

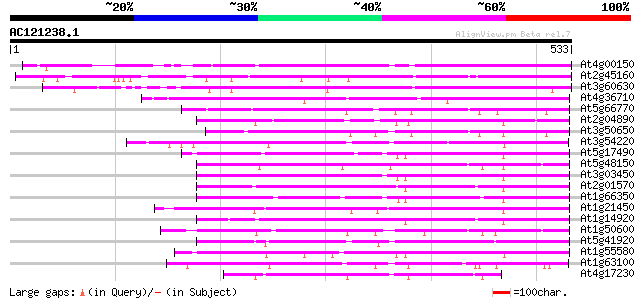
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121238.1 + phase: 0
(533 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g00150 scarecrow-like 6 (SCL6) 389 e-108
At2g45160 putative SCARECROW gene regulator 356 2e-98
At3g60630 scarecrow - like protein 331 7e-91
At4g36710 SCARECROW-like protein 182 4e-46
At5g66770 SCARECROW gene regulator 145 7e-35
At2g04890 putative SCARECROW gene regulator 135 4e-32
At3g50650 scarecrow-like 7 (SCL7) 127 1e-29
At3g54220 SCARECROW1 127 2e-29
At5g17490 RGA-like protein 122 6e-28
At5g48150 SCARECROW gene regulator-like 121 8e-28
At3g03450 RGA1-like protein 120 1e-27
At2g01570 putative RGA1, giberellin repsonse modulation protein 120 1e-27
At1g66350 gibberellin regulatory protein like 117 2e-26
At1g21450 scarecrow-like 1 (SCL1) 115 8e-26
At1g14920 signal response protein (GAI) 114 1e-25
At1g50600 scarecrow-like 5 (SCL5) 112 7e-25
At5g41920 SCARECROW gene regulator-like protein 108 1e-23
At1g55580 unknown protein 102 4e-22
At1g63100 transcription factor SCARECROW, putative 100 3e-21
At4g17230 scarecrow-like 13 (SCL13) 90 3e-18
>At4g00150 scarecrow-like 6 (SCL6)
Length = 558
Score = 389 bits (998), Expect = e-108
Identities = 239/530 (45%), Positives = 313/530 (58%), Gaps = 66/530 (12%)
Query: 13 DDNSEEKCGGGGMRMEDWEGQ-----DQSLLRLIMGDVEDPSAGLNKILQNSGYGSQNVD 67
D ++E+CG G+ DWE Q +QS+L LIMGD DPS LN ILQ S +
Sbjct: 83 DATTDEQCGAIGLG--DWEEQVPHDHEQSILGLIMGDSTDPSLELNSILQTSPTFHDSDY 140
Query: 68 FHGGFGVLDHQQQQGLTMMDASVQQGNYNVFPFIPENYNVLPLLDSGQEVF-ARRHQQQE 126
GFGV+D F ++++V P SG + ++ H Q
Sbjct: 141 SSPGFGVVDTG---------------------FGLDHHSVPPSHVSGLLINQSQTHYTQN 179
Query: 127 TQLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIE 186
+ HH+ P AK +++ G I +QL K AE+IE
Sbjct: 180 PAAIFYGHHH----------HTPPPAK--RLNPGPVG----------ITEQLVKAAEVIE 217
Query: 187 AGNPVQAQGILARLNHQLS-PIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKI 245
+ + AQGILARLN QLS P+G P +RA+FY KEAL +LH+ L +P S IFKI
Sbjct: 218 SDTCL-AQGILARLNQQLSSPVGKPLERAAFYFKEALNNLLHNVSQTL---NPYSLIFKI 273
Query: 246 GAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNG 305
AYKSFSEISPVLQFANFT NQ+L+E+ F R+H+IDFDIG+G QW+S MQE+VLR N
Sbjct: 274 AAYKSFSEISPVLQFANFTSNQALLESFHGFHRLHIIDFDIGYGGQWASLMQELVLRDNA 333
Query: 306 KP-SLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHF 364
P SLKIT SP++ +++EL FTQ+NL +A ++NI + VL+++ L S P
Sbjct: 334 APLSLKITVFASPANHDQLELGFTQDNLKHFASEINISLDIQVLSLDLLGSISWPNS--- 390
Query: 365 FDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVL 424
EA+ VN +SF S P+ L F+K L P I+V D+ C+R D+P + H L
Sbjct: 391 -SEKEAVAVNISAASF----SHLPLVLRFVKHLSPTIIVCSDRGCERTDLPFSQQLAHSL 445
Query: 425 QCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQRDK-LPPWRNMFLQSGFS 483
