

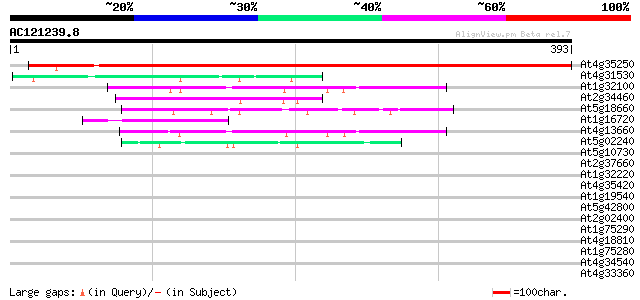
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.8 + phase: 0
(393 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g35250 unknown protein 636 0.0
At4g31530 unknown protein 55 9e-08
At1g32100 pinoresinol-lariciresinol reductase like protein 54 1e-07
At2g34460 unknown protein 53 3e-07
At5g18660 unknown protein 51 1e-06
At1g16720 unknown protein (At1g16720) 50 2e-06
At4g13660 isoflavone reductase-like protein 48 8e-06
At5g02240 unknown protein 42 6e-04
At5g10730 unknown protein 41 0.001
At2g37660 unknown protein 40 0.002
At1g32220 unknown protein 40 0.002
At4g35420 unknown protein 38 0.008
At1g19540 2'-hydroxyisoflavone reductase like protein 38 0.008
At5g42800 dihydroflavonol 4-reductase 37 0.019
At2g02400 putative cinnamoyl-CoA reductase 37 0.019
At1g75290 NADPH oxidoreductase, putative 36 0.042
At4g18810 unknown protein 35 0.055
At1g75280 putative NADPH oxidoreductase 35 0.055
At4g34540 isoflavone reductase - like protein 34 0.12
At4g33360 putative dihydrokaempferol 4-reductase 33 0.21
>At4g35250 unknown protein
Length = 395
Score = 636 bits (1640), Expect = 0.0
Identities = 324/392 (82%), Positives = 339/392 (85%), Gaps = 15/392 (3%)
Query: 14 PGRVVTRTREAFHALSPS------------PSPSPPHFVEFCKGGRRGRRIIRVGVKCNS 61
P ++VTR H S S P + P F R I V V C
Sbjct: 7 PAQLVTRGNLIHHNSSSSSSGRLSWRRSLTPENTIPLFPSSSSSSLNRERSIVVPVTC-- 64
Query: 62 NSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW 121
S AVNL PGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW
Sbjct: 65 -SAAAVNLAPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW 123
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
GATVVNADLSKPETIPATLVG+HTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY
Sbjct: 124 GATVVNADLSKPETIPATLVGIHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 183
Query: 182 VFYSIHNCDKHPEVPLMEIKYCTENFLRDSGLNHIVIRLCGFMQGLIGQYAVPILEEKSV 241
VFYSIHNCDKHPEVPLMEIKYCTE FL++SGLNHI IRLCGFMQGLIGQYAVPILEEKSV
Sbjct: 184 VFYSIHNCDKHPEVPLMEIKYCTEKFLQESGLNHITIRLCGFMQGLIGQYAVPILEEKSV 243
Query: 242 WGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDA 301
WGTDAPTR+AYMDTQDIARLT IALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDA
Sbjct: 244 WGTDAPTRVAYMDTQDIARLTLIALRNEKINGKLLTFAGPRAWTTQEVITLCERLAGQDA 303
Query: 302 NVTTVPVSVLRLTRQLTRFFEWTNDVADRLAFSEVLTSDTVFSVPMAETYNLLGVDTKDI 361
