

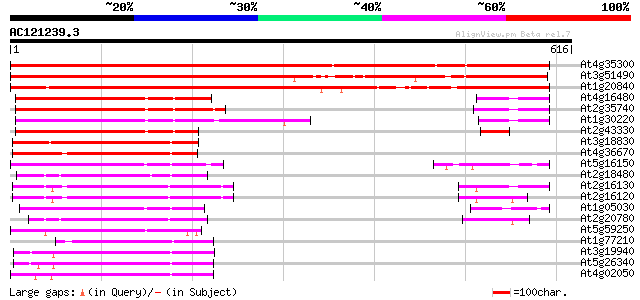
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121239.3 + phase: 0 /pseudo
(616 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g35300 sugar transporter like protein 871 0.0
At3g51490 sugar transporter-like protein 669 0.0
At1g20840 putative sugar transporter protein 646 0.0
At4g16480 membrane transporter like protein 154 1e-37
At2g35740 putative sugar transporter 154 2e-37
At1g30220 unknown protein 154 2e-37
At2g43330 membrane transporter like protein 146 4e-35
At3g18830 sugar transporter like protein 136 4e-32
At4g36670 sugar transporter like protein 135 9e-32
At5g16150 sugar transporter like protein 130 3e-30
At2g18480 putative sugar transporter 127 2e-29
At2g16130 putative sugar transporter 122 6e-28
At2g16120 putative sugar transporter 120 2e-27
At1g05030 sugar transporter like protein 119 4e-27
At2g20780 putative sugar transporter 114 1e-25
At5g59250 D-xylose-H+ symporter - like protein 107 1e-23
At1g77210 unknown protein 105 1e-22
At3g19940 putative monosaccharide transport protein, STP4 104 1e-22
At5g26340 hexose transporter - like protein 101 1e-21
At4g02050 putative hexose transporter 100 3e-21
>At4g35300 sugar transporter like protein
Length = 739
Score = 871 bits (2251), Expect = 0.0
Identities = 436/594 (73%), Positives = 506/594 (84%), Gaps = 5/594 (0%)
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
MSGAV+VA+AAA+GNLLQGWDNATIAG++LYIK+EF L+S P+VEGLIVAMSLIGAT++T
Sbjct: 1 MSGAVLVAIAAAVGNLLQGWDNATIAGAVLYIKKEFNLESNPSVEGLIVAMSLIGATLIT 60
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
TCSG ++D GRRPMLI+SS+LYF+ SLVM WSPNVY+LL RLLDG G+GL VTLVP+Y
Sbjct: 61 TCSGGVADWLGRRPMLILSSILYFVGSLVMLWSPNVYVLLLGRLLDGFGVGLVVTLVPIY 120
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL 180
ISE APPEIRG LNTLPQF GS GMF SYCMVFGMSL +PSWRLMLGVL IPSL++F L
Sbjct: 121 ISETAPPEIRGLLNTLPQFTGSGGMFLSYCMVFGMSLMPSPSWRLMLGVLFIPSLVFFFL 180
Query: 181 TLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGP 240
T+ LPESPRWLVSKGRMLEAK+VLQRLRG +DV+GEMALLVEGLG+GG+T+IEEYIIGP
Sbjct: 181 TVFFLPESPRWLVSKGRMLEAKRVLQRLRGREDVSGEMALLVEGLGIGGETTIEEYIIGP 240
Query: 241 DNELADEEDPSTGKDQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGS-MANPSG-LVDPL 298
+E+ D+ D + KDQIKLYG E G SWVARPV G S++ ++SR GS M+ G L+DPL
Sbjct: 241 ADEVTDDHDIAVDKDQIKLYGAEEGLSWVARPVKGGSTMSVLSRHGSTMSRRQGSLIDPL 300
Query: 299 VTLFGSVHEKLPETGSMRSTLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAAAG 358
VTLFGSVHEK+P+TGSMRS LFPHFGSMFSVGGNQPR+EDWDEE+L EG+DY SD
Sbjct: 301 VTLFGSVHEKMPDTGSMRSALFPHFGSMFSVGGNQPRHEDWDEENLVGEGEDYPSD-HGD 359
Query: 359 DSDDNLQSPLISRQTTSMDKDMPLPAQGSLSNMRQGSLLQGNAGEPVGSTGIGGGWQLAW 418
DS+D+L SPLISRQTTSM+KDMP A G+LS R GS +QG GE GS GIGGGWQ+AW
Sbjct: 360 DSEDDLHSPLISRQTTSMEKDMPHTAHGTLSTFRHGSQVQGAQGEGAGSMGIGGGWQVAW 419
Query: 419 KWSEQEGPGGKKEGGFKRIYLHQEGGPGSIRASVVSLPGGDVPTDGDVVQQAAALVSQPA 478
KW+E+E G+KEGGFKRIYLHQEG PGS R S+VSLPGGD + D V QA+ALVSQPA
Sbjct: 420 KWTEREDESGQKEGGFKRIYLHQEGFPGSRRGSIVSLPGGDGTGEADFV-QASALVSQPA 478
Query: 479 LYNKELMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGINGVL 538
LY+K+L+ + +GPAM+HPSET KG W+DL +PGVK AL VGVGLQILQQFSGINGVL
Sbjct: 479 LYSKDLLKEHTIGPAMVHPSET-TKGSIWHDLHDPGVKRALVVGVGLQILQQFSGINGVL 537
Query: 539 YYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR 592
YYTPQILEQAGVG LLSN+G+SS+S+S LISA+TT +MLP IAVAMRLMD+SGR
Sbjct: 538 YYTPQILEQAGVGILLSNMGISSSSASLLISALTTFVMLPAIAVAMRLMDLSGR 591
>At3g51490 sugar transporter-like protein
Length = 729
Score = 669 bits (1725), Expect = 0.0
Identities = 367/603 (60%), Positives = 439/603 (71%), Gaps = 33/603 (5%)
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
M V+VA+AAAIGN+LQGWDNATIAG+++YIK+EF L+ EP +EGLIVAMSLIGAT++T
Sbjct: 1 MRSVVLVALAAAIGNMLQGWDNATIAGAVIYIKKEFHLEKEPKIEGLIVAMSLIGATLIT 60
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
T SG +SD GRR MLI+SS+LYFLSS+VMFWSPNVY+LLFARLLDG GIGLAVTLVP+Y
