

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.7 - phase: 0
(83 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
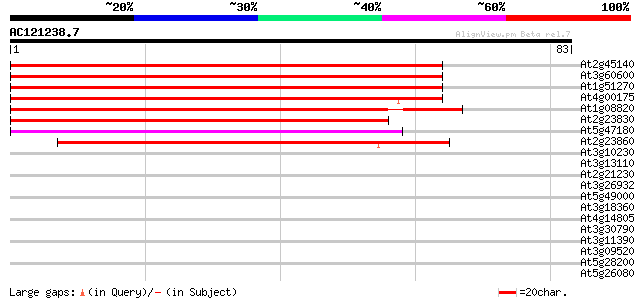
Score E
Sequences producing significant alignments: (bits) Value
At2g45140 putative VAMP-associated protein 88 7e-19
At3g60600 unknown protein 85 6e-18
At1g51270 unknown protein 74 1e-14
At4g00175 putative proline-rich protein 70 2e-13
At1g08820 unknown protein 67 2e-12
At2g23830 unknown protein 63 2e-11
At5g47180 VAMP (vesicle-associated membrane protein)-associated ... 59 6e-10
At2g23860 pseudogene 50 2e-07
At3g10230 lycopene beta cyclase 28 1.1
At3g13110 serine acetyltransferase (Sat-1) 27 2.4
At2g21230 bZIP transcription factor AtbZip30 27 2.4
At3g26932 putative protein 26 3.1
At5g49000 unknown protein 26 4.0
At3g18360 hypothetical protein 26 4.0
At4g14805 putative protein 25 5.2
At3g30790 hypothetical protein 25 5.2
At3g11390 unknown protein 25 5.2
At3g09520 hypothetical protein 25 5.2
At5g28200 putative protein 25 6.9
At5g26080 extensin - like protein 25 6.9
>At2g45140 putative VAMP-associated protein
Length = 239
Score = 88.2 bits (217), Expect = 7e-19
Identities = 40/64 (62%), Positives = 51/64 (79%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAP D+QCKDKFLLQ +VA PGAT KD+T +MF K++G+ V+E+K R V VAPP
Sbjct: 70 MQAQKEAPADLQCKDKFLLQCVVASPGATPKDVTHEMFSKEAGHRVEETKLRVVYVAPPR 129
Query: 61 SPNP 64
P+P
Sbjct: 130 PPSP 133
>At3g60600 unknown protein
Length = 256
Score = 85.1 bits (209), Expect = 6e-18
Identities = 37/64 (57%), Positives = 49/64 (75%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +EAP DMQCKDKFLLQ ++A PG T K++T +MF K++G+ V+E+K R VAPP
Sbjct: 87 MQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEMFSKEAGHRVEETKLRVTYVAPPR 146
Query: 61 SPNP 64
P+P
Sbjct: 147 PPSP 150
>At1g51270 unknown protein
Length = 289
Score = 74.3 bits (181), Expect = 1e-14
Identities = 32/64 (50%), Positives = 45/64 (70%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQAL+EAP DMQC+DK L Q V P T KD+TS+MF K++G+ +E++ + + V PP
Sbjct: 193 MQALKEAPADMQCRDKLLFQCKVVEPETTAKDVTSEMFSKEAGHPAEETRLKVMYVTPPQ 252
Query: 61 SPNP 64
P+P
Sbjct: 253 PPSP 256
Score = 73.2 bits (178), Expect = 2e-14
Identities = 33/64 (51%), Positives = 45/64 (69%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQAL+EAP D QCKDK L Q V PG K++TS+MF K++G+ V+E+ F+ + VAPP
Sbjct: 23 MQALKEAPADRQCKDKLLFQCKVVEPGTMDKEVTSEMFSKEAGHRVEETIFKIIYVAPPQ 82
Query: 61 SPNP 64
+P
Sbjct: 83 PQSP 86
>At4g00175 putative proline-rich protein
Length = 239
Score = 70.1 bits (170), Expect = 2e-13
Identities = 33/65 (50%), Positives = 48/65 (73%), Gaps = 1/65 (1%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV-APP 59
MQA +EAP DMQCKDKFL+Q++V G T+K++ ++MF K++G +++ K R V + A P