NVTTVPVSVLR+TRQLTRFF+WTNDVADRLAFSEVL+SDTVFS PM ET +LLGVD KD+
Sbjct: 304 NVTTVPVSVLRVTRQLTRFFQWTNDVADRLAFSEVLSSDTVFSAPMTETNSLLGVDQKDM 363
Query: 362 ITLEKYLQDYFTNILKKLKDLKAQSKQSDIFF 393
+TLEKYLQDYF+NILKKLKDLKAQSKQSDI+F
Sbjct: 364 VTLEKYLQDYFSNILKKLKDLKAQSKQSDIYF 395
>At4g31530 unknown protein
Length = 324
Score = 54.7 bits (130), Expect = 9e-08
Identities = 68/240 (28%), Positives = 91/240 (37%), Gaps = 29/240 (12%)
Query: 3 MATLMRLPTATPG--RVVTRTREAFHALSPSPSPSPPHFVEFCKGGRRGRRIIRVGVKCN 60
MAT L A P R + + L SP P P F I V
Sbjct: 1 MATTTNLSFAPPSYSRFAATKSQIRNPLFTSPLPLPSSFFLVRNEASLSSSITPV----Q 56
Query: 61 SNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFL-- 118
+ +E AV+ +LVVG TG +G+ +V L R L+R
Sbjct: 57 AFTEEAVDTSDLASSSSKLVLVVGGTGGVGQLVVASLLKRNIRSRLLLRDLDKATKLFGK 116
Query: 119 -RDWGATVVNADLSKPETI-PATLVGVHTVIDCATGRPEEPIK---------TVDWEGKV 167
++ VV D E + P+ GV VI C TG P K VDWEG
Sbjct: 117 QDEYSLQVVKGDTRNAEDLDPSMFEGVTHVI-CTTGTTAFPSKRWNEENTPEKVDWEGVK 175
Query: 168 ALIQCAKAMGIQKYVFYSIHNCDKHPEVP--------LMEIKYCTENFLRDSGLNHIVIR 219
LI A +++ V S K E+P +++ K E+FLRDSGL +IR
Sbjct: 176 NLIS-ALPSSVKRVVLVSSVGVTKSNELPWSIMNLFGVLKYKKMGEDFLRDSGLPFTIIR 234
>At1g32100 pinoresinol-lariciresinol reductase like protein
Length = 317
Score = 53.9 bits (128), Expect = 1e-07
Identities = 60/258 (23%), Positives = 110/258 (42%), Gaps = 24/258 (9%)
Query: 69 LGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPR-----PAPADFL--RDW 121
+G T +LVVGATG +G++IVR L EG++ L RP FL +
Sbjct: 1 MGESKRTEKTRVLVVGATGYIGKRIVRACLAEGHETYVLQRPEIGLEIEKVQLFLSFKKL 60
Query: 122 GATVVNADLSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKY 181
GA +V S +++ + + V V+ +G ++ + ++ L++ K G K
Sbjct: 61 GARIVEGSFSDHQSLVSAVKLVDVVVSAMSG---VHFRSHNILVQLKLVEAIKEAGNVKR 117
Query: 182 VFYSIHNCDK-------HPEVPLMEIKYCTENFLRDSGLNH-IVIRLC--GFMQGLIGQY 231
S D P + K + +G+ + V+ C + G + Q
Sbjct: 118 FLPSEFGMDPPRMGHALPPGRETFDQKMEVRQAIEAAGIPYTYVVGACFAAYFAGNLSQM 177
Query: 232 A--VPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGPRAWTTQ-E 288
+P E+ +++G D ++ + D DIA+ T L + + K + P TQ E
Sbjct: 178 VTLLPPKEKVNIYG-DGNVKVVFADEDDIAKYTAKTLNDPRTLNKTVNIRPPDNVLTQLE 236
Query: 289 VITLCERLAGQDANVTTV 306
++ + E+L G++ T +
Sbjct: 237 LVQIWEKLTGKELEKTNI 254
>At2g34460 unknown protein
Length = 280
Score = 53.1 bits (126), Expect = 3e-07
