

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121245.10 - phase: 0
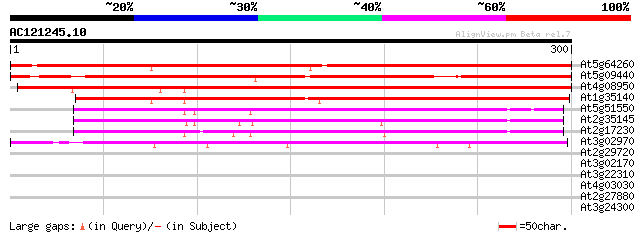
(300 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g64260 phi-1-like protein 437 e-123
At5g09440 unknown protein 358 3e-99
At4g08950 putative phi-1-like phosphate-induced protein 354 3e-98
At1g35140 phosphate-induced (phi-1) protein, putative 338 2e-93
At5g51550 unknown protein 190 7e-49
At2g35145 unknown protein 171 3e-43
At2g17230 unknown protein 164 7e-41
At3g02970 phi-1-like protein 163 9e-41
At2g29720 Hydroxylase/Oxygenase (CTF2B) 32 0.32
At3g02170 unknown protein 32 0.42
At3g22310 putative RNA helicase 30 1.6
At4g03030 OR23peptide 29 2.7
At2g27880 Argonaute (AGO1)-like protein 29 2.7
At3g24300 ammonium transporter 28 6.0
>At5g64260 phi-1-like protein
Length = 305
Score = 437 bits (1123), Expect = e-123
Identities = 221/309 (71%), Positives = 246/309 (79%), Gaps = 13/309 (4%)
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
MASNY FAI L+ L S LV+EQPLV+KYHNG LLKG +TVNL+WYG FTPI
Sbjct: 1 MASNYRFAIFLT--LFFATAGFSAAALVEEQPLVMKYHNGVLLKGNITVNLVWYGKFTPI 58
Query: 61 QRSIIVDFINSLST----TGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNL 116
QRS+IVDFI+SL++ + AA+PS ++WWKTTEKYK GSS L VGKQ L Y LGK+L
Sbjct: 59 QRSVIVDFIHSLNSKDVASSAAVPSVASWWKTTEKYKGGSSTLVVGKQLLLENYPLGKSL 118
Query: 117 KGKDLLALATKFNE-LSSITVVLTAKDVNVEGFCMSRCGTHGSV----RRGSGGARTPYI 171
K L AL+TK N L SITVVLTAKDV VE FCMSRCGTHGS RR + GA Y+
Sbjct: 119 KNPYLRALSTKLNGGLRSITVVLTAKDVTVERFCMSRCGTHGSSGSNPRRAANGAA--YV 176
Query: 172 WVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNG 231
WVGN+ET CPG CAWPFHQPIYGPQTPPLVAPNGDVGVDGM+INLATLLA TVTNPFNNG
Sbjct: 177 WVGNSETQCPGYCAWPFHQPIYGPQTPPLVAPNGDVGVDGMIINLATLLANTVTNPFNNG 236
Query: 232 YFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDP 291
Y+QGP APLEAV+AC G+FGSG+YPGYAGRVLVD +GSSYNA G GRKYLLPAMWDP
Sbjct: 237 YYQGPPTAPLEAVSACPGIFGSGSYPGYAGRVLVDKTTGSSYNARGLAGRKYLLPAMWDP 296
Query: 292 QTSACKTLV 300
Q+S CKTLV
Sbjct: 297 QSSTCKTLV 305
>At5g09440 unknown protein
Length = 278
Score = 358 bits (918), Expect = 3e-99
Identities = 190/303 (62%), Positives = 219/303 (71%), Gaps = 28/303 (9%)
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
MA NY FAI+L +L+ T+ F + P+ + L G +T+NLIWYG FTPI
Sbjct: 1 MAYNYRFAILL----VLLSATVGFTAAALKPPV-------RTLNGNITLNLIWYGKFTPI 49
Query: 61 QRSIIVDFINSLSTTGAAL-PSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGK 119
QRSIIVDFI S+S+ AA PS ++WWKTTEKYK G S L VGKQ L Y LGK+LK
Sbjct: 50 QRSIIVDFIRSISSVTAAKGPSVASWWKTTEKYKTGVSTLVVGKQLLLENYPLGKSLKSP 109
Query: 120 DLLALATKFNE--LSSITVVLTAKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAE 177
