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF------------------- 254
+NL+ N G V S+ R S+ SL++S N F G++ +
Sbjct: 217 HINLSRNWFSGDVPSDIGRCSSLKSLDLSENYFSGNLPDSMKSLGSCSSIRLRGNSLIGE 276
Query: 255 -------VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLK 307
+ LE LDLS N F G + N L L+LS N L+GE+ Q L+N NL
Sbjct: 277 IPDWIGDIATLEILDLSANNFTGTVPFSLGNLEFLKDLNLSANMLAGELPQTLSNCSNLI 336
Query: 308 HLSLACN---------------------RFSRQK-------FPKIEMLLGLEYLNLSKTS 339
+ ++ N RFS K P + L GL L+LS
Sbjct: 337 SIDVSKNSFTGDVLKWMFTGNSESSSLSRFSLHKRSGNDTIMPIVGFLQGLRVLDLSSNG 396
Query: 340 LVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFILKS 397
G +P I L +L L++S N L G IP + K +++D S N L+G +PS I +
Sbjct: 397 FTGELPSNIWILTSLLQLNMSTNSLFGSIPTGIGGLKVAEILDLSSNLLNGTLPSEIGGA 456
Query: 398 LPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVN 432
+ +K+ + N L + +I I S ++N
Sbjct: 457 V-SLKQLHLHRNRL---SGQIPAKISNCSALNTIN 487
Score = 113 bits (282), Expect = 5e-25
Identities = 88/288 (30%), Positives = 135/288 (46%), Gaps = 22/288 (7%)
Query: 149 SFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGD 208
S S I L L L L L +N ++ SL +D S N LSG +P GF +
Sbjct: 79 SLSGHIGRGLLRLQFLHTLVLSNNNLTGTLNPEFPHLGSLQVVDFSGNNLSGRIPDGFFE 138
Query: 209 AFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGNSFQGSI-IEVFVLK-LEALDLSR 265
LR+++LA N + G + + S ++ LN+S N G + +++ LK L++LD S
Sbjct: 139 QCGSLRSVSLANNKLTGSIPVSLSYCSTLTHLNLSSNQLSGRLPRDIWFLKSLKSLDFSH 198
Query: 266 NQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIE 325
N QG I L +++LS N SG++ ++ +LK L L+ N FS ++
Sbjct: 199 NFLQGDIPDGLGGLYDLRHINLSRNWFSGDVPSDIGRCSSLKSLDLSENYFSGNLPDSMK 258
Query: 326 MLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSH 383
L + L SL+G IPD I + L LDLS N+ G +P L + L+ ++ S
Sbjct: 259 SLGSCSSIRLRGNSLIGEIPDWIGDIATLEILDLSANNFTGTVPFSLGNLEFLKDLNLSA 318
Query: 384 NNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSV 431
N L+G +P L+ C++ I D+ K SF G V
Sbjct: 319 NMLAGELP-----------------QTLSNCSNLISIDVSKNSFTGDV 349
>At5g56040 receptor protein kinase-like protein
Length = 1090
Score = 288 bits (736), Expect = 1e-77
Identities = 245/817 (29%), Positives = 382/817 (45%), Gaps = 124/817 (15%)
Query: 78 LTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGN 136
L G IP T +G +L +DLS N +T +P F +L +L+ L LS N +SG++ + N
Sbjct: 301 LVGKIP-TELGTCPELFLVDLSENLLTGNIPRSFGNLPNLQELQLSVNQLSGTIPEELAN 359
Query: 137 FGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSN 196
L + ++ N S EIP + L SL + N IP + +CQ L +IDLS N
Sbjct: 360 CTKLTHLEIDNNQISGEIPPLIGKLTSLTMFFAWQNQLTGIIPESLSQCQELQAIDLSYN 419
Query: 197 QLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGNSFQGSI-IEVF 254
LSG++P+G + L L L N + G + + ++ L ++GN G+I E+
Sbjct: 420 NLSGSIPNGIFE-IRNLTKLLLLSNYLSGFIPPDIGNCTNLYRLRLNGNRLAGNIPAEIG 478
Query: 255 VLK-LEALDLSRNQFQGHI-------SQVKYNWSH---------------LVYLDLSENQ 291
LK L +D+S N+ G+I + +++ H L ++DLS+N
Sbjct: 479 NLKNLNFIDISENRLIGNIPPEISGCTSLEFVDLHSNGLTGGLPGTLPKSLQFIDLSDNS 538
Query: 292 LSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHL 351
L+G + + + L L+LA NRFS + +I L+ LNL G IP+E+ +
Sbjct: 539 LTGSLPTGIGSLTELTKLNLAKNRFSGEIPREISSCRSLQLLNLGDNGFTGEIPNELGRI 598
Query: 352 GNLN-ALDLSMNHLDGKIPLLKNK--HLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSY 408
+L +L+LS NH G+IP + +L +D SHN L+G + +L L + N S+
Sbjct: 599 PSLAISLNLSCNHFTGEIPSRFSSLTNLGTLDVSHNKLAGNLN--VLADLQNLVSLNISF 656
Query: 409 NNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFFKKRR-----DVGHRGMKLALVLTLS 463
N E ++ T FF + + +N F R HR A+ +T+S