Identities = 45/165 (27%), Positives = 76/165 (45%), Gaps = 20/165 (12%)
Query: 75 VRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRP-RPAPADFLRDWGATVVNADLSK- 132
V+ + V GATG G++IV + L G+ V+ VR A F D +V AD+++
Sbjct: 44 VKTKKVFVAGATGQTGKRIVEQLLSRGFAVKAGVRDVEKAKTSFKDDPSLQIVRADVTEG 103
Query: 133 PETIPATLVGVHTVIDCATG-RPEEPIKT---VDWEGKVALIQCAKAMGIQKYVFYSIHN 188
P+ + + + CATG RP I T VD G V L+ + G++K+V S
Sbjct: 104 PDKLAEVIGDDSQAVICATGFRPGFDIFTPWKVDNFGTVNLVDACRKQGVEKFVLVSSIL 163
Query: 189 CD-------KHPEVPLMEI-------KYCTENFLRDSGLNHIVIR 219
+ +P + + K E +++ SG+N+ ++R
Sbjct: 164 VNGAAMGQILNPAYLFLNLFGLTLVAKLQAEKYIKKSGINYTIVR 208
>At5g18660 unknown protein
Length = 417
Score = 50.8 bits (120), Expect = 1e-06
Identities = 64/263 (24%), Positives = 116/263 (43%), Gaps = 40/263 (15%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPA------PADFLRD-WGATVVNADLS 131
++LVVG+TG +GR +V+ + G++V + R + + L+ GA V +D++
Sbjct: 85 NVLVVGSTGYIGRFVVKEMIKRGFNVIAVAREKSGIRGKNDKEETLKQLQGANVCFSDVT 144
Query: 132 KPETIPATL----VGVHTVIDCATGRPEEPIK---TVDWEGKVALIQCAKAMGIQKYVFY 184
+ + + ++ GV V+ C R IK +D+E + K G + +V
Sbjct: 145 ELDVLEKSIENLGFGVDVVVSCLASR-NGGIKDSWKIDYEATKNSLVAGKKFGAKHFVLL 203
Query: 185 SIHNCDKHPEVPLMEIKYCTENF---------LRDSGLNHIVIRLCGFMQGLIGQYAVPI 235
S K PL+E + F +DS + ++R F + L GQ V I
Sbjct: 204 SAICVQK----PLLEFQRAKLKFEAELMDLAEQQDSSFTYSIVRPTAFFKSLGGQ--VEI 257
Query: 236 LEEKS---VWGTDAPTRIAYMDTQDIARLTFIA---LRNEKINGKLLTFAGP-RAWTTQE 288
+++ ++G + QD+A FIA L KIN ++L GP +A T E
Sbjct: 258 VKDGKPYVMFGDGKLCACKPISEQDLA--AFIADCVLEENKIN-QVLPIGGPGKALTPLE 314
Query: 289 VITLCERLAGQDANVTTVPVSVL 311
+ ++ G++ VP+ ++
Sbjct: 315 QGEILFKILGREPKFLKVPIEIM 337
>At1g16720 unknown protein (At1g16720)
Length = 598
Score = 50.4 bits (119), Expect = 2e-06
Identities = 31/102 (30%), Positives = 49/102 (47%), Gaps = 9/102 (8%)
Query: 52 IIRVGVKCNSNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPR 111
+IR G C A N+ ++LVVGAT +GR +VR+ + GY V+ LVR +
Sbjct: 146 LIREGPMCEFAVPGAQNV---------TVLVVGATSRIGRIVVRKLMLRGYTVKALVRKQ 196
Query: 112 PAPADFLRDWGATVVNADLSKPETIPATLVGVHTVIDCATGR 153
+ +V D+ +P T+ + + +I CAT R
Sbjct: 197 DEEVMSMLPRSVDIVVGDVGEPSTLKSAVESCSKIIYCATAR 238
>At4g13660 isoflavone reductase-like protein
Length = 317
Score = 48.1 bits (113), Expect = 8e-06
Identities = 56/250 (22%), Positives = 104/250 (41%), Gaps = 26/250 (10%)