Sbjct: 61 TFSGPVSDKVGRRSMLILSSVLYFLSSIVMFWSPNVYVLLFARLLDGFGIGLAVTLVPIY 120
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL 180
ISE AP EIRG LNT PQF GS GMF SYC+VFGMSL ++PSWRLMLGVLSIPS+ YF L
Sbjct: 121 ISETAPSEIRGLLNTFPQFCGSGGMFLSYCLVFGMSLQESPSWRLMLGVLSIPSIAYFVL 180
Query: 181 TLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGP 240
LPESPRWLVSKGRM EA++VLQRLRG +DV+GE+ALLVEGLGVG DTSIEEY+IGP
Sbjct: 181 AAFFLPESPRWLVSKGRMDEARQVLQRLRGREDVSGELALLVEGLGVGKDTSIEEYVIGP 240
Query: 241 DNELADEEDPSTGKDQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSG-LVDPLV 299
DNE + + KDQIKLYGPE GQSW+A+PV GQSS+ L SR+GSM G L+DPLV
Sbjct: 241 DNEENEGGNELPRKDQIKLYGPEDGQSWMAKPVKGQSSLALASRQGSMLPRGGSLMDPLV 300
Query: 300 TLFGSVHEKLPE---TGSMRSTLFPHFGSMFSVGGNQPRNEDWDEESLAREGDDYISDAA 356
TLFGS+HE LP S RS LFP+ GS+ + G Q WD E R +D
Sbjct: 301 TLFGSIHENLPSENMNASSRSMLFPNMGSILGMMGRQ--ESQWDPE---RNNED------ 349
Query: 357 AGDSDDNLQSPLISRQTTSMDKDMPLPAQGSLSNMRQGSLLQGNAGEPVGSTGIGGGWQL 416
+ D D+NL SPL+S QTT D D G++ + RQ SL N GE +T IGGGWQL
Sbjct: 350 SSDQDENLNSPLLSPQTTEPD-DYHQRTVGTM-HRRQSSLFMANVGETATATSIGGGWQL 407
Query: 417 AWKWSEQEGPGGKK-EGGFKRIYLHQEGG-------PGSIRASVVSL-PGGDVPTDGDVV 467
AWK++++ G GK+ GG +R+Y+H+E P S R S++S P GD +
Sbjct: 408 AWKYNDKVGADGKRVNGGLQRMYIHEETANNNTNNIPFSRRGSLLSFHPEGDGHDQVNGY 467
Query: 468 QQAAALVSQPALYNKELMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQI 527
QAAALVSQ + M G + P E GP W +L EPGVK AL VGVGLQI
Sbjct: 468 VQAAALVSQAS------MMPGGKGETAMLPKE-VKDGPGWRELKEPGVKRALMVGVGLQI 520
Query: 528 LQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLM 587
LQQF+GINGV+YYTPQILE+ GV LL+NLG+S+ S+S LISA+TTLLMLPCI V+MR +
Sbjct: 521 LQQFAGINGVMYYTPQILEETGVSSLLTNLGISAESASLLISALTTLLMLPCILVSMRSL 580
Query: 588 DIS 590
+S
Sbjct: 581 MLS 583
>At1g20840 putative sugar transporter protein
Length = 734
Score = 646 bits (1666), Expect = 0.0
Identities = 357/605 (59%), Positives = 441/605 (72%), Gaps = 34/605 (5%)
Query: 1 MSGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
M GA +VA+AA IGN LQGWDNATIAG+++YI ++ L + +V+GL+VAMSLIGATV+T
Sbjct: 1 MKGATLVALAATIGNFLQGWDNATIAGAMVYINKDLNLPT--SVQGLVVAMSLIGATVIT 58
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
TCSG +SD GRRPMLI+SS++YF+ L+M WSPNVY+L FARLL+G G GLAVTLVP+Y
Sbjct: 59 TCSGPISDWLGRRPMLILSSVMYFVCGLIMLWSPNVYVLCFARLLNGFGAGLAVTLVPVY 118
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLIYFAL 180
ISE APPEIRG LNTLPQF GS GMF SYCMVF MSL+ +PSWR MLGVLSIPSL+Y L
Sbjct: 119 ISETAPPEIRGQLNTLPQFLGSGGMFLSYCMVFTMSLSDSPSWRAMLGVLSIPSLLYLFL 178
Query: 181 TLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYIIGP 240
T+ LPESPRWLVSKGRM EAK+VLQ+L G +DV EMALLVEGL +GG+ ++E+ ++
Sbjct: 179 TVFYLPESPRWLVSKGRMDEAKRVLQQLCGREDVTDEMALLVEGLDIGGEKTMEDLLVTL 238
Query: 241 DNELADEEDPSTGKD-QIKLYGPEHGQSWVARPVTGQ-SSVGLVSRKGSMANPSGLV-DP 297
++ D+ + +D Q++LYG QS++ARPV Q SS+GL SR GS+AN S ++ DP
Sbjct: 239 EDHEGDDTLETVDEDGQMRLYGTHENQSYLARPVPEQNSSLGLRSRHGSLANQSMILKDP 298
Query: 298 LVTLFGSVHEKLPET-GSMRSTLFPHFGSMFSVGGNQPRNE--DWD---EESLAREGDDY 351
LV LFGS+HEK+PE G+ RS +FPHFGSMFS + P + W+ E ++ DDY
Sbjct: 299 LVNLFGSLHEKMPEAGGNTRSGIFPHFGSMFSTTADAPHGKPAHWEKDIESHYNKDNDDY 358
Query: 352 ISDAAAGDSDD---NLQSPLISRQTTSMDKDM-PLPAQGSLSNMRQGSLLQGNAGEPVGS 407
+D AGD DD +L+SPL+SRQTTSMDKDM P P GS +MR+ S L GE S
Sbjct: 359 ATDDGAGDDDDSDNDLRSPLMSRQTTSMDKDMIPHPTSGSTLSMRRHSTLMQGNGE--SS 416
Query: 408 TGIGGGWQLAWKWSEQEGPGGKKEGGFKRIYLHQEGGPGSIRASVVSLPGGDVPTDGDVV 467
GIGGGW + +++ E +KR YL +E G S R S++S+PGG P G
Sbjct: 417 MGIGGGWHMGYRYENDE---------YKRYYL-KEDGAESRRGSIISIPGG--PDGGGSY 464
Query: 468 QQAAALVSQPALYNKELMHQQPVGPAMIHPSETAAKGPSWNDLFEPGVKHALFVGVGLQI 527
A+ALVS+ L K + G AM+ P + AA GP W+ L EPGVK AL VGVG+QI
Sbjct: 465 IHASALVSRSVLGPKSVH-----GSAMVPPEKIAASGPLWSALLEPGVKRALVVGVGIQI 519
Query: 528 LQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLM 587
LQQFSGINGVLYYTPQILE+AGV LLS+LGLSS S+SFLIS +TTLLMLP I VAMRLM