Sbjct: 657 N-------EFSGELPNTLFFRKLPLSVLESNKGLFISTRPENGIQTRHRS---AVKVTMS 706
Query: 464 LIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPV 523
++ A + +L L + + + Q I+G Q + +W V
Sbjct: 707 ILVAASVVLVL------------MAVYTLVKAQRITGK---QEELDSW----------EV 741
Query: 524 VIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVLVVGSTLTD 583
+++K L+ + D++ N ++ G G VYR +P +AVK + S +
Sbjct: 742 TLYQK--LDFSIDDIVK---NLTSANVIGTGSSGVVYRVTIPSGETLAVKKM--WSKEEN 794
Query: 584 EEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDW 643
E+ LG I+H N++ L G+C + ++ YDY+ NG+L +LL+ G D
Sbjct: 795 RAFNSEINTLGSIRHRNIIRLLGWCSNRNLKLLFYDYLPNGSLSSLLHGAGKGSGGAD-- 852
Query: 644 STDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVY 703
W R+ + LG A ALA+LHH C PPI+H VKA +V
Sbjct: 853 -----------------------WEARYDVVLGVAHALAYLHHDCLPPILHGDVKAMNVL 889
Query: 704 LDYDLEPRLSDFGLAKIF-GSGLDEEIA---------RGSPGYVPPEFSQPEFESPTPKS 753
L E L+DFGLAKI G G+ + + GS GY+ PE + + T KS
Sbjct: 890 LGSRFESYLADFGLAKIVSGEGVTDGDSSKLSNRPPLAGSYGYMAPEHA--SMQHITEKS 947
Query: 754 DVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRG-LVRKNQTSRAIDPKICDTGSD 812
DVY +GVVL E+LTGK P+ D LV WVR L K +DP++ +D
Sbjct: 948 DVYSYGVVLLEVLTGKHPLDPDLPG---GAHLVQWVRDHLAGKKDPREILDPRLRGR-AD 1003
Query: 813 EQIEEALK--EVGYLCTADLPFKRPTMQQIVGLLKDI 847
+ E L+ V +LC ++ RP M+ IV +LK+I
Sbjct: 1004 PIMHEMLQTLAVSFLCVSNKASDRPMMKDIVAMLKEI 1040
Score = 149 bits (377), Expect = 5e-36
Identities = 127/414 (30%), Positives = 202/414 (48%), Gaps = 17/414 (4%)
Query: 3 LGVFGSVLVLTFLFKHLASQQPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVYCD 62
LG +++ LT LA + P T + E LK + + + N + W+ C+
Sbjct: 164 LGNLVNLIELTLFDNKLAGEIPRT----IGE-LKNLEIFRAGGNKNLRGEL-PWEIGNCE 217
Query: 63 SNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNL 121
S +V L L+ L+G +P +IG L K+ ++ L + ++ +P + + T L++L L
Sbjct: 218 S----LVTLGLAETSLSGRLP-ASIGNLKKVQTIALYTSLLSGPIPDEIGNCTELQNLYL 272
Query: 122 SSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSG 181
N ISGS+ ++G L++ L +N+ +IP L + L ++ L N+ +IP
Sbjct: 273 YQNSISGSIPVSMGRLKKLQSLLLWQNNLVGKIPTELGTCPELFLVDLSENLLTGNIPRS 332
Query: 182 ILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNF-SRLKSIVSLN 240
+L + LS NQLSGT+P + KL L + N I G + +L S+
Sbjct: 333 FGNLPNLQELQLSVNQLSGTIPEELANC-TKLTHLEIDNNQISGEIPPLIGKLTSLTMFF 391
Query: 241 ISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQ 298
N G I E +L+A+DLS N G I + +L L L N LSG I
Sbjct: 392 AWQNQLTGIIPESLSQCQELQAIDLSYNNLSGSIPNGIFEIRNLTKLLLLSNYLSGFIPP 451
Query: 299 NLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALD 358
++ N NL L L NR + +I L L ++++S+ L+G+IP EIS +L +D
Sbjct: 452 DIGNCTNLYRLRLNGNRLAGNIPAEIGNLKNLNFIDISENRLIGNIPPEISGCTSLEFVD 511
Query: 359 LSMNHLDGKIPLLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
L N L G +P K LQ ID S N+L+G +P+ I SL ++ K N + N +
Sbjct: 512 LHSNGLTGGLPGTLPKSLQFIDLSDNSLTGSLPTGI-GSLTELTKLNLAKNRFS 564
Score = 126 bits (317), Expect = 4e-29
Identities = 114/406 (28%), Positives = 177/406 (43%), Gaps = 79/406 (19%)
Query: 51 SSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDF 110
S+ C W G+ C+ + V E+ L + GP+P T + ++ L
Sbjct: 57 SNPCQWVGIKCNERGQ-VSEIQLQVMDFQGPLPATNLRQIKSL----------------- 98
Query: 111 WSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLD 170
+L SL S+NL+ GS+ +G+ LE DL+ NS S EIP + L LK+L L+
Sbjct: 99 -TLLSLTSVNLT-----GSIPKELGDLSELEVLDLADNSLSGEIPVDIFKLKKLKILSLN 152
Query: 171 HNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDA--------------------- 209
N IPS + +L+ + L N+L+G +P G+