L AL++K N SITVVLTAKDV VEG CM+RCGTHGS + S Y+WVGN+E
Sbjct: 110 YLRALSSKLNAGGARSITVVLTAKDVTVEGLCMNRCGTHGS--KSSSVNSGAYVWVGNSE 167
Query: 178 TLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPA 237
T CPG CAWPFHQPIYGPQ+PPLVAPNGDVGVDGM+IN+ATLL TVTNP
Sbjct: 168 TQCPGYCAWPFHQPIYGPQSPPLVAPNGDVGVDGMIINIATLLVNTVTNP---------- 217
Query: 238 AAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACK 297
+P EAV+ACTG+FGSGAYPGYAGRVLVD SG+SYNA G GRKYLLPA+WDPQTS CK
Sbjct: 218 -SP-EAVSACTGIFGSGAYPGYAGRVLVDKTSGASYNALGLAGRKYLLPALWDPQTSTCK 275
Query: 298 TLV 300
T+V
Sbjct: 276 TMV 278
>At4g08950 putative phi-1-like phosphate-induced protein
Length = 314
Score = 354 bits (909), Expect = 3e-98
Identities = 178/313 (56%), Positives = 220/313 (69%), Gaps = 17/313 (5%)
Query: 5 YYFAIVLSAVLLLVLPTLSFGELVQEQP---LVLKYHNGQLLKGKLTVNLIWYGTFTPIQ 61
Y L L L+ ++S L ++P +LKYH G LL GK++VNLIWYG F P Q
Sbjct: 2 YLLVFKLFLFLSLLQISVSARNLASQEPNQFQLLKYHKGALLSGKISVNLIWYGKFKPSQ 61
Query: 62 RSIIVDFINSLSTTGAAL-----PSASAWWKTTEKY--------KVGSSALTVGKQFLHP 108
R+II DFI SL+ T PS + WWKTTEKY +LT+GKQ +
Sbjct: 62 RAIISDFITSLTHTSPTSKTLHQPSVATWWKTTEKYYKLATPSKNSSPLSLTLGKQIIDE 121
Query: 109 AYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVNVEGFCMSRCGTHGSVRR-GSGGAR 167
+ +LGK+L K + LA+K ++ ++I VVLT+ DV V GF MSRCGTHG R G G++
Sbjct: 122 SCSLGKSLTDKKIQTLASKGDQRNAINVVLTSADVTVTGFGMSRCGTHGHARGLGKRGSK 181
Query: 168 TPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINLATLLAGTVTNP 227
YIWVGN+ET CPGQCAWPFH P+YGPQ+PPLVAPN DVG+DGMVINLA+LLAGT TNP
Sbjct: 182 FAYIWVGNSETQCPGQCAWPFHAPVYGPQSPPLVAPNNDVGLDGMVINLASLLAGTATNP 241
Query: 228 FNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPA 287
F NGY+QGP APLEA +AC GV+G GAYPGYAG +LVD +G S+NA+GANGRK+LLPA
Sbjct: 242 FGNGYYQGPQNAPLEAASACPGVYGKGAYPGYAGDLLVDTTTGGSFNAYGANGRKFLLPA 301
Query: 288 MWDPQTSACKTLV 300
++DP TSAC T+V
Sbjct: 302 LYDPTTSACSTMV 314
>At1g35140 phosphate-induced (phi-1) protein, putative
Length = 309
Score = 338 bits (867), Expect = 2e-93
Identities = 161/277 (58%), Positives = 204/277 (73%), Gaps = 14/277 (5%)
Query: 36 KYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLST----TGAALPSASAWWKTTEK 91
+YH G LL G +++NLIWYG F P QR+I+ DF+ SLS+ T A PS + WWKT EK
Sbjct: 33 QYHKGALLTGDVSINLIWYGKFKPSQRAIVTDFVASLSSSRRSTMAQNPSVATWWKTVEK 92
Query: 92 Y-------KVGSSALTVGKQFLHPAYTLGKNLKGKDLLALATKFNELSSITVVLTAKDVN 144
Y +L++G+Q L Y++GK+L K+L LA K + ++ VVLT+ DV
Sbjct: 93 YYQFRKMTTTRGLSLSLGEQILDQGYSMGKSLTEKNLKDLAAKGGQSYAVNVVLTSADVT 152
Query: 145 VEGFCMSRCGTHGSVRRGSG--GARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVA 202