Sbjct: 520 LQQFSGINGVLYYTPQILERAGVDILLSSLGLSSISASFLISGLTTLLMLPAIVVAMRLM 579
Query: 588 DISGR 592
D+SGR
Sbjct: 580 DVSGR 584
>At4g16480 membrane transporter like protein
Length = 582
Score = 154 bits (389), Expect = 1e-37
Identities = 80/217 (36%), Positives = 136/217 (61%), Gaps = 5/217 (2%)
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREF-QLQSEPTVEGLIVAMSLIGATVVTTCSGA 65
+A++A IG LL G+D I+G++L+IK +F ++ + ++ IV+M++ GA V G
Sbjct: 30 LALSAGIGGLLFGYDTGVISGALLFIKEDFDEVDKKTWLQSTIVSMAVAGAIVGAAVGGW 89
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
++D FGRR ++I+ +L+ + ++VM ++P ++++ R+ G G+G+A PLYISE +
Sbjct: 90 INDKFGRRMSILIADVLFLIGAIVMAFAPAPWVIIVGRIFVGFGVGMASMTSPLYISEAS 149
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLL 184
P IRG+L + + G FFSY + ++ P +WR MLGV +P+++ F L +L
Sbjct: 150 PARIRGALVSTNGLLITGGQFFSY--LINLAFVHTPGTWRWMLGVAGVPAIVQFVL-MLS 206
Query: 185 LPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALL 221
LPESPRWL K R+ E++ +L+R+ +V EM L
Sbjct: 207 LPESPRWLYRKDRIAESRAILERIYPADEVEAEMEAL 243
Score = 45.4 bits (106), Expect = 9e-05
Identities = 27/80 (33%), Positives = 43/80 (53%), Gaps = 9/80 (11%)
Query: 513 PGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVT 572
P V+ L G+ +Q+ QQF GIN V+YY+P I++ A G +S ++ +S +T
Sbjct: 272 PVVRRGLAAGITVQVAQQFVGINTVMYYSPSIVQFA---------GYASNKTAMALSLIT 322
Query: 573 TLLMLPCIAVAMRLMDISGR 592
+ L V+M +D GR
Sbjct: 323 SGLNALGSIVSMMFVDRYGR 342
>At2g35740 putative sugar transporter
Length = 580
Score = 154 bits (388), Expect = 2e-37
Identities = 90/233 (38%), Positives = 144/233 (61%), Gaps = 7/233 (3%)
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREF-QLQSEPTVEGLIVAMSLIGATVVTTCSGA 65
+A++A IG LL G++ IAG++LYIK EF ++ ++ ++ +IV+M++ GA V G
Sbjct: 29 LALSAGIGGLLFGYNTGVIAGALLYIKEEFGEVDNKTWLQEIIVSMTVAGAIVGAAIGGW 88
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
+D FGRR ++I+ +L+ L +LVM + ++++ RLL G G+G+A PLYISE++
Sbjct: 89 YNDKFGRRMSVLIADVLFLLGALVMVIAHAPWVIILGRLLVGFGVGMASMTSPLYISEMS 148
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLL 184
P IRG+L + + G F SY + ++ P +WR MLGV +IP++I F L +L
Sbjct: 149 PARIRGALVSTNGLLITGGQFLSY--LINLAFVHTPGTWRWMLGVSAIPAIIQFCL-MLT 205
Query: 185 LPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIEEYI 237
LPESPRWL R E++ +L+R+ + V E+A L E V +T+ E+ I
Sbjct: 206 LPESPRWLYRNDRKAESRDILERIYPAEMVEAEIAALKE--SVRAETADEDII 256
Score = 49.7 bits (117), Expect = 5e-06
Identities = 29/83 (34%), Positives = 45/83 (53%), Gaps = 9/83 (10%)
Query: 510 LFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLIS 569
L P V+H L G+ +Q+ QQF GIN V+YY+P IL+ A G +S ++ ++
Sbjct: 268 LSNPVVRHGLAAGITVQVAQQFVGINTVMYYSPTILQFA---------GYASNKTAMALA 318
Query: 570 AVTTLLMLPCIAVAMRLMDISGR 592
+T+ L V+M +D GR
Sbjct: 319 LITSGLNAVGSVVSMMFVDRYGR 341
>At1g30220 unknown protein
Length = 580
Score = 154 bits (388), Expect = 2e-37
Identities = 110/333 (33%), Positives = 177/333 (53%), Gaps = 16/333 (4%)
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-VEGLIVAMSLIGATVVTTCSGA 65
+A +A IG LL G+D I+G++LYI+ +F+ T ++ +IV+M++ GA V G
Sbjct: 31 LAFSAGIGGLLFGYDTGVISGALLYIRDDFKSVDRNTWLQEMIVSMAVAGAIVGAAIGGW 90
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
+D GRR ++++ L+ L +++M +PN +L+ R+ GLG+G+A PLYISE +
Sbjct: 91 ANDKLGRRSAILMADFLFLLGAIIMAAAPNPSLLVVGRVFVGLGVGMASMTAPLYISEAS 150
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLL 184
P +IRG+L + F + G F SY + ++ T +WR MLG+ IP+L+ F L +
Sbjct: 151 PAKIRGALVSTNGFLITGGQFLSY--LINLAFTDVTGTWRWMLGIAGIPALLQFVL-MFT 207
Query: 185 LPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSI-EEYIIGPDNE 243
LPESPRWL KGR EAK +L+R+ +DV E+ L + + +T I EE N
Sbjct: 208 LPESPRWLYRKGREEEAKAILRRIYSAEDVEQEIRALKDSV----ETEILEEGSSEKINM 263
Query: 244 LADEEDPSTGKDQIKLYGPEHGQSWVARPVTGQSSVGLVSRKGSMANPSGLVDPLVT--- 300
+ + + + I G + Q +V S +V G +N + L+ LVT