Sbjct: 153 TNNLEGVIPSELGNLVNLIELTLFDNKLAGEIPRTIGELKNLEIFRAGGNKNLRGELPWE 212
Query: 210 ---FPKLRTLNLAENNIYG----GVSNFSRLKSIV---------------------SLNI 241
L TL LAE ++ G + N ++++I +L +
Sbjct: 213 IGNCESLVTLGLAETSLSGRLPASIGNLKKVQTIALYTSLLSGPIPDEIGNCTELQNLYL 272
Query: 242 SGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
NS GSI + KL++L L +N G I L +DLSEN L+G I ++
Sbjct: 273 YQNSISGSIPVSMGRLKKLQSLLLWQNNLVGKIPTELGTCPELFLVDLSENLLTGNIPRS 332
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
N NL+ L L+ N+ S ++ L +L + + G IP I L +L
Sbjct: 333 FGNLPNLQELQLSVNQLSGTIPEELANCTKLTHLEIDNNQISGEIPPLIGKLTSLTMFFA 392
Query: 360 SMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPS--FILKSLPKM 401
N L G IP L + + LQ ID S+NNLSG +P+ F +++L K+
Sbjct: 393 WQNQLTGIIPESLSQCQELQAIDLSYNNLSGSIPNGIFEIRNLTKL 438
>At5g62230 unknown protein
Length = 966
Score = 287 bits (734), Expect = 2e-77
Identities = 256/829 (30%), Positives = 382/829 (45%), Gaps = 104/829 (12%)
Query: 30 YVSEFLKKMGLTSSSKVYNFSSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGK 89
Y +E L+ +GL + SS +C G++ ++ G LTG IP++ IG
Sbjct: 188 YWNEVLQYLGLRGNMLTGTLSSDMCQLTGLWY---------FDVRGNNLTGTIPES-IGN 237
Query: 90 LNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKN 148
LD+S N+IT +P + L + +L+L N ++G + IG L DLS N
Sbjct: 238 CTSFQILDISYNQITGEIPYNIGFL-QVATLSLQGNRLTGRIPEVIGLMQALAVLDLSDN 296
Query: 149 SFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGD 208
IP L +L L L NM IPS + L + L+ N+L GT+P G
Sbjct: 297 ELVGPIPPILGNLSFTGKLYLHGNMLTGPIPSELGNMSRLSYLQLNDNKLVGTIPPELGK 356
Query: 209 AFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSR 265
+L LNLA N + G + SN S ++ N+ GN GSI F + L L+LS
Sbjct: 357 -LEQLFELNLANNRLVGPIPSNISSCAALNQFNVHGNLLSGSIPLAFRNLGSLTYLNLSS 415
Query: 266 NQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIE 325
N F+G I + +L LDLS N SG I L + +L L+L+ N S Q +
Sbjct: 416 NNFKGKIPVELGHIINLDKLDLSGNNFSGSIPLTLGDLEHLLILNLSRNHLSGQLPAEFG 475
Query: 326 MLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSH 383
L ++ +++S L G IP E+ L NLN+L L+ N L GKIP L L ++ S
Sbjct: 476 NLRSIQMIDVSFNLLSGVIPTELGQLQNLNSLILNNNKLHGKIPDQLTNCFTLVNLNVSF 535
Query: 384 NNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDIMKTSFFGSVNSCPIAANPSFF 443
NNLSG VP +K+ + +F N LC + + GS+ P+ + F
Sbjct: 536 NNLSGIVPP--MKNFSRFAPASF-VGNPYLCGNWV----------GSICG-PLPKSRVF- 580
Query: 444 KKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCRRKNKMWEVKQGSYREEQNISGPFS 503
RG + +VL + L ++FLA + K ++ QGS ++ + ++
Sbjct: 581 -------SRGALICIVLG---VITLLCMIFLAVYKSMQQK--KILQGSSKQAEGLT---- 624
Query: 504 FQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGF 563
+VI + TF D++ T N + ++ G VY+
Sbjct: 625 ------------------KLVILHMDMAIHTFDDIMRVTENLNEKFIIGYGASSTVYKCA 666
Query: 564 LPGNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMEN 623
L + +A+K L E ELE +G I+H N+V L GY ++ + YDYMEN
Sbjct: 667 LKSSRPIAIKRLYNQYPHNLREFETELETIGSIRHRNIVSLHGYALSPTGNLLFYDYMEN 726
Query: 624 GNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAF 683
G+L +LL+ V+ DW T R KIA+G A+ LA+
Sbjct: 727 GSLWDLLHGSLKKVKL--DWET------------------------RLKIAVGAAQGLAY 760
Query: 684 LHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKIFGSGLD--EEIARGSPGYVPPEF 741
LHH C+P IIHR +K+S++ LD + E LSDFG+AK + G+ GY+ PE+
Sbjct: 761 LHHDCTPRIIHRDIKSSNILLDENFEAHLSDFGIAKSIPASKTHASTYVLGTIGYIDPEY 820
Query: 742 SQPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRA 801
++ KSD+Y FG+VL ELLTGKK V D EA L + N A
Sbjct: 821 ART--SRINEKSDIYSFGIVLLELLTGKKAV------DNEA-NLHQLILSKADDNTVMEA 871
Query: 802 IDPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDIEPT 850