V+GFCM+RCG+HG+ GSG G+R YIWVGN+ET CPGQCAWPFH P+YGPQ+PPLVA
Sbjct: 153 VQGFCMNRCGSHGT-GSGSGKKGSRFAYIWVGNSETQCPGQCAWPFHAPVYGPQSPPLVA 211
Query: 203 PNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGSGAYPGYAGR 262
PN DVG+DGMVINLA+L+A T TNPF +GY+QGP APLEA +ACTGV+G G+YPGYAG
Sbjct: 212 PNNDVGLDGMVINLASLMAATATNPFGDGYYQGPKTAPLEAGSACTGVYGKGSYPGYAGE 271
Query: 263 VLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKTL 299
+LVD +G SYN G NGRKYLLPA++DP+T +C TL
Sbjct: 272 LLVDATTGGSYNVKGLNGRKYLLPALFDPKTDSCSTL 308
>At5g51550 unknown protein
Length = 337
Score = 190 bits (483), Expect = 7e-49
Identities = 107/278 (38%), Positives = 154/278 (54%), Gaps = 18/278 (6%)
Query: 35 LKYHNGQLLKGKLTVNLIWYGTFTPIQRSIIVDFINSLSTTGAALPSASAWWKTTEKY-- 92
L+YH G +L +TV+ IWYGT+ Q+ II +FINS+S G+ PS S WWKT + Y
Sbjct: 55 LRYHMGPVLTNNITVHPIWYGTWQKSQKKIIREFINSISAVGSKHPSVSGWWKTVQLYTD 114
Query: 93 KVGSS---ALTVGKQFLHPAYTLGKNLKGKDLLALATK----------FNELSSITVVLT 139
+ GS+ + +G++ Y+ GK+L + ++ N S + ++LT
Sbjct: 115 QTGSNITGTVRLGEEKNDRFYSHGKSLTRLSIQSVIKSAVTSRSRPLPVNPKSGLYLLLT 174
Query: 140 AKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPP 199
A DV V+ FC CG H G PY WVGN+ LCPG CA+PF P + P P
Sbjct: 175 ADDVYVQDFCGQVCGFHYFTFPSIVGFTLPYAWVGNSAKLCPGVCAYPFAVPAFIPGLKP 234
Query: 200 LVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQGP-AAAPLEAVTACTGVFGSGAYPG 258
+ +PNGDVGVDGM+ +A +A TNP N ++ GP AP+E C G++G+G
Sbjct: 235 VKSPNGDVGVDGMISVIAHEIAELATNPLVNAWYAGPDPVAPVEIADLCEGIYGTGGGGS 294
Query: 259 YAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
Y G++L D SG++YN +G R+YL+ +W S C
Sbjct: 295 YTGQMLND-HSGATYNVNGIR-RRYLIQWLWSHVVSYC 330
>At2g35145 unknown protein
Length = 323
Score = 171 bits (434), Expect = 3e-43
Identities = 107/282 (37%), Positives = 149/282 (51%), Gaps = 21/282 (7%)
Query: 35 LKYHNGQLLKGKLT-VNLIWYGTFTPIQRSIIVDFINSLSTTGAA-LPSASAWWKTTEKY 92
L+YH G ++ +T + +IWYG + P +SII DF+ S+S A PS S WWKT Y
Sbjct: 36 LQYHLGPVISSPVTSLYIIWYGRWNPTHQSIIRDFLYSVSAPAPAQYPSVSNWWKTVRLY 95
Query: 93 K--VGSS---ALTVGKQFLHPAYTLGKNLKGKDL-----LALATKF--NELSSITVVLTA 140
+ GS+ L + +F Y+ G +L + AL +K N ++ + +VLT+
Sbjct: 96 RDQTGSNITDTLVLSGEFHDSTYSHGSHLTRFSVQSVIRTALTSKLPLNAVNGLYLVLTS 155
Query: 141 KDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQT--- 197
DV ++ FC + CG H GA PY WVGN+E CP CA+PF QP P +
Sbjct: 156 DDVEMQEFCRAICGFHYFTFPSVVGATVPYAWVGNSERQCPEMCAYPFAQPKPFPGSGFV 215
Query: 198 --PPLVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQG-PAAAPLEAVTACTGVFGSG 254
+ PNG+VG+DGM+ +A LA +NP NG++ G A AP E C GV+GSG
Sbjct: 216 AREKMKPPNGEVGIDGMISVIAHELAEVSSNPMLNGWYGGEDATAPTEIADLCLGVYGSG 275