Sbjct: 264 IKLCKAKTVRRGLIAGVGLQVFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLVTAGL 323
Query: 301 -LFGSVHE--KLPETGSMRSTLFPHFGSMFSVG 330
FGS+ + G + + FG + S+G
Sbjct: 324 NAFGSIISIYFIDRIGRKKLLIISLFGVIISLG 356
Score = 48.9 bits (115), Expect = 8e-06
Identities = 29/78 (37%), Positives = 43/78 (54%), Gaps = 9/78 (11%)
Query: 515 VKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTL 574
V+ L GVGLQ+ QQF GIN V+YY+P I++ A G +S ++ L+S VT
Sbjct: 272 VRRGLIAGVGLQVFQQFVGINTVMYYSPTIVQLA---------GFASNRTALLLSLVTAG 322
Query: 575 LMLPCIAVAMRLMDISGR 592
L +++ +D GR
Sbjct: 323 LNAFGSIISIYFIDRIGR 340
>At2g43330 membrane transporter like protein
Length = 509
Score = 146 bits (368), Expect = 4e-35
Identities = 78/203 (38%), Positives = 127/203 (62%), Gaps = 5/203 (2%)
Query: 7 VAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPT-VEGLIVAMSLIGATVVTTCSGA 65
+ V A IG LL G+D I+G++LYIK +F++ + + ++ IV+M+L+GA + G
Sbjct: 34 LTVTAGIGGLLFGYDTGVISGALLYIKDDFEVVKQSSFLQETIVSMALVGAMIGAAAGGW 93
Query: 66 LSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIA 125
++D +GR+ + + +++ ++VM +P+ Y+L+ RLL GLG+G+A P+YI+E +
Sbjct: 94 INDYYGRKKATLFADVVFAAGAIVMAAAPDPYVLISGRLLVGLGVGVASVTAPVYIAEAS 153
Query: 126 PPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SWRLMLGVLSIPSLIYFALTLLL 184
P E+RG L + + G F SY + + T+ P +WR MLGV +P++I F L +L
Sbjct: 154 PSEVRGGLVSTNVLMITGGQFLSY--LVNSAFTQVPGTWRWMLGVSGVPAVIQFIL-MLF 210
Query: 185 LPESPRWLVSKGRMLEAKKVLQR 207
+PESPRWL K R EA +VL R
Sbjct: 211 MPESPRWLFMKNRKAEAIQVLAR 233
Score = 42.7 bits (99), Expect = 6e-04
Identities = 18/32 (56%), Positives = 23/32 (71%)
Query: 518 ALFVGVGLQILQQFSGINGVLYYTPQILEQAG 549
A G GLQ QQF+GIN V+YY+P I++ AG
Sbjct: 275 AFLAGAGLQAFQQFTGINTVMYYSPTIVQMAG 306
>At3g18830 sugar transporter like protein
Length = 539
Score = 136 bits (342), Expect = 4e-32
Identities = 70/205 (34%), Positives = 125/205 (60%), Gaps = 4/205 (1%)
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCS 63
A A+ A++ ++L G+D ++G+++YIKR+ ++ G++ I + + + +
Sbjct: 36 AFACAILASMTSILLGYDIGVMSGAMIYIKRDLKINDLQI--GILAGSLNIYSLIGSCAA 93
Query: 64 GALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISE 123
G SD GRR ++++ ++F +++M SPN L+F R + G+G+G A+ + P+Y +E
Sbjct: 94 GRTSDWIGRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRFIAGIGVGYALMIAPVYTAE 153
Query: 124 IAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMS-LTKAPSWRLMLGVLSIPSLIYFALTL 182
++P RG LN+ P+ +AG+ Y S L WRLMLG+ ++PS+I A+ +
Sbjct: 154 VSPASSRGFLNSFPEVFINAGIMLGYVSNLAFSNLPLKVGWRLMLGIGAVPSVI-LAIGV 212
Query: 183 LLLPESPRWLVSKGRMLEAKKVLQR 207
L +PESPRWLV +GR+ +AK+VL +
Sbjct: 213 LAMPESPRWLVMQGRLGDAKRVLDK 237
Score = 39.7 bits (91), Expect = 0.005
Identities = 21/75 (28%), Positives = 39/75 (52%), Gaps = 7/75 (9%)
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV----GYLLSNLGL 559
W +L P V+ + +G+ QQ SGI+ V+ ++P+I + AG+ LL+ + +
Sbjct: 280 WRELLIRPTPAVRRVMIAAIGIHFFQQASGIDAVVLFSPRIFKTAGLKTDHQQLLATVAV 339
Query: 560 SSTSSSFLISAVTTL 574
+SF++ A L
Sbjct: 340 GVVKTSFILVATFLL 354
>At4g36670 sugar transporter like protein
Length = 493
Score = 135 bits (339), Expect = 9e-32
Identities = 71/206 (34%), Positives = 125/206 (60%), Gaps = 8/206 (3%)
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEP--TVEGLIVAMSLIGATVVTT 61
A+ A+ A+I +++ G+D ++G++++I+ + + + G++ +L+G+ +
Sbjct: 17 ALQCAIVASIVSIIFGYDTGVMSGAMVFIEEDLKTNDVQIEVLTGILNLCALVGSLL--- 73
Query: 62 CSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYI 121
+G SD+ GRR ++++S+L+ L S++M W PN +LL R GLG+G A+ + P+Y
Sbjct: 74 -AGRTSDIIGRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLGVGFALMVAPVYS 132
Query: 122 SEIAPPEIRGSLNTLPQFAGSAGMFFSYCM-VFGMSLTKAPSWRLMLGVLSIPSLIYFAL 180
+EIA RG L +LP S G+ Y + F L WRLMLG+ ++PSL+ A
Sbjct: 133 AEIATASHRGLLASLPHLCISIGILLGYIVNYFFSKLPMHIGWRLMLGIAAVPSLV-LAF 191
Query: 181 TLLLLPESPRWLVSKGRMLEAKKVLQ 206
+L +PESPRWL+ +GR+ E K++L+
Sbjct: 192 GILKMPESPRWLIMQGRLKEGKEILE 217