+DP++ T D ++ LCT P +RPTM ++ +L + P+
Sbjct: 872 VDPEVTVTCMDLGHIRKTFQLALLCTKRNPLERPTMLEVSRVLLSLVPS 920
Score = 139 bits (350), Expect = 7e-33
Identities = 119/415 (28%), Positives = 172/415 (40%), Gaps = 78/415 (18%)
Query: 50 SSSVCSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSD 109
+S +CSW+GV+CD+ VV LNLS + L G I IG L L S+DL NK+
Sbjct: 55 NSDLCSWRGVFCDNVSYSVVSLNLSSLNLGGEI-SPAIGDLRNLQSIDLQGNKL------ 107
Query: 110 FWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKL 169
+G + + IGN L DLS+N +IP ++S L L+ L L
Sbjct: 108 -----------------AGQIPDEIGNCASLVYLDLSENLLYGDIPFSISKLKQLETLNL 150
Query: 170 DHNMFVRSIPSGILKCQSLVSIDLSS---------------------------------- 195
+N +P+ + + +L +DL+
Sbjct: 151 KNNQLTGPVPATLTQIPNLKRLDLAGNHLTGEISRLLYWNEVLQYLGLRGNMLTGTLSSD 210
Query: 196 --------------NQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSNFSRLKSIVSLNI 241
N L+GT+P G+ + L+++ N I G + + +L++
Sbjct: 211 MCQLTGLWYFDVRGNNLTGTIPESIGNC-TSFQILDISYNQITGEIPYNIGFLQVATLSL 269
Query: 242 SGNSFQGSIIEVFVL--KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQN 299
GN G I EV L L LDLS N+ G I + N S L L N L+G I
Sbjct: 270 QGNRLTGRIPEVIGLMQALAVLDLSDNELVGPIPPILGNLSFTGKLYLHGNMLTGPIPSE 329
Query: 300 LNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDL 359
L N L +L L N+ P++ L L LNL+ LVG IP IS LN ++
Sbjct: 330 LGNMSRLSYLQLNDNKLVGTIPPELGKLEQLFELNLANNRLVGPIPSNISSCAALNQFNV 389
Query: 360 SMNHLDGKIPLLKNK--HLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLT 412
N L G IPL L ++ S NN G +P L + + K + S NN +
Sbjct: 390 HGNLLSGSIPLAFRNLGSLTYLNLSSNNFKGKIP-VELGHIINLDKLDLSGNNFS 443
>At5g53890 receptor protein kinase-like protein
Length = 1036
Score = 286 bits (733), Expect = 3e-77
Identities = 261/885 (29%), Positives = 389/885 (43%), Gaps = 171/885 (19%)
Query: 68 VVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHI 126
+ +L++ LTG +PD + + +L L LS N ++ L + +L+ LKSL +S N
Sbjct: 210 IQQLHIDSNRLTGQLPDY-LYSIRELEQLSLSGNYLSGELSKNLSNLSGLKSLLISENRF 268
Query: 127 SGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQ 186
S + + GN LE+ D+S N FS P +LS L+VL L +N SI
Sbjct: 269 SDVIPDVFGNLTQLEHLDVSSNKFSGRFPPSLSQCSKLRVLDLRNNSLSGSINLNFTGFT 328
Query: 187 SLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVSN-FSRLKS---------- 235
L +DL+SN SG LP G PK++ L+LA+N G + + F L+S
Sbjct: 329 DLCVLDLASNHFSGPLPDSLGHC-PKMKILSLAKNEFRGKIPDTFKNLQSLLFLSLSNNS 387
Query: 236 --------------------IVSLNISGNSFQGSI-----IEVFVL-------------- 256
I+S N G ++ + + L
Sbjct: 388 FVDFSETMNVLQHCRNLSTLILSKNFIGEEIPNNVTGFDNLAILALGNCGLRGQIPSWLL 447
Query: 257 ---KLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLS--- 310
KLE LDLS N F G I L Y+D S N L+G I + NL L+
Sbjct: 448 NCKKLEVLDLSWNHFYGTIPHWIGKMESLFYIDFSNNTLTGAIPVAITELKNLIRLNGTA 507
Query: 311 -----------------------------------LACNRFSRQKFPKIEMLLGLEYLNL 335
L NR + P+I L L L+L
Sbjct: 508 SQMTDSSGIPLYVKRNKSSNGLPYNQVSRFPPSIYLNNNRLNGTILPEIGRLKELHMLDL 567
Query: 336 SKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIPLLKNKHLQVIDFS--HNNLSGPVPSF 393
S+ + G IPD IS L NL LDLS NHL G IPL + FS +N L+G +PS
Sbjct: 568 SRNNFTGTIPDSISGLDNLEVLDLSYNHLYGSIPLSFQSLTFLSRFSVAYNRLTGAIPS- 626
Query: 394 ILKSLPKMKKYNFSYN----NLTLCASEIKP-DIMKTSFFGSVNSCPIAANPSFFKKRRD 448
+ Y+F ++ NL LC + P D++ ++ NP +R +
Sbjct: 627 ------GGQFYSFPHSSFEGNLGLCRAIDSPCDVLMSNML----------NPKGSSRRNN 670
Query: 449 VGHR-GMKLALVLTLSLIFALAGIL-FLAFGCRRKNKMWEVKQGSYREEQNISGPFSFQT 506
G + G +VLT+SL + +L + RK+ + +E+ ISG
Sbjct: 671 NGGKFGRSSIVVLTISLAIGITLLLSVILLRISRKDVDDRINDV---DEETISGVS---- 723