Query: 255 AYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
GY G V D + YN G GRKYL+ +WD + C
Sbjct: 276 GGGGYMGSVYKD-RWRNVYNVKGVKGRKYLIQWVWDLNRNRC 316
>At2g17230 unknown protein
Length = 363
Score = 164 bits (414), Expect = 7e-41
Identities = 99/280 (35%), Positives = 147/280 (52%), Gaps = 20/280 (7%)
Query: 35 LKYHNGQLLKGK-LTVNLIWYGTFTPIQRSIIVDFINSLSTTGAALPSASAWWKTTEKY- 92
L+YH G +L + + +IWYG ++ +S+I DF+NS+S A PS S WW+T Y
Sbjct: 79 LRYHMGPVLSSSPINIYVIWYGQWSRPHKSLIRDFLNSISDAKAPSPSVSEWWRTASLYT 138
Query: 93 -----KVGSSALTVGKQFLHPAYTLGKNLKG---KDLLALATK-----FNELSSITVVLT 139
V S L G ++ Y+ G++L ++++A A + + + + +VLT
Sbjct: 139 DQTGSNVSRSVLIAG-EYSDSKYSHGQHLTRLTIQEVIASAARSASFPVDHKNGMYLVLT 197
Query: 140 AKDVNVEGFCMSRCGTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPP 199
+ DV ++ FC + CG H G PY WVG + CP CA+PF P Y P
Sbjct: 198 SHDVTMQDFCRAVCGFHYFTFPSMVGYTMPYAWVGQSGKQCPEVCAYPFALPGYMGHGGP 257
Query: 200 --LVAPNGDVGVDGMVINLATLLAGTVTNPFNNGYFQG-PAAAPLEAVTACTGVFGSGAY 256
L PNG+ GVDGMV + LA V+NP N ++ G AP E C G++GSG
Sbjct: 258 GELRPPNGETGVDGMVSVIGHELAEVVSNPLINAWYAGEDPTAPTEIGDLCEGLYGSGGG 317
Query: 257 PGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSAC 296
GY G+V+ D G ++N +G GRK+L+ +W+P AC
Sbjct: 318 GGYIGQVMRD-REGKTFNMNGKGGRKFLVQWIWNPNLKAC 356
>At3g02970 phi-1-like protein
Length = 332
Score = 163 bits (413), Expect = 9e-41
Identities = 110/338 (32%), Positives = 158/338 (46%), Gaps = 50/338 (14%)
Query: 1 MASNYYFAIVLSAVLLLVLPTLSFGELVQEQPLVLKYHNGQLLKGKLTVNLIWYGTFTPI 60
MAS + +S ++ L+L L L +E P Q+ G L ++L+WYG FTP
Sbjct: 3 MASASSSSSSISVIIFLLLAPLC---LSREPP-------SQIPNGTLDLSLLWYGQFTPT 52
Query: 61 QRSIIVDFINSLSTTG--AALPSASAWWKTTEKYKVGSSALTVGKQ-------------- 104
Q+ + DFI SL+ P SAWWK E Y+ + +Q
Sbjct: 53 QKERVHDFIESLNFDAKEGLDPKVSAWWKVVESYQERFEVKDIYRQKKSNRTVAPRIKVK 112
Query: 105 ----FLHPAYTLGKNLK-GKDLLALATKFNELSSIT-VVLTAKDVNVEG--FCMSRCGTH 156
++ GK L G + T +S + VVL + V G FC C +
Sbjct: 113 VVRSYVDEKMKYGKELTMGNGEKLVETAIGNMSKVVPVVLLSAQVRAHGVGFCDGTCQHN 172
Query: 157 GSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNGDVGVDGMVINL 216
+ YI V N E CPG+CAWPFH GP+ +G+VG D +VI L
Sbjct: 173 ALAKIKGQKEPRRYIMVSNPEVECPGECAWPFHTADKGPRGMTYQPASGEVGADALVIQL 232
Query: 217 ATLLAGTVTNP-------------FNNGYFQGPAAAPLEAV---TACTGVFGSGAYPGYA 260
AT LA TNP +N+ + ++ + V T CT VFGSGA+PG+
Sbjct: 233 ATGLADLATNPDLTKSLFKSETTPYNDDVKKNHESSSMYIVDPATKCTRVFGSGAFPGFT 292
Query: 261 GRVLVDGASGSSYNAHGANGRKYLLPAMWDPQTSACKT 298
GR+ VD +G ++N+HG N K+L+P++WDP+T +C T
Sbjct: 293 GRIRVDPITGGAFNSHGINHLKFLIPSIWDPKTKSCWT 330
>At2g29720 Hydroxylase/Oxygenase (CTF2B)
Length = 427
Score = 32.3 bits (72), Expect = 0.32