Score = 39.7 bits (91), Expect = 0.005
Identities = 22/75 (29%), Positives = 36/75 (47%), Gaps = 7/75 (9%)
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV----GYLLSNLGL 559
W +L P V+ L +G+ Q SGI VL Y P+I ++AG+ L +G+
Sbjct: 261 WKELILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGPRIFKKAGITTKDKLFLVTIGV 320
Query: 560 SSTSSSFLISAVTTL 574
++F+ +A L
Sbjct: 321 GIMKTTFIFTATLLL 335
>At5g16150 sugar transporter like protein
Length = 546
Score = 130 bits (326), Expect = 3e-30
Identities = 82/235 (34%), Positives = 132/235 (55%), Gaps = 9/235 (3%)
Query: 2 SGAVIVAVAAA-IGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVT 60
SG V+ V A +G +L G+ + G++ Y+ ++ + ++G IV+ L GATV +
Sbjct: 102 SGTVLPFVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENTVLQGWIVSSLLAGATVGS 161
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
GAL+D FGR + ++ + + + + +V ++ RLL G+GIG++ +VPLY
Sbjct: 162 FTGGALADKFGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLY 221
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPS-WRLMLGVLSIPSLIYFA 179
ISEI+P EIRG+L ++ Q G+ + ++ G+ L P WR M GV IPS++ A
Sbjct: 222 ISEISPTEIRGALGSVNQLFICIGILAA--LIAGLPLAANPLWWRTMFGVAVIPSVL-LA 278
Query: 180 LTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEGLGVGGDTSIE 234
+ + PESPRWLV +G++ EA+K ++ L G + V LV L G S E
Sbjct: 279 IGMAFSPESPRWLVQQGKVSEAEKAIKTLYGKERVVE----LVRDLSASGQGSSE 329
Score = 43.1 bits (100), Expect = 5e-04
Identities = 39/135 (28%), Positives = 58/135 (42%), Gaps = 26/135 (19%)
Query: 466 VVQQAAALVSQPA---LYNKELMHQQPVGPAMIHPSETAAKGPS-----WNDLFEPGVKH 517
+VQQ ++ A LY KE + + ++ + +G S W DLF
Sbjct: 291 LVQQGKVSEAEKAIKTLYGKERVVE------LVRDLSASGQGSSEPEAGWFDLFSSRYWK 344
Query: 518 ALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLML 577
+ VG L + QQ +GIN V+YY+ + AG+ S ++S L+ A
Sbjct: 345 VVSVGAALFLFQQLAGINAVVYYSTSVFRSAGI--------QSDVAASALVGASNVF--- 393
Query: 578 PCIAVAMRLMDISGR 592
AVA LMD GR
Sbjct: 394 -GTAVASSLMDKMGR 407
>At2g18480 putative sugar transporter
Length = 508
Score = 127 bits (319), Expect = 2e-29
Identities = 72/212 (33%), Positives = 129/212 (59%), Gaps = 6/212 (2%)
Query: 8 AVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCSGALS 67
A+ A+I +++ G+D ++G+ ++I+ + ++ ++ +E L ++L A V + +G S
Sbjct: 26 AIVASIISIIFGYDTGVMSGAQIFIRDDLKI-NDTQIEVLAGILNLC-ALVGSLTAGKTS 83
Query: 68 DLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPP 127
D+ GRR + +S++++ + S++M + PN +L+ R + G+G+G A+ + P+Y +EI+
Sbjct: 84 DVIGRRYTIALSAVIFLVGSVLMGYGPNYPVLMVGRCIAGVGVGFALMIAPVYSAEISSA 143
Query: 128 EIRGSLNTLPQFAGSAGMFFSYC--MVFGMSLTKAPSWRLMLGVLSIPSLIYFALTLLLL 185
RG L +LP+ S G+ Y FG LT WRLMLG+ + PSLI A + +
Sbjct: 144 SHRGFLTSLPELCISLGILLGYVSNYCFG-KLTLKLGWRLMLGIAAFPSLI-LAFGITRM 201
Query: 186 PESPRWLVSKGRMLEAKKVLQRLRGCQDVAGE 217
PESPRWLV +GR+ EAKK++ + ++ A E
Sbjct: 202 PESPRWLVMQGRLEEAKKIMVLVSNTEEEAEE 233
Score = 40.8 bits (94), Expect = 0.002
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 7/75 (9%)
Query: 507 WNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV----GYLLSNLGL 559
W +L P V+ L VG+ + +GI V+ Y+P+I ++AGV LL+ +G+
Sbjct: 266 WRELVIKPRPAVRLILIAAVGIHFFEHATGIEAVVLYSPRIFKKAGVVSKDKLLLATVGV 325
Query: 560 SSTSSSFLISAVTTL 574
T + F+I A L
Sbjct: 326 GLTKAFFIIIATFLL 340
>At2g16130 putative sugar transporter
Length = 511
Score = 122 bits (306), Expect = 6e-28
Identities = 73/247 (29%), Positives = 142/247 (56%), Gaps = 12/247 (4%)
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVE---GLIVAMSLIGATVVT 60
A A+ A++ +++ G+D ++G+ ++IK + +L S+ +E G++ SLIG+
Sbjct: 26 AFACAILASMTSIILGYDIGVMSGAAIFIKDDLKL-SDVQLEILMGILNIYSLIGSGA-- 82
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
+G SD GRR ++++ +F +L+M ++ N ++ R + G+G+G A+ + P+Y
Sbjct: 83 --AGRTSDWIGRRYTIVLAGFFFFCGALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVY 140
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCM-VFGMSLTKAPSWRLMLGVLSIPSLIYFA 179