Query: 507 DSTTWVADVKQATSVPVVIFEK-PLLNITFADLLSATSNFDRGTLLAEGKFGPVYRGFLP 565
K +V+F +++ +LL +T+NF + ++ G FG VY+ P
Sbjct: 724 ---------KALGPSKIVLFHSCGCKDLSVEELLKSTNNFSQANIIGCGGFGLVYKANFP 774
Query: 566 GNIHVAVKVLVVGSTLTDEEAARELEFLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGN 625
AVK L + E E+E L R +H NLV L GYC G+ R+ IY +MENG+
Sbjct: 775 DGSKAAVKRLSGDCGQMEREFQAEVEALSRAEHKNLVSLQGYCKHGNDRLLIYSFMENGS 834
Query: 626 LQNLLYDLPLGVQSTDDWSTDTWEEADNGIQNVGSEGLLTTWRFRHKIALGTARALAFLH 685
L L+ E D + + W R KIA G AR LA+LH
Sbjct: 835 LDYWLH-----------------ERVDGNMTLI--------WDVRLKIAQGAARGLAYLH 869
Query: 686 HGCSPPIIHRAVKASSVYLDYDLEPRLSDFGLAKI---FGSGLDEEIARGSPGYVPPEFS 742
C P +IHR VK+S++ LD E L+DFGLA++ + + + ++ G+ GY+PPE+S
Sbjct: 870 KVCEPNVIHRDVKSSNILLDEKFEAHLADFGLARLLRPYDTHVTTDLV-GTLGYIPPEYS 928
Query: 743 QPEFESPTPKSDVYCFGVVLFELLTGKKPVGDDYTDDKEATTLVSWVRGLVRKNQTSRAI 802
Q T + DVY FGVVL EL+TG++PV + K LVS V + + + + I
Sbjct: 929 QSLI--ATCRGDVYSFGVVLLELVTGRRPV--EVCKGKSCRDLVSRVFQMKAEKREAELI 984
Query: 803 DPKICDTGSDEQIEEALKEVGYLCTADLPFKRPTMQQIVGLLKDI 847
D I + ++ + E L E+ C P +RP ++++V L+D+
Sbjct: 985 DTTIRENVNERTVLEML-EIACKCIDHEPRRRPLIEEVVTWLEDL 1028
Score = 139 bits (350), Expect = 7e-33
Identities = 141/489 (28%), Positives = 215/489 (43%), Gaps = 90/489 (18%)
Query: 9 VLVLTFLFKHLASQ--QPNTDEFYVSEFLKKMGLTSSSKVYNFSSSVCSWKGVYCDSN-- 64
+L+L F SQ PN D + E + S ++ + S C W GV+C+ +
Sbjct: 4 ILLLVFFVGSSVSQPCHPN-DLSALRELAGALKNKSVTESWLNGSRCCEWDGVFCEGSDV 62
Query: 65 KEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSS 123
V +L L GL G I ++G+L +L LDLS N++ +P++ L L+ L+LS
Sbjct: 63 SGRVTKLVLPEKGLEGVI-SKSLGELTELRVLDLSRNQLKGEVPAEISKLEQLQVLDLSH 121
Query: 124 NHISGSLTNNIGNFGLLENFDLSKNSFSDEI------------------------PEALS 159
N +SGS+ + L+++ ++S NS S ++ PE S
Sbjct: 122 NLLSGSVLGVVSGLKLIQSLNISSNSLSGKLSDVGVFPGLVMLNVSNNLFEGEIHPELCS 181
Query: 160 SLVSLKVLKLDHNMFVRSIPSGILKC-QSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNL 218
S ++VL L N V ++ G+ C +S+ + + SN+L+G LP + + +L L+L
Sbjct: 182 SSGGIQVLDLSMNRLVGNL-DGLYNCSKSIQQLHIDSNRLTGQLP-DYLYSIRELEQLSL 239
Query: 219 AENNIYGGVS-NFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQV 275
+ N + G +S N S L + SL IS N F I +VF + +LE LD+S N+F G
Sbjct: 240 SGNYLSGELSKNLSNLSGLKSLLISENRFSDVIPDVFGNLTQLEHLDVSSNKFSGRFPPS 299
Query: 276 KYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNL 335
S L LDL N LSG I N +L L LA N FS + ++ L+L
Sbjct: 300 LSQCSKLRVLDLRNNSLSGSINLNFTGFTDLCVLDLASNHFSGPLPDSLGHCPKMKILSL 359
Query: 336 SKTSLVGHIPDE--------------------------ISHLGNLNALDLSMNHLDGKIP 369
+K G IPD + H NL+ L LS N + +IP
Sbjct: 360 AKNEFRGKIPDTFKNLQSLLFLSLSNNSFVDFSETMNVLQHCRNLSTLILSKNFIGEEIP 419
Query: 370 --------------------------LLKNKHLQVIDFSHNNLSGPVPSFILKSLPKMKK 403
LL K L+V+D S N+ G +P +I K + +
Sbjct: 420 NNVTGFDNLAILALGNCGLRGQIPSWLLNCKKLEVLDLSWNHFYGTIPHWIGK-MESLFY 478
Query: 404 YNFSYNNLT 412
+FS N LT
Sbjct: 479 IDFSNNTLT 487
Score = 132 bits (333), Expect = 6e-31
Identities = 106/344 (30%), Positives = 164/344 (46%), Gaps = 11/344 (3%)
Query: 68 VVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSLT-SLKSLNLSSNHI 126
+V LN+S G I + LDLS N++ ++ + S++ L++ SN +
Sbjct: 161 LVMLNVSNNLFEGEIHPELCSSSGGIQVLDLSMNRLVGNLDGLYNCSKSIQQLHIDSNRL 220
Query: 127 SGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQ 186
+G L + + + LE LS N S E+ + LS+L LK L + N F IP
Sbjct: 221 TGQLPDYLYSIRELEQLSLSGNYLSGELSKNLSNLSGLKSLLISENRFSDVIPDVFGNLT 280
Query: 187 SLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGNS 245
L +D+SSN+ SG P KLR L+L N++ G ++ NF+ + L+++ N