Sbjct: 72 MQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLRVVYIPANP 131
Query: 60 PSPNP 64
PSP P
Sbjct: 132 PSPVP 136
>At1g08820 unknown protein
Length = 386
Score = 67.0 bits (162), Expect = 2e-12
Identities = 34/67 (50%), Positives = 42/67 (61%), Gaps = 2/67 (2%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPP 60
MQA +E PPDM CKDKFL+QS T +DIT+ MF K G ++E+K R V PP
Sbjct: 70 MQAFKEPPPDMVCKDKFLIQSTAVSAETTDEDITASMFSKAEGKHIEENKLRVTLV--PP 127
Query: 61 SPNPPLS 67
S +P LS
Sbjct: 128 SDSPELS 134
>At2g23830 unknown protein
Length = 149
Score = 63.2 bits (152), Expect = 2e-11
Identities = 28/56 (50%), Positives = 41/56 (73%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCV 56
MQA +E P DMQ +KF++QS++A PG T K++T +MF K+SG+ V+E+K R V
Sbjct: 71 MQAQKEVPSDMQSFEKFMIQSVLASPGVTAKEVTREMFSKESGHVVEETKLRVTYV 126
>At5g47180 VAMP (vesicle-associated membrane protein)-associated
protein-like
Length = 220
Score = 58.5 bits (140), Expect = 6e-10
Identities = 27/58 (46%), Positives = 34/58 (58%)
Query: 1 MQALEEAPPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAP 58
+QA E PPDMQCKDKFLLQS + P ++ D F KDSG + E K + + P
Sbjct: 74 LQAQREYPPDMQCKDKFLLQSTIVPPHTDVDELPQDTFTKDSGKTLTECKLKVSYITP 131
>At2g23860 pseudogene
Length = 110
Score = 50.4 bits (119), Expect = 2e-07
Identities = 24/60 (40%), Positives = 36/60 (60%), Gaps = 2/60 (3%)
Query: 8 PPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRA--VCVAPPPSPNPP 65
P DMQ +KF++Q + A P T K++T + F K+SG+ V+E+K R VC +PP
Sbjct: 31 PSDMQSNEKFMIQRVKASPAVTAKEVTLETFNKESGHLVEETKLRVTYVCSTTTNITSPP 90
>At3g10230 lycopene beta cyclase
Length = 501
Score = 27.7 bits (60), Expect = 1.1
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 2/48 (4%)
Query: 21 SIVARPGATTKDITSDMF--YKDSGYEVKESKFRAVCVAPPPSPNPPL 66
S+VARPG +DI M K G VK + CV P P P L
Sbjct: 299 SLVARPGLRMEDIQERMAARLKHLGINVKRIEEDERCVIPMGGPLPVL 346
>At3g13110 serine acetyltransferase (Sat-1)
Length = 391
Score = 26.6 bits (57), Expect = 2.4
Identities = 10/17 (58%), Positives = 13/17 (75%)
Query: 49 SKFRAVCVAPPPSPNPP 65
SKF+ ++PPPSP PP
Sbjct: 37 SKFKHHTLSPPPSPPPP 53
>At2g21230 bZIP transcription factor AtbZip30
Length = 519
Score = 26.6 bits (57), Expect = 2.4
Identities = 18/79 (22%), Positives = 31/79 (38%), Gaps = 10/79 (12%)
Query: 15 DKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPPSPNPPLSVHA---- 70
D ++Q + + G ++ I + + FR PP P PP+S ++
Sbjct: 9 DTNMMQRVNSSSGTSSSSIPKHNLHLNPALIRSHHHFRHPFTGAPPPPIPPISPYSQIPA 68
Query: 71 ------SESLDQVSFYFIF 83
S S+ Q S +F F
Sbjct: 69 TLQPRHSRSMSQPSSFFSF 87
>At3g26932 putative protein
Length = 357
Score = 26.2 bits (56), Expect = 3.1
Identities = 9/11 (81%), Positives = 9/11 (81%)
Query: 54 VCVAPPPSPNP 64
VC APPP PNP
Sbjct: 323 VCSAPPPKPNP 333
>At5g49000 unknown protein