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADF--------LRDWGATVVNAD 129
T +LVVG TG+LGR+IV L EG++ L RP D + GA +V
Sbjct: 10 TRVLVVGGTGSLGRRIVSACLAEGHETYVLQRPE-IGVDIEKVQLLLSFKRLGAHLVEGS 68
Query: 130 LSKPETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSIHNC 189
S +++ + + V V+ +G +T + ++ L+ K G K S
Sbjct: 69 FSDHQSLVSAVKQVDVVVSAMSG---VHFRTHNIPVQLKLVAAIKEAGNVKRFLPSEFGM 125
Query: 190 DKH-------PEVPLMEIKYCTENFLRDSGLNH-IVIRLC--GFMQGLIGQYAV--PILE 237
D P + K N ++ +G++H ++ C + G + Q P
Sbjct: 126 DPSRMGHAMPPGSETFDQKMEIRNAIKAAGISHTYLVGACFAAYFGGNLSQMGTLFPPKN 185
Query: 238 EKSVWGTDAPTRIAYMDTQDIARLTFIALRNEK-INGKLLTFAGPRAWTTQEVITLCERL 296
+ ++G D ++ ++D D+A+ T L + + +N + T E++ + E+L
Sbjct: 186 KVDIYG-DGNVKVVFVDEDDMAKYTAKTLNDPRTLNKTVYVRPTDNILTQMELVQIWEKL 244
Query: 297 AGQDANVTTV 306
++ T V
Sbjct: 245 TEKELEKTYV 254
>At5g02240 unknown protein
Length = 253
Score = 42.0 bits (97), Expect = 6e-04
Identities = 56/227 (24%), Positives = 92/227 (39%), Gaps = 39/227 (17%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYD---VRCLVRPRPAPADFLRDWGATVVNADLSKPET 135
++LV GA+G G QIV + L EG D + LVR + A V D++ ++
Sbjct: 6 TVLVTGASGRTG-QIVYKKLKEGSDKFVAKGLVRSAQGKEKIGGE--ADVFIGDITDADS 62
Query: 136 IPATLVGVHTVIDCAT--------------GRPE------EPIKTVDWEGKVALIQCAKA 175
I G+ ++ + GRPE + + VDW G+ I AK
Sbjct: 63 INPAFQGIDALVILTSAVPKMKPGFDPTKGGRPEFIFEDGQYPEQVDWIGQKNQIDAAKV 122
Query: 176 MGIQKYVFYSIHNCDKHPEVPLMEI--------KYCTENFLRDSGLNHIVIRLCGFMQGL 227
G++ V +P+ PL ++ K E +L DSG + +IR G +
Sbjct: 123 AGVKHIVVVGSMG-GTNPDHPLNKLGNGNILVWKRKAEQYLADSGTPYTIIRAGGLLDKE 181
Query: 228 IGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKINGK 274
G + + ++ + TD T + D+A + AL E+ K
Sbjct: 182 GGVRELLVGKDDELLQTDTKT----VPRADVAEVCIQALLFEEAKNK 224
>At5g10730 unknown protein
Length = 287
Score = 41.2 bits (95), Expect = 0.001
Identities = 30/116 (25%), Positives = 51/116 (43%), Gaps = 6/116 (5%)
Query: 74 PVRPTS-ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVV--NADL 130
P PT +LV+G G +G + + ALD G V L R + W + V +L
Sbjct: 52 PPPPTEKLLVLGGNGFVGSHVCKEALDRGLSVSSL--SRSGRSSLQESWASRVTWHQGNL 109
Query: 131 SKPETIPATLVGVHTVIDCATG-RPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYS 185
+ + L GV +VI C G + ++ + I+ A G++++V+ S
Sbjct: 110 LSSDLLKDALEGVTSVISCVGGFGSNSYMYKINGTANINAIRAASEKGVKRFVYIS 165
>At2g37660 unknown protein