+E+AP RG L++ P+ + G+ Y F L + WR MLG+ ++PS ++ A
Sbjct: 141 TTEVAPASSRGFLSSFPEIFINIGILLGYVSNYFFAKLPEHIGWRFMLGIGAVPS-VFLA 199
Query: 180 LTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVA-GEMALLVEGLGVGGDTSIEEYII 238
+ +L +PESPRWLV +GR+ +A KVL + ++ A + + +G+ D + ++ I+
Sbjct: 200 IGVLAMPESPRWLVMQGRLGDAFKVLDKTSNTKEEAISRLNDIKRAVGIPDDMT-DDVIV 258
Query: 239 GPDNELA 245
P+ + A
Sbjct: 259 VPNKKSA 265
Score = 43.9 bits (102), Expect = 3e-04
Identities = 30/102 (29%), Positives = 47/102 (45%), Gaps = 12/102 (11%)
Query: 494 MIHPSETAAKGPSWNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV 550
++ P++ +A W DL P V+H L +G+ QQ SGI+ V+ Y+P I +A
Sbjct: 257 IVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFSQQASGIDAVVLYSPTIFSRA-- 314
Query: 551 GYLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRLMDISGR 592
GL S + L + ++ I V L+D GR
Sbjct: 315 -------GLKSKNDQLLATVAVGVVKTLFIVVGTCLVDRFGR 349
>At2g16120 putative sugar transporter
Length = 511
Score = 120 bits (302), Expect = 2e-27
Identities = 73/247 (29%), Positives = 141/247 (56%), Gaps = 12/247 (4%)
Query: 4 AVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVE---GLIVAMSLIGATVVT 60
A A+ A++ +++ G+D ++G+ ++IK + +L S+ +E G++ SL+G+
Sbjct: 26 AFACAILASMTSIILGYDIGVMSGASIFIKDDLKL-SDVQLEILMGILNIYSLVGSGA-- 82
Query: 61 TCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLY 120
+G SD GRR ++++ +F +L+M ++ N ++ R + G+G+G A+ + P+Y
Sbjct: 83 --AGRTSDWLGRRYTIVLAGAFFFCGALLMGFATNYPFIMVGRFVAGIGVGYAMMIAPVY 140
Query: 121 ISEIAPPEIRGSLNTLPQFAGSAGMFFSYCM-VFGMSLTKAPSWRLMLGVLSIPSLIYFA 179
+E+AP RG L + P+ + G+ Y F L + WR MLGV ++PS ++ A
Sbjct: 141 TAEVAPASSRGFLTSFPEIFINIGILLGYVSNYFFSKLPEHLGWRFMLGVGAVPS-VFLA 199
Query: 180 LTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVA-GEMALLVEGLGVGGDTSIEEYII 238
+ +L +PESPRWLV +GR+ +A KVL + ++ A + + +G+ D + ++ I+
Sbjct: 200 IGVLAMPESPRWLVLQGRLGDAFKVLDKTSNTKEEAISRLDDIKRAVGIPDDMT-DDVIV 258
Query: 239 GPDNELA 245
P+ + A
Sbjct: 259 VPNKKSA 265
Score = 43.1 bits (100), Expect = 5e-04
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 7/82 (8%)
Query: 494 MIHPSETAAKGPSWNDLF---EPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV 550
++ P++ +A W DL P V+H L +G+ QQ SGI+ V+ Y+P I +AG+
Sbjct: 257 IVVPNKKSAGKGVWKDLLVRPTPSVRHILIACLGIHFAQQASGIDAVVLYSPTIFSKAGL 316
Query: 551 ----GYLLSNLGLSSTSSSFLI 568
LL+ + + + F++
Sbjct: 317 KSKNDQLLATVAVGVVKTLFIV 338
>At1g05030 sugar transporter like protein
Length = 524
Score = 119 bits (299), Expect = 4e-27
Identities = 76/205 (37%), Positives = 114/205 (55%), Gaps = 4/205 (1%)
Query: 11 AAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCSGALSDLF 70
A++ N L G+ + G I+ I RE + +EGL+V++ + GA + + +G L D F
Sbjct: 86 ASMANFLFGYHIGVMNGPIVSIARELGFEGNSILEGLVVSIFIAGAFIGSIVAGPLVDKF 145
Query: 71 GRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIR 130
G R I ++ L +LV + ++ +L R L GLGIG+ LVP+YISE+AP + R
Sbjct: 146 GYRRTFQIFTIPLILGALVSAQAHSLDEILCGRFLVGLGIGVNTVLVPIYISEVAPTKYR 205
Query: 131 GSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPS-WRLMLGVLSIPSLIYFALTLLLLPESP 189
GSL TL Q G+ FS ++ G+ P WR ML V S+P + AL + ESP
Sbjct: 206 GSLGTLCQIGTCLGIIFS--LLLGIPAEDDPHWWRTMLYVASMPGFL-LALGMQFAVESP 262
Query: 190 RWLVSKGRMLEAKKVLQRLRGCQDV 214
RWL GR+ +AK V++ + G +V
Sbjct: 263 RWLCKVGRLDDAKVVIRNIWGGSEV 287
Score = 43.1 bits (100), Expect = 5e-04
Identities = 28/86 (32%), Positives = 41/86 (47%), Gaps = 12/86 (13%)
Query: 507 WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSF 566
W +L + F+G L +LQQF+GINGVLY++ N+G++S + +
Sbjct: 309 WLELLDKPHSRVAFIGGSLFVLQQFAGINGVLYFS---------SLTFQNVGITSGAQAS 359
Query: 567 LISAVTTLLMLPCIAVAMRLMDISGR 592
L VT C A L+D GR
Sbjct: 360 LYVGVTNFAGALC---ASYLIDKQGR 382
>At2g20780 putative sugar transporter
Length = 547
Score = 114 bits (286), Expect = 1e-25
Identities = 67/198 (33%), Positives = 116/198 (57%), Gaps = 4/198 (2%)
Query: 21 DNATIAGSILYIKREFQLQSEPTVEGLIVAMSLIGATVVTTCSGALSDLFGRRPMLIISS 80