Sbjct: 281 QLEHLDVSSNKFSGRFPPSLSQC-SKLRVLDLRNNSLSGSINLNFTGFTDLCVLDLASNH 339
Query: 246 FQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQL--SGEIFQNLN 301
F G + + K++ L L++N+F+G I N L++L LS N E L
Sbjct: 340 FSGPLPDSLGHCPKMKILSLAKNEFRGKIPDTFKNLQSLLFLSLSNNSFVDFSETMNVLQ 399
Query: 302 NSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSM 361
+ NL L L+ N + + L L L L G IP + + L LDLS
Sbjct: 400 HCRNLSTLILSKNFIGEEIPNNVTGFDNLAILALGNCGLRGQIPSWLLNCKKLEVLDLSW 459
Query: 362 NHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI--LKSLPKM 401
NH G IP + K + L IDFS+N L+G +P I LK+L ++
Sbjct: 460 NHFYGTIPHWIGKMESLFYIDFSNNTLTGAIPVAITELKNLIRL 503
>At1g34110 putative protein
Length = 1049
Score = 281 bits (720), Expect = 8e-76
Identities = 249/797 (31%), Positives = 374/797 (46%), Gaps = 105/797 (13%)
Query: 70 ELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISG 128
+L LS TG IP + + L +L L NK++ ++PS +L SL+S L N ISG
Sbjct: 317 QLQLSDNMFTGQIP-WELSNCSSLIALQLDKNKLSGSIPSQIGNLKSLQSFFLWENSISG 375
Query: 129 SLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSL 188
++ ++ GN L DLS+N + IPE L SL L L L N +P + KCQSL
Sbjct: 376 TIPSSFGNCTDLVALDLSRNKLTGRIPEELFSLKRLSKLLLLGNSLSGGLPKSVAKCQSL 435
Query: 189 VSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGNSFQ 247
V + + NQLSG +P G+ L L+L N+ GG+ S + + L++ N
Sbjct: 436 VRLRVGENQLSGQIPKEIGE-LQNLVFLDLYMNHFSGGLPYEISNITVLELLDVHNNYIT 494
Query: 248 GSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMN 305
G I ++ LE LDLSRN F G+I N S+L L L+ N L+G+I +++ N
Sbjct: 495 GDIPAQLGNLVNLEQLDLSRNSFTGNIPLSFGNLSYLNKLILNNNLLTGQIPKSIKNLQK 554
Query: 306 LKHLSLACNRFSRQKFPKIEMLLGLEY-LNLSKTSLVGHIPDEISHLGNLNALDLSMNHL 364
L L L+ N S + ++ + L L+LS + G+IP+ S L L +LDLS N L
Sbjct: 555 LTLLDLSYNSLSGEIPQELGQVTSLTINLDLSYNTFTGNIPETFSDLTQLQSLDLSSNSL 614
Query: 365 DGKIPLLKN-KHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNLTLCASEIKPDIM 423
G I +L + L ++ S NN SGP+PS Y N LC S I
Sbjct: 615 HGDIKVLGSLTSLASLNISCNNFSGPIPSTPFFKTISTTSY---LQNTNLCHS--LDGIT 669
Query: 424 KTSFFGSVNSCPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAFGCRRKNK 483
+S G N +P + L V+ S+ A+ L R N
Sbjct: 670 CSSHTGQNNG---VKSPKI-----------VALTAVILASITIAILAAWLLIL---RNNH 712
Query: 484 MWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFADLLSATS 543
+++ Q S S P + + S W T +P F+K L IT +++++ +
Sbjct: 713 LYKTSQNS------SSSPSTAEDFSYPW-------TFIP---FQK--LGITVNNIVTSLT 754
Query: 544 NFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVL-------VVGSTLTDEEAARELEFLGRI 596
+ ++ +G G VY+ +P VAVK L G + D AA E++ LG I
Sbjct: 755 D---ENVIGKGCSGIVYKAEIPNGDIVAVKKLWKTKDNNEEGESTIDSFAA-EIQILGNI 810
Query: 597 KHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEADNGIQ 656
+H N+V L GYC ++ +Y+Y NGNLQ LL G +
Sbjct: 811 RHRNIVKLLGYCSNKSVKLLLYNYFPNGNLQQLL----------------------QGNR 848
Query: 657 NVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPRLSDFG 716
N+ W R+KIA+G A+ LA+LHH C P I+HR VK +++ LD E L+DFG
Sbjct: 849 NL-------DWETRYKIAIGAAQGLAYLHHDCVPAILHRDVKCNNILLDSKYEAILADFG 901
Query: 717 LAKIFGSGLDEEIARGSPGYVPPEFSQPEF---ESPTPKSDVYCFGVVLFELLTGKKPVG 773
LAK+ SP Y E+ + T KSDVY +GVVL E+L+G+ V
Sbjct: 902 LAKLM---------MNSPNYHNAMSRVAEYGYTMNITEKSDVYSYGVVLLEILSGRSAVE 952
Query: 774 DDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVG--YLCTADLP 831
D +V WV+ + + + ++ D+ ++E L+ +G C P
Sbjct: 953 PQIGD---GLHIVEWVKKKMGTFEPALSVLDVKLQGLPDQIVQEMLQTLGIAMFCVNPSP 1009
Query: 832 FKRPTMQQIVGLLKDIE 848
+RPTM+++V LL +++
Sbjct: 1010 VERPTMKEVVTLLMEVK 1026
Score = 123 bits (309), Expect = 4e-28