Length = 325
Score = 40.4 bits (93), Expect = 0.002
Identities = 52/247 (21%), Positives = 96/247 (38%), Gaps = 35/247 (14%)
Query: 62 NSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALD--EGYDVRCLVRPRPAPADFLR 119
N +V + P ++LV GA G G+ + ++ + E + R LVR + +
Sbjct: 61 NRRVSVTVSAAATTEPLTVLVTGAGGRTGQIVYKKLKERSEQFVARGLVRTKESKEKING 120
Query: 120 DWGATVVNADLSKPETIPATLVGVHTVIDCATGRPE-----EPIK--------------- 159
+ V D+ +I + G+ ++ + P+ +P K
Sbjct: 121 E--DEVFIGDIRDTASIAPAVEGIDALVILTSAVPQMKPGFDPSKGGRPEFFFDDGAYPE 178
Query: 160 TVDWEGKVALIQCAKAMGIQKYVFYSI-------HNCDKHPEVPLMEIKYCTENFLRDSG 212
VDW G+ I AKA G+++ V H + ++ K E +L DSG
Sbjct: 179 QVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTNINHPLNSIGNANILVWKRKAEQYLADSG 238
Query: 213 LNHIVIRLCGFMQGLIGQYAVPILEEKSVWGTDAPTRIAYMDTQDIARLTFIALRNEKIN 272
+ + +IR G G + + ++ + T+ T + D+A + AL+ E+
Sbjct: 239 IPYTIIRAGGLQDKDGGIRELLVGKDDELLETETRT----IARADVAEVCVQALQLEEAK 294
Query: 273 GKLLTFA 279
K L A
Sbjct: 295 FKALDLA 301
>At1g32220 unknown protein
Length = 296
Score = 40.4 bits (93), Expect = 0.002
Identities = 38/151 (25%), Positives = 66/151 (43%), Gaps = 10/151 (6%)
Query: 51 RIIRVGVKCN------SNSERAVNLGPGTPVRPTSILVVGATGTLGRQIVRRALDEGYDV 104
R+ + VKC S++ + + V+ ++V+G G +G I + A+ G +V
Sbjct: 30 RLFKSCVKCTYAEAGLSSASWSAPIDIVADVKSERVVVLGGNGFVGSAICKAAISNGIEV 89
Query: 105 RCLVRP-RPAPADFLRDWGATVVNADLSKPETIPATLVGVHTVIDCATG-RPEEPIKTVD 162
+ R RP D D T V D+ L+G V+ G EE +K ++
Sbjct: 90 VSVSRSGRPNFEDSWLDQ-VTWVTGDVFYLNW-DEVLLGATAVVSTIGGFGNEEQMKRIN 147
Query: 163 WEGKVALIQCAKAMGIQKYVFYSIHNCDKHP 193
E V + AK G+ K+V ++H+ + P
Sbjct: 148 GEANVTAVNAAKDFGVPKFVLITVHDYNLPP 178
>At4g35420 unknown protein
Length = 326
Score = 38.1 bits (87), Expect = 0.008
Identities = 40/139 (28%), Positives = 60/139 (42%), Gaps = 19/139 (13%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDW---GA----TVVNADLSK 132
+ V GA+G L +V+R L EGY+V VR W GA +V ADL +
Sbjct: 8 VCVTGASGFLASWLVKRLLLEGYEVIGTVRDPGNEKKLAHLWKLEGAKERLRLVKADLME 67
Query: 133 PETIPATLVGVHTVIDCA------TGRPEEPIKTVDWEGKVALIQ-CAKAMGIQKYVFYS 185
+ ++G V A T PEE I EG + +++ C K +++ V S
Sbjct: 68 EGSFDNAIMGCQGVFHTASPVLKPTSNPEEEILRPAIEGTLNVLRSCRKNPSLKRVVLTS 127
Query: 186 IHNC-----DKHPEVPLME 199
+ D P++PL E
Sbjct: 128 SSSTVRIRDDFDPKIPLDE 146
>At1g19540 2'-hydroxyisoflavone reductase like protein
Length = 310