D ++G++L+I+++ ++ +E E LI ++S+I + + G SD GR+ + +++
Sbjct: 93 DVGVMSGAVLFIQQDLKI-TEVQTEVLIGSLSII-SLFGSLAGGRTSDSIGRKWTMALAA 150
Query: 81 LLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFA 140
L++ + VM +P+ +L+ R L G+GIGL V + P+YI+EI+P RG + P+
Sbjct: 151 LVFQTGAAVMAVAPSFEVLMIGRTLAGIGIGLGVMIAPVYIAEISPTVARGFFTSFPEIF 210
Query: 141 GSAGMFFSYCMVFGMS-LTKAPSWRLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRML 199
+ G+ Y + S L+ SWR+ML V +PS ++ L ++PESPRWLV KGR+
Sbjct: 211 INLGILLGYVSNYAFSGLSVHISWRIMLAVGILPS-VFIGFALCVIPESPRWLVMKGRVD 269
Query: 200 EAKKVLQRLRGCQDVAGE 217
A++VL + D A E
Sbjct: 270 SAREVLMKTNERDDEAEE 287
Score = 50.4 bits (119), Expect = 3e-06
Identities = 26/79 (32%), Positives = 45/79 (56%), Gaps = 6/79 (7%)
Query: 498 SETAAKGPSWNDLFEPG--VKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGV----G 551
+E + P W +L P V+ L VG G+Q QQ +GI+ +YY+P+IL++AG+
Sbjct: 299 TEGSEDRPVWRELLSPSPVVRKMLIVGFGIQCFQQITGIDATVYYSPEILKEAGIQDETK 358
Query: 552 YLLSNLGLSSTSSSFLISA 570
L + + + T + F++ A
Sbjct: 359 LLAATVAVGVTKTVFILFA 377
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 107 bits (268), Expect = 1e-23
Identities = 76/223 (34%), Positives = 114/223 (51%), Gaps = 16/223 (7%)
Query: 2 SGAVIVAVAAAIGNLLQGWDNATIAGSILYIKREFQ-----LQSEPTVEGLIVAMSLIGA 56
S ++ + A+G LL G+D +G+ L ++ P GL+V+ SL GA
Sbjct: 96 SSVILPFIFPALGGLLFGYDIGATSGATLSLQSPALSGTTWFNFSPVQLGLVVSGSLYGA 155
Query: 57 TVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGIGLAVTL 116
+ + ++D GRR LII+++LY L SL+ +P++ ILL RLL G GIGLA+
Sbjct: 156 LLGSISVYGVADFLGRRRELIIAAVLYLLGSLITGCAPDLNILLVGRLLYGFGIGLAMHG 215
Query: 117 VPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVLSIPSLI 176
PLYI+E P +IRG+L +L + G+ + V + WR M G P +
Sbjct: 216 APLYIAETCPSQIRGTLISLKELFIVLGILLGF-SVGSFQIDVVGGWRYMYG-FGTPVAL 273
Query: 177 YFALTLLLLPESPRWLV-----SKGRMLEAKK----VLQRLRG 210
L + LP SPRWL+ KG++ E K+ L +LRG
Sbjct: 274 LMGLGMWSLPASPRWLLLRAVQGKGQLQEYKEKAMLALSKLRG 316
>At1g77210 unknown protein
Length = 504
Score = 105 bits (261), Expect = 1e-22
Identities = 59/173 (34%), Positives = 101/173 (58%), Gaps = 8/173 (4%)
Query: 51 MSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARLLDGLGI 110
+S GA+ VT ++GRR +++ S+ +FL ++ + N+ +L+ R+ G+GI
Sbjct: 98 ISTFGASYVTR-------IYGRRGSILVGSVSFFLGGVINAAAKNILMLILGRIFLGIGI 150
Query: 111 GLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWRLMLGVL 170
G VPLY+SE+AP +IRG++N L Q G+ + + + WRL LG+
Sbjct: 151 GFGNQAVPLYLSEMAPAKIRGTVNQLFQLTTCIGILVANLINYKTEQIHPWGWRLSLGLA 210
Query: 171 SIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVE 223
++P+++ F L L+LPE+P LV +G++ +AK VL ++RG ++ E LVE
Sbjct: 211 TVPAILMF-LGGLVLPETPNSLVEQGKLEKAKAVLIKVRGTNNIEAEFQDLVE 262
Score = 40.0 bits (92), Expect = 0.004
Identities = 19/74 (25%), Positives = 38/74 (50%)
Query: 501 AAKGPSWNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLS 560
A K P N L + +GL QQ +G+N +L+Y P + + G G S + +
Sbjct: 269 AVKNPFRNLLARRNRPQLVIGAIGLPAFQQLTGMNSILFYAPVMFQSLGFGGSASLISST 328
Query: 561 STSSSFLISAVTTL 574
T+++ +++A+ ++
Sbjct: 329 ITNAALVVAAIMSM 342
>At3g19940 putative monosaccharide transport protein, STP4
Length = 514
Score = 104 bits (260), Expect = 1e-22
Identities = 66/241 (27%), Positives = 127/241 (52%), Gaps = 23/241 (9%)
Query: 5 VIVAVAAAIGNLLQGWDNATIAGSILYIKREFQLQSEPTVEG------------------ 46
++ + AA+G LL G+D I+G + ++ EF + P VE
Sbjct: 26 IMTCIVAAMGGLLFGYDLG-ISGGVTSME-EFLTKFFPQVESQMKKAKHDTAYCKFDNQM 83
Query: 47 --LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARL 104
L + + A V + + ++ GR+ + I L + + +L ++ NV +L+ RL
Sbjct: 84 LQLFTSSLYLAALVASFMASVITRKHGRKVSMFIGGLAFLIGALFNAFAVNVSMLIIGRL 143
Query: 105 LDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWR 164
L G+G+G A P+Y+SE+AP +IRG+LN Q A + G+ + + +G S WR
Sbjct: 144 LLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMAITIGILVANLINYGTSKMAQHGWR 203