Length = 328
Score = 25.8 bits (55), Expect = 4.0
Identities = 14/44 (31%), Positives = 21/44 (46%), Gaps = 2/44 (4%)
Query: 21 SIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPPSPNP 64
S+V++ T +TS YK + + VC+ PP PNP
Sbjct: 46 SLVSKSSRTL--VTSPELYKTRSFFNRTESCLYVCLDFPPDPNP 87
>At3g18360 hypothetical protein
Length = 285
Score = 25.8 bits (55), Expect = 4.0
Identities = 12/52 (23%), Positives = 24/52 (46%)
Query: 14 KDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPPSPNPP 65
K F ++++ ++ IT++ + G + AV + P P P+PP
Sbjct: 158 KGIFFNNTMISEDSESSSVITTEENIGEHGQVNSSLPYSAVAIPPQPPPHPP 209
>At4g14805 putative protein
Length = 219
Score = 25.4 bits (54), Expect = 5.2
Identities = 11/24 (45%), Positives = 15/24 (61%)
Query: 58 PPPSPNPPLSVHASESLDQVSFYF 81
PPP P P L +S+SL +F+F
Sbjct: 180 PPPPPLPVLQYFSSDSLKIRNFWF 203
>At3g30790 hypothetical protein
Length = 953
Score = 25.4 bits (54), Expect = 5.2
Identities = 15/47 (31%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Query: 8 PPDMQCKDKFLLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAV 54
PPDM + +FL SI+ PG D+F + +E+K+ + V
Sbjct: 288 PPDMYMQSEFLFLSILV-PGPKHPKRALDVFLQPLIHELKKLWYEGV 333
>At3g11390 unknown protein
Length = 710
Score = 25.4 bits (54), Expect = 5.2
Identities = 9/11 (81%), Positives = 10/11 (90%)
Query: 56 VAPPPSPNPPL 66
V PPPSP+PPL
Sbjct: 81 VPPPPSPHPPL 91
>At3g09520 hypothetical protein
Length = 628
Score = 25.4 bits (54), Expect = 5.2
Identities = 12/37 (32%), Positives = 20/37 (53%), Gaps = 3/37 (8%)
Query: 47 KESKFRAVCVAPPPS---PNPPLSVHASESLDQVSFY 80
+++ CVA PS P PPL++ +SL+ + Y
Sbjct: 4 RKAMINICCVATTPSKYPPPPPLTLTPRQSLESAAIY 40
>At5g28200 putative protein
Length = 528
Score = 25.0 bits (53), Expect = 6.9
Identities = 18/58 (31%), Positives = 26/58 (44%)
Query: 18 LLQSIVARPGATTKDITSDMFYKDSGYEVKESKFRAVCVAPPPSPNPPLSVHASESLD 75
+L + + G T +D F K+S E + SK + P P P +V AS S D
Sbjct: 301 MLDAKLETQGKTMISTLTDWFAKNSFIEGEHSKDPDCGINRPKEPVPASNVGASNSDD 358
>At5g26080 extensin - like protein
Length = 141
Score = 25.0 bits (53), Expect = 6.9
Identities = 11/26 (42%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Query: 57 APPPSP-NPPLSVHASESLDQVSFYF 81
+PPP+P +PP VH Q +FY+
Sbjct: 100 SPPPTPISPPPKVHHPAPQAQKAFYY 125
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,054,244
Number of Sequences: 26719
Number of extensions: 73633
Number of successful extensions: 540
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 498
Number of HSP's gapped (non-prelim): 53
length of query: 83
length of database: 11,318,596
effective HSP length: 59
effective length of query: 24
effective length of database: 9,742,175
effective search space: 233812200
effective search space used: 233812200
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC121238.7