Score = 38.1 bits (87), Expect = 0.008
Identities = 27/83 (32%), Positives = 41/83 (48%), Gaps = 8/83 (9%)
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRPR----PAPADFL---RDWGATVVNADL 130
+ ILV+GATG +G+ +V + G+ LVR P A + +D G T++ L
Sbjct: 3 SKILVIGATGLIGKVLVEESAKSGHATFALVREASLSDPVKAQLVERFKDLGVTILYGSL 62
Query: 131 SKPETIPATLVGVHTVIDCATGR 153
S E++ + V VI A GR
Sbjct: 63 SDKESLVKAIKQVDVVIS-AVGR 84
>At5g42800 dihydroflavonol 4-reductase
Length = 382
Score = 37.0 bits (84), Expect = 0.019
Identities = 34/121 (28%), Positives = 55/121 (45%), Gaps = 14/121 (11%)
Query: 79 SILVVGATGTLGRQIVRRALDEGYDVRCLVRP--RPAPADFLRDWG-----ATVVNADLS 131
++ V GA+G +G +V R L+ GY VR VR L D T+ ADLS
Sbjct: 7 TVCVTGASGFIGSWLVMRLLERGYFVRATVRDPGNLKKVQHLLDLPNAKTLLTLWKADLS 66
Query: 132 KPETIPATLVG------VHTVIDCATGRPEEPIKTVDWEGKVALIQ-CAKAMGIQKYVFY 184
+ + + G V T +D + PE + G + +++ C KA ++++VF
Sbjct: 67 EEGSYDDAINGCDGVFHVATPMDFESKDPENEVIKPTVNGMLGIMKACVKAKTVRRFVFT 126
Query: 185 S 185
S
Sbjct: 127 S 127
>At2g02400 putative cinnamoyl-CoA reductase
Length = 318
Score = 37.0 bits (84), Expect = 0.019
Identities = 33/148 (22%), Positives = 63/148 (42%), Gaps = 17/148 (11%)
Query: 79 SILVVGATGTLGRQIVRRALDEGY-DVRCLVRPRPAPADFLR----DWGATVVNADLSKP 133
++ V GA G +G I+R +++GY + + P P L+ D + ADL
Sbjct: 5 TVCVTGANGFIGSWIIRTLIEKGYTKIHASIYPGSDPTHLLQLPGSDSKIKIFEADLLDS 64
Query: 134 ETIPATLVG----VHTVIDCATGRPEEPIKTV---DWEGKVALIQCAKAMGIQKYVFYS- 185
+ I + G H C P +P K + +G + +++ AK +++ V S
Sbjct: 65 DAISRAIDGCAGVFHVASPCTLDPPVDPEKELVEPAVKGTINVLEAAKRFNVRRVVITSS 124
Query: 186 ----IHNCDKHPEVPLMEIKYCTENFLR 209
+ N + +VP+ E + +F +
Sbjct: 125 ISALVPNPNWPEKVPVDESSWSDLDFCK 152
>At1g75290 NADPH oxidoreductase, putative
Length = 323
Score = 35.8 bits (81), Expect = 0.042
Identities = 20/77 (25%), Positives = 38/77 (48%), Gaps = 7/77 (9%)
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP-------RPAPADFLRDWGATVVNADL 130
+ ILV+G TG +G+ I+ ++ G+ LVR + +D+G T+++ DL
Sbjct: 6 SKILVIGGTGHIGKLIIEASVKAGHSTLALVREASLSDPNKGKTVQNFKDFGVTLLHGDL 65
Query: 131 SKPETIPATLVGVHTVI 147
+ E++ + VI
Sbjct: 66 NDHESLVKAIKQADVVI 82
>At4g18810 unknown protein
Length = 596
Score = 35.4 bits (80), Expect = 0.055
Identities = 26/73 (35%), Positives = 36/73 (48%), Gaps = 2/73 (2%)
Query: 80 ILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADFLRDWGATVVNADLSKPET-IPA 138