Query: 165 LMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVEG 224
+ LG+ ++P+++ + +LP++P ++ +G+ EAK++L+++RG +V E L++
Sbjct: 204 VSLGLAAVPAVV-MVIGSFILPDTPNSMLERGKNEEAKQMLKKIRGADNVDHEFQDLIDA 262
Query: 225 L 225
+
Sbjct: 263 V 263
Score = 34.3 bits (77), Expect = 0.21
Identities = 19/90 (21%), Positives = 41/90 (45%), Gaps = 10/90 (11%)
Query: 507 WNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSF 566
W ++ E + AL + QQ +GIN +++Y P + + G G ++
Sbjct: 273 WKNIMESKYRPALIFCSAIPFFQQITGINVIMFYAPVLFKTLGFG----------DDAAL 322
Query: 567 LISAVTTLLMLPCIAVAMRLMDISGRSISF 596
+ + +T ++ + V++ +D GR + F
Sbjct: 323 MSAVITGVVNMLSTFVSIYAVDRYGRRLLF 352
>At5g26340 hexose transporter - like protein
Length = 526
Score = 101 bits (252), Expect = 1e-21
Identities = 71/240 (29%), Positives = 121/240 (49%), Gaps = 23/240 (9%)
Query: 5 VIVAVAAAIGNLLQGWDNATIAGSIL----YIKREFQLQSEPTVEG-------------- 46
+I + AA G L+ G+D ++G + ++++ F + V G
Sbjct: 24 IISCIMAATGGLMFGYD-VGVSGGVTSMPDFLEKFFPVVYRKVVAGADKDSNYCKYDNQG 82
Query: 47 --LIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARL 104
L + + T + + GRR ++I+ + + + + + ++ +L+ R+
Sbjct: 83 LQLFTSSLYLAGLTATFFASYTTRTLGRRLTMLIAGVFFIIGVALNAGAQDLAMLIAGRI 142
Query: 105 LDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAP-SW 163
L G G+G A VPL++SEIAP IRG LN L Q + G+ F+ + +G + K W
Sbjct: 143 LLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNVTIGILFANLVNYGTAKIKGGWGW 202
Query: 164 RLMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVE 223
RL LG+ IP+L+ + LL+ E+P LV +GR+ E K VL+R+RG +V E A L+E
Sbjct: 203 RLSLGLAGIPALL-LTVGALLVTETPNSLVERGRLDEGKAVLRRIRGTDNVEPEFADLLE 261
Score = 37.4 bits (85), Expect = 0.025
Identities = 19/59 (32%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query: 519 LFVGVGLQILQQFSGINGVLYYTPQILEQAGVGYLLSNLGLSSTSSSFLISAVTTLLML 577
L + V LQI QQ +GIN +++Y P + G G S+ L S + ++ ++TL+ +
Sbjct: 285 LVIAVALQIFQQCTGINAIMFYAPVLFSTLGFG---SDASLYSAVVTGAVNVLSTLVSI 340
>At4g02050 putative hexose transporter
Length = 513
Score = 100 bits (248), Expect = 3e-21
Identities = 70/239 (29%), Positives = 119/239 (49%), Gaps = 18/239 (7%)
Query: 2 SGAVIVAVAAAIGNLLQGWDNATIAG------------SILYIKREFQLQSEPTV---EG 46
S +I + AAIG + G+D G +Y K++ +S +G
Sbjct: 25 SYVIIACLVAAIGGSIFGYDIGISGGVTSMDEFLEEFFHTVYEKKKQAHESNYCKYDNQG 84
Query: 47 LIVAMS--LIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPNVYILLFARL 104
L S + V T + ++ +GRR ++ + + + S + + N+ +LL R+
Sbjct: 85 LAAFTSSLYLAGLVSTLVASPITRNYGRRASIVCGGISFLIGSGLNAGAVNLAMLLAGRI 144
Query: 105 LDGLGIGLAVTLVPLYISEIAPPEIRGSLNTLPQFAGSAGMFFSYCMVFGMSLTKAPSWR 164
+ G+GIG VPLY+SE+AP +RG LN + Q A + G+F + + +G K WR
Sbjct: 145 MLGVGIGFGNQAVPLYLSEVAPTHLRGGLNMMFQLATTIGIFTANMVNYGTQQLKPWGWR 204
Query: 165 LMLGVLSIPSLIYFALTLLLLPESPRWLVSKGRMLEAKKVLQRLRGCQDVAGEMALLVE 223
L LG+ + P+L+ L LPE+P LV +G ++VL +LRG ++V E+ +V+
Sbjct: 205 LSLGLAAFPALL-MTLGGYFLPETPNSLVERGLTERGRRVLVKLRGTENVNAELQDMVD 262
Score = 32.7 bits (73), Expect = 0.61
Identities = 27/95 (28%), Positives = 44/95 (45%), Gaps = 9/95 (9%)
Query: 494 MIHPSETA--AKGPSWNDLFEPGVKHALFVGVGLQILQQFSGINGVLYYTPQILEQAGVG 551
M+ SE A K P + ++ + + L + + + + Q +GIN +L+Y P + + G G
Sbjct: 260 MVDASELANSIKHP-FRNILQKRHRPQLVMAICMPMFQILTGINSILFYAPVLFQTMGFG 318
Query: 552 YLLSNLGLSSTSSSFLISAVTTLLMLPCIAVAMRL 586
G +S SS L AV L I + RL
Sbjct: 319 ------GNASLYSSALTGAVLVLSTFISIGLVDRL 347
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,174,112
Number of Sequences: 26719
Number of extensions: 659497
Number of successful extensions: 2001
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 1735
Number of HSP's gapped (non-prelim): 171
length of query: 616
length of database: 11,318,596
effective HSP length: 105
effective length of query: 511
effective length of database: 8,513,101
effective search space: 4350194611
effective search space used: 4350194611
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC121239.3