Identities = 112/353 (31%), Positives = 161/353 (44%), Gaps = 34/353 (9%)
Query: 54 CSWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITTLPSDFWSL 113
CSW G+ C ++ V+ ++ IPDT + ++++P D SL
Sbjct: 37 CSWYGITCSADNR-VISVS---------IPDTFLN--------------LSSIP-DLSSL 71
Query: 114 TSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNM 173
+SL+ LNLSS ++SG + + G L DLS NS S IP L L +L+ L L+ N
Sbjct: 72 SSLQFLNLSSTNLSGPIPPSFGKLTHLRLLDLSSNSLSGPIPSELGRLSTLQFLILNANK 131
Query: 174 FVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGG--VSNFS 231
SIPS I +L + L N L+G++P FG + L+ L N GG +
Sbjct: 132 LSGSIPSQISNLFALQVLCLQDNLLNGSIPSSFG-SLVSLQQFRLGGNTNLGGPIPAQLG 190
Query: 232 RLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSE 289
LK++ +L + + GSI F ++ L+ L L + G I S L L L
Sbjct: 191 FLKNLTTLGFAASGLSGSIPSTFGNLVNLQTLALYDTEISGTIPPQLGLCSELRNLYLHM 250
Query: 290 NQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEIS 349
N+L+G I + L + L L N S P+I L ++S L G IP ++
Sbjct: 251 NKLTGSIPKELGKLQKITSLLLWGNSLSGVIPPEISNCSSLVVFDVSANDLTGDIPGDLG 310
Query: 350 HLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPVPSFI--LKSL 398
L L L LS N G+IP L L + N LSG +PS I LKSL
Sbjct: 311 KLVWLEQLQLSDNMFTGQIPWELSNCSSLIALQLDKNKLSGSIPSQIGNLKSL 363
Score = 108 bits (269), Expect = 2e-23
Identities = 74/215 (34%), Positives = 112/215 (51%), Gaps = 4/215 (1%)
Query: 66 EHVVELNLSGIGLTGPIPDTTIGKLNKLHSLDLSNNKITT-LPSDFWSLTSLKSLNLSSN 124
+ +V L + L+G IP IG+L L LDL N + LP + ++T L+ L++ +N
Sbjct: 433 QSLVRLRVGENQLSGQIPKE-IGELQNLVFLDLYMNHFSGGLPYEISNITVLELLDVHNN 491
Query: 125 HISGSLTNNIGNFGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILK 184
+I+G + +GN LE DLS+NSF+ IP + +L L L L++N+ IP I
Sbjct: 492 YITGDIPAQLGNLVNLEQLDLSRNSFTGNIPLSFGNLSYLNKLILNNNLLTGQIPKSIKN 551
Query: 185 CQSLVSIDLSSNQLSGTLPHGFGDAFPKLRTLNLAENNIYGGV-SNFSRLKSIVSLNISG 243
Q L +DLS N LSG +P G L+L+ N G + FS L + SL++S
Sbjct: 552 LQKLTLLDLSYNSLSGEIPQELGQVTSLTINLDLSYNTFTGNIPETFSDLTQLQSLDLSS 611
Query: 244 NSFQGSI-IEVFVLKLEALDLSRNQFQGHISQVKY 277
NS G I + + L +L++S N F G I +
Sbjct: 612 NSLHGDIKVLGSLTSLASLNISCNNFSGPIPSTPF 646
>At4g08850 receptor protein kinase like protein
Length = 1045
Score = 273 bits (697), Expect = 4e-73
Identities = 232/793 (29%), Positives = 366/793 (45%), Gaps = 104/793 (13%)
Query: 78 LTGPIPDTTIGKLNKLHSLDLSNNKIT-TLPSDFWSLTSLKSLNLSSNHISGSLTNNIGN 136
L G IP +G++ + L++S NK+T +P F LT+L+ L L N +SG + I N
Sbjct: 322 LNGSIPPE-LGEMESMIDLEISENKLTGPVPDSFGKLTALEWLFLRDNQLSGPIPPGIAN 380
Query: 137 FGLLENFDLSKNSFSDEIPEALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSN 196
L + N+F+ +P+ + L+ L LD N F +P + C+SL+ + N
Sbjct: 381 STELTVLQVDTNNFTGFLPDTICRGGKLENLTLDDNHFEGPVPKSLRDCKSLIRVRFKGN 440
Query: 197 QLSGTLPHGFGDAFPKLRTLNLAENNIYGGVS-NFSRLKSIVSLNISGNSFQGSII-EVF 254
SG + FG +P L ++L+ NN +G +S N+ + + +V+ +S NS G+I E++
Sbjct: 441 SFSGDISEAFG-VYPTLNFIDLSNNNFHGQLSANWEQSQKLVAFILSNNSITGAIPPEIW 499
Query: 255 -VLKLEALDLSRNQFQGHISQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLAC 313
+ +L LDLS N+ G + + N + + L L+ N+LSG+I + NL++L L+
Sbjct: 500 NMTQLSQLDLSSNRITGELPESISNINRISKLQLNGNRLSGKIPSGIRLLTNLEYLDLSS 559
Query: 314 NRFSRQKFPKIEMLLGLEYLNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKI--PLL 371
NRFS + P + L L Y+NLS+ L IP+ ++ L L LDLS N LDG+I
Sbjct: 560 NRFSSEIPPTLNNLPRLYYMNLSRNDLDQTIPEGLTKLSQLQMLDLSYNQLDGEISSQFR 619
Query: 372 KNKHLQVIDFSHNNLSGPVPSFILKSLPKMKKYNFSYNNL------TLCASEIKPDIMK- 424
++L+ +D SHNNLSG +P K + + + S+NNL PD +
Sbjct: 620 SLQNLERLDLSHNNLSGQIPP-SFKDMLALTHVDVSHNNLQGPIPDNAAFRNAPPDAFEG 678