ILV GATG +GR+IV G V+ LVR L ++ AD++K T +P
Sbjct: 125 ILVAGATGGVGRRIVDILRKRGLPVKALVRNEEKARKMLGP-EIDLIVADITKENTLVPE 183
Query: 139 TLVGVHTVIDCAT 151
GV VI+ +
Sbjct: 184 KFKGVRKVINAVS 196
>At1g75280 putative NADPH oxidoreductase
Length = 310
Score = 35.4 bits (80), Expect = 0.055
Identities = 21/77 (27%), Positives = 37/77 (47%), Gaps = 7/77 (9%)
Query: 78 TSILVVGATGTLGRQIVRRALDEGYDVRCLVRP-------RPAPADFLRDWGATVVNADL 130
+ ILV+G TG +G+ +V + G+ LVR + +D G T+++ DL
Sbjct: 6 SKILVIGGTGYIGKFLVEASAKAGHSTFALVREATLSDPVKGKTVQSFKDLGVTILHGDL 65
Query: 131 SKPETIPATLVGVHTVI 147
+ E++ + V VI
Sbjct: 66 NDHESLVKAIKQVDVVI 82
>At4g34540 isoflavone reductase - like protein
Length = 306
Score = 34.3 bits (77), Expect = 0.12
Identities = 49/219 (22%), Positives = 93/219 (42%), Gaps = 22/219 (10%)
Query: 76 RPTSILVVGATGTLGRQIVRRALDEGYDVRCLVRPRPAPADF--LRDWGATVVNADLSKP 133
+ + +L++GATG LG + R +++ G+ L+R L D G T++ L
Sbjct: 6 KKSRVLIIGATGRLGNYLTRFSIESGHPTFALIRNTTLSDKLKSLSDAGVTLLKGSLEDE 65
Query: 134 ETIPATLVGVHTVIDCATGRPEEPIKTVDWEGKVALIQCAKAMGIQKYVFYSI-HNCDKH 192
++ + V VI P K V + K+ + +A I++++ N DK
Sbjct: 66 GSLAEAVSKVDVVISAI------PSKHV-LDQKLLVRVIKQAGSIKRFIPAEYGANPDKT 118
Query: 193 PEVPLMEIKYCTENFLR----DSGLNHIVIRLCG-FMQGLIGQYAVPIL-----EEKSVW 242
L Y ++ +R G+ + I CG FM+ L+ P L ++ +V+
Sbjct: 119 QVSDLDHDFYSKKSEIRHMIESEGIPYTYI-CCGLFMRVLLPSLVQPGLQSPPTDKVTVF 177
Query: 243 GTDAPTRIAYMDTQDIARLTFIALRNEKINGKLLTFAGP 281
G D + +++ D+A T + + + K L + P
Sbjct: 178 G-DGNVKAVFVNDVDVAAFTIKTIDDPRTLNKTLYLSPP 215
>At4g33360 putative dihydrokaempferol 4-reductase
Length = 344
Score = 33.5 bits (75), Expect = 0.21
Identities = 17/39 (43%), Positives = 21/39 (53%)
Query: 71 PGTPVRPTSILVVGATGTLGRQIVRRALDEGYDVRCLVR 109
P T ILV G+TG LG ++ L G+ VR LVR
Sbjct: 6 PNTETENMKILVTGSTGYLGARLCHVLLRRGHSVRALVR 44
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,752,260
Number of Sequences: 26719
Number of extensions: 385095
Number of successful extensions: 1002
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 975
Number of HSP's gapped (non-prelim): 37
length of query: 393
length of database: 11,318,596
effective HSP length: 101
effective length of query: 292
effective length of database: 8,619,977
effective search space: 2517033284
effective search space used: 2517033284
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC121239.8