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 3/71 (4%)
Query: 62 RSIIVDFINSLSTTGAALPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNLKGKDL 121
RS++++ SL T GA+L + W+ + +G + KQFL + K G++L
Sbjct: 67 RSVVLEQAESLRTGGASLTLSKNGWRVLDAISIGPQ---LRKQFLEMQGVVLKKEDGREL 123
Query: 122 LALATKFNELS 132
+ K N+ S
Sbjct: 124 RSFKFKDNDQS 134
>At3g02170 unknown protein
Length = 905
Score = 32.0 bits (71), Expect = 0.42
Identities = 26/68 (38%), Positives = 32/68 (46%), Gaps = 4/68 (5%)
Query: 58 TPIQRSIIVDFINSLSTTGAA-LPSASAWWKTTEKYKVGSSALTVGKQFLHPAYTLGKNL 116
T +QRS VD I + + A+ P A WK + K G SALTV + L
Sbjct: 307 TALQRSRSVDSIKRIPASAASKFPMEPAPWK---QMKAGDSALTVYGEIQKRLTQLEFKK 363
Query: 117 KGKDLLAL 124
GKDL AL
Sbjct: 364 SGKDLRAL 371
>At3g22310 putative RNA helicase
Length = 610
Score = 30.0 bits (66), Expect = 1.6
Identities = 13/36 (36%), Positives = 17/36 (47%)
Query: 248 TGVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKY 283
+G G G+Y GY G G G SY G + +Y
Sbjct: 527 SGRSGGGSYGGYGGSSGRSGGGGGSYGGSGGSSSRY 562
>At4g03030 OR23peptide
Length = 434
Score = 29.3 bits (64), Expect = 2.7
Identities = 16/42 (38%), Positives = 21/42 (49%), Gaps = 1/42 (2%)
Query: 249 GVFGSGAYPGYAGRVLVDGASGSSYNAHGANGRKYLLPAMWD 290
G F A PG GR++V G GS + GA G + M+D
Sbjct: 183 GSFACAAMPGSCGRIIVAG-GGSRHTLFGAAGSRMSSVEMYD 223
>At2g27880 Argonaute (AGO1)-like protein
Length = 997
Score = 29.3 bits (64), Expect = 2.7
Identities = 18/52 (34%), Positives = 21/52 (39%), Gaps = 4/52 (7%)
Query: 154 GTHGSVRRGSGGARTPYIWVGNAETLCPGQCAWPFHQPIYGPQTPPLVAPNG 205
G HG RG GG R + GQ AWP Q YG + + A G
Sbjct: 7 GGHGGASRGRGGGRRS----DQRQDQSSGQVAWPGLQQSYGGRGGSVSAGRG 54
>At3g24300 ammonium transporter
Length = 498
Score = 28.1 bits (61), Expect = 6.0
Identities = 24/82 (29%), Positives = 33/82 (39%), Gaps = 8/82 (9%)
Query: 208 GVDGMVINLATLLAGTVTNPFNNGYFQGPAAAPLEAVTACTGVFGS--------GAYPGY 259
GV MV +A L + P + +G A L +A V G+ G PG
Sbjct: 207 GVVHMVGGIAGLWGALIEGPRRGRFEKGGRAIALRGHSASLVVLGTFLLWFGWYGFNPGS 266
Query: 260 AGRVLVDGASGSSYNAHGANGR 281
++LV SGS+Y GR
Sbjct: 267 FTKILVPYNSGSNYGQWSGIGR 288
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,901,376
Number of Sequences: 26719
Number of extensions: 292311
Number of successful extensions: 642
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 604
Number of HSP's gapped (non-prelim): 14
length of query: 300
length of database: 11,318,596
effective HSP length: 99
effective length of query: 201
effective length of database: 8,673,415
effective search space: 1743356415
effective search space used: 1743356415
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC121245.10