Query: 425 -TSFFGSVNS------CPIAANPSFFKKRRDVGHRGMKLALVLTLSLIFALAGILFLAFG 477
GSVN+ C I ++ K R + + L ++ +I ++ +F+ F
Sbjct: 679 NKDLCGSVNTTQGLKPCSITSSKKSHKDRNLIIY---ILVPIIGAIIILSVCAGIFICF- 734
Query: 478 CRRKNKMWEVKQGSYREEQNISGPFSFQTDSTTWVADVKQATSVPVVIFEKPLLNITFAD 537
R++ K E S + +S FSF + + +
Sbjct: 735 -RKRTKQIEEHTDSESGGETLS-IFSFDG-------------------------KVRYQE 767
Query: 538 LLSATSNFDRGTLLAEGKFGPVYRGFLPGNIHVAVKVL------VVGSTLTDEEAARELE 591
++ AT FD L+ G G VY+ LP N +AVK L + + T +E E+
Sbjct: 768 IIKATGEFDPKYLIGTGGHGKVYKAKLP-NAIMAVKKLNETTDSSISNPSTKQEFLNEIR 826
Query: 592 FLGRIKHPNLVPLTGYCVAGDQRIAIYDYMENGNLQNLLYDLPLGVQSTDDWSTDTWEEA 651
L I+H N+V L G+C +Y+YME G+L+ +L DD
Sbjct: 827 ALTEIRHRNVVKLFGFCSHRRNTFLVYEYMERGSLRKVL--------ENDD--------- 869
Query: 652 DNGIQNVGSEGLLTTWRFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEPR 711
E W R + G A AL+++HH SP I+HR + + ++ L D E +
Sbjct: 870 ---------EAKKLDWGKRINVVKGVAHALSYMHHDRSPAIVHRDISSGNILLGEDYEAK 920
Query: 712 LSDFGLAKIF-GSGLDEEIARGSPGYVPPEFSQPEFESPTPKSDVYCFGVVLFELLTGKK 770
+SDFG AK+ + G+ GYV PE + T K DVY FGV+ E++ G+
Sbjct: 921 ISDFGTAKLLKPDSSNWSAVAGTYGYVAPELAYA--MKVTEKCDVYSFGVLTLEVIKGEH 978
Query: 771 P---VGDDYTDDKEATTLVSWVRGLVRKNQTSRAIDPKICDTGSDEQIEEALKEVGYLCT 827
P V + +AT + + T P+I E++ E LK V LC
Sbjct: 979 PGDLVSTLSSSPPDATLSLKSISDHRLPEPT-----PEI-----KEEVLEILK-VALLCL 1027
Query: 828 ADLPFKRPTMQQI 840
P RPTM I
Sbjct: 1028 HSDPQARPTMLSI 1040
Score = 130 bits (327), Expect = 3e-30
Identities = 122/410 (29%), Positives = 178/410 (42%), Gaps = 45/410 (10%)
Query: 41 TSSSKVYNF----SSSVC-SWKGVYCDSNKEHVVELNLSGIGLTGPIPDTTIGKLNKLHS 95
TSSSK+ ++ +SS C SW GV C ++ LNL+ G+ G D
Sbjct: 65 TSSSKLSSWVNPNTSSFCTSWYGVACSLGS--IIRLNLTNTGIEGTFEDFP--------- 113
Query: 96 LDLSNNKITTLPSDFWSLTSLKSLNLSSNHISGSLTNNIGNFGLLENFDLSKNSFSDEIP 155
F SL +L ++LS N SG+++ G F LE FDLS N EIP
Sbjct: 114 --------------FSSLPNLTFVDLSMNRFSGTISPLWGRFSKLEYFDLSINQLVGEIP 159
Query: 156 EALSSLVSLKVLKLDHNMFVRSIPSGILKCQSLVSIDLSSNQLSGTLPHGFGDAFPKLRT 215
L L +L L L N SIPS I + + I + N L+G +P FG+ KL
Sbjct: 160 PELGDLSNLDTLHLVENKLNGSIPSEIGRLTKVTEIAIYDNLLTGPIPSSFGN-LTKLVN 218
Query: 216 LNLAENNIYGGV-SNFSRLKSIVSLNISGNSFQGSIIEVF--VLKLEALDLSRNQFQGHI 272
L L N++ G + S L ++ L + N+ G I F + + L++ NQ G I
Sbjct: 219 LYLFINSLSGSIPSEIGNLPNLRELCLDRNNLTGKIPSSFGNLKNVTLLNMFENQLSGEI 278
Query: 273 SQVKYNWSHLVYLDLSENQLSGEIFQNLNNSMNLKHLSLACNRFSRQKFPKIEMLLGLEY 332
N + L L L N+L+G I L N L L L N+ + P++ + +
Sbjct: 279 PPEIGNMTALDTLSLHTNKLTGPIPSTLGNIKTLAVLHLYLNQLNGSIPPELGEMESMID 338
Query: 333 LNLSKTSLVGHIPDEISHLGNLNALDLSMNHLDGKIP--LLKNKHLQVIDFSHNNLSGPV 390
L +S+ L G +PD L L L L N L G IP + + L V+ NN +G +
Sbjct: 339 LEISENKLTGPVPDSFGKLTALEWLFLRDNQLSGPIPPGIANSTELTVLQVDTNNFTGFL 398
Query: 391 PSFILKSLPKMKKYNFSYN--------NLTLCASEIKPDIMKTSFFGSVN 432
P I + K++ N +L C S I+ SF G ++
Sbjct: 399 PDTICRG-GKLENLTLDDNHFEGPVPKSLRDCKSLIRVRFKGNSFSGDIS 447
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,773,838
Number of Sequences: 26719
Number of extensions: 908250
Number of successful extensions: 17996
Number of sequences better than 10.0: 1168
Number of HSP's better than 10.0 without gapping: 906
Number of HSP's successfully gapped in prelim test: 263
Number of HSP's that attempted gapping in prelim test: 2336
Number of HSP's gapped (non-prelim): 3735
length of query: 852
length of database: 11,318,596
effective HSP length: 108
effective length of query: 744
effective length of database: 8,432,944
effective search space: 6274110336
effective search space used: 6274110336
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC122160.2