++AL ESLDAVN NLD +QKIER IQP I K+VL ++ + W+ MFLQ GFS
Sbjct: 446 HSHTALFESLDAVNANLDAMQKIERFLIQPEIEKLVLDRSRPIERPMMTWQAMFLQMGFS 505
Query: 484 PFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
P + SNFTE+QAECLVQR PVRGF VE+K +SL+LCWQR EL+ VS WRC
Sbjct: 506 PVTHSNFTESQAECLVQRTPVRGFHVEKKHNSLLLCWQRTELVGVSAWRC 555
>At2g45160 putative SCARECROW gene regulator
Length = 640
Score = 356 bits (914), Expect = 2e-98
Identities = 242/573 (42%), Positives = 315/573 (54%), Gaps = 66/573 (11%)
Query: 6 NLAHLSNDDNSEEKCGGGGMRMEDW------EGQDQSLLRLIMGD-----VEDPSAGLNK 54
N + DDN+ KC G+ D GQ+QS+LRLIM V DP G
Sbjct: 89 NTTVTAGDDNNNNKCSQMGLDDLDGVLSASSPGQEQSILRLIMDPGSAFGVFDPGFGFG- 147
Query: 55 ILQNSGYGSQNVDFHGGFGVLDHQQQQGLTM-MDASVQQGNYNVF---PFIP--ENYN-- 106
SG G + +L + Q +T +A + N+ +F P P + +N
Sbjct: 148 ----SGSGPVSAPVSDNSNLLCNFPFQEITNPAEALINPSNHCLFYNPPLSPPAKRFNSG 203
Query: 107 -----VLPLLDS--GQEVFARRHQQQETQLPLFP-HHYLQHQQQQQQSSVVPFAKQQKVS 158
V PL D G + R+HQ Q FP +H Q QQ SS A S
Sbjct: 204 SLHQPVFPLSDPDPGHDPVRRQHQFQ------FPFYHNNQQQQFPSSSSSTAVAMVPVPS 257
Query: 159 SSTTGDDASIQLQQSIFDQLFKTAELI------EAGNPVQAQGILARLNHQLSPIGN--- 209
GDD S+ I +QLF AELI + V AQGILARLNH L+ N
Sbjct: 258 PGMAGDDQSV-----IIEQLFNAAELIGTTGNNNGDHTVLAQGILARLNHHLNTSSNHKS 312
Query: 210 PFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSL 269
PFQRA+ ++ EAL ++H N ++ +P + I +I AY+SFSE SP LQF NFT NQS+
Sbjct: 313 PFQRAASHIAEALLSLIH-NESSPPLITPENLILRIAAYRSFSETSPFLQFVNFTANQSI 371
Query: 270 IEALER--FDRIHVIDFDIGFGVQWSSFMQEIVL-----RSNGKPSLKITAVVSPSSC-- 320
+E+ FDRIH+IDFD+G+G QWSS MQE+ R N SLK+T P S
Sbjct: 372 LESCNESGFDRIHIIDFDVGYGGQWSSLMQELASGVGGRRRNRASSLKLTVFAPPPSTVS 431
Query: 321 NEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFPVSSF 380
+E EL FT+ENL +A ++ I FE +L++E L P+ EAI VN PV+S
Sbjct: 432 DEFELRFTEENLKTFAGEVKIPFEIELLSVELLLNPAYWPLSLRSSEKEAIAVNLPVNSV 491
Query: 381 ISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVN 440
S P+ L FLKQL P IVV D+ CDR D P P V+H LQ +++LLESLDA N N
Sbjct: 492 ASG--YLPLILRFLKQLSPNIVVCSDRGCDRNDAPFPNAVIHSLQYHTSLLESLDA-NQN 548
Query: 441 LDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ 500
D IER ++QP+I K+++ H ++ PPWR +F Q GFSP S S EAQAECL+Q
Sbjct: 549 QDD-SSIERFWVQPSIEKLLMKRHRWIERSPPWRILFTQCGFSPASLSQMAEAQAECLLQ 607
Query: 501 RAPVRGFQVERKPSSLVLCWQRKELISVSTWRC 533
R PVRGF VE++ SSLV+CWQRKEL++VS W+C
Sbjct: 608 RNPVRGFHVEKRQSSLVMCWQRKELVTVSAWKC 640
>At3g60630 scarecrow - like protein
Length = 623
Score = 331 bits (848), Expect = 7e-91
Identities = 220/531 (41%), Positives = 307/531 (57%), Gaps = 53/531 (9%)
Query: 32 GQDQSLLRLIM-GDVEDPSAGLNKILQNSG----YGSQNVDFHGGFGVLDHQQQQGLTMM 86
GQ+QS+ RLIM GDV DP + SG + N F GF + +++ +
Sbjct: 117 GQEQSIFRLIMAGDVVDPGSEFVGFDIGSGSDPVIDNPNPLFGYGFPFQNAPEEEKF-QI 175
Query: 87 DASVQQGNYNVFPFIPENYNVLPLLDSGQEVFARRHQQQETQLPLFPHHYLQHQQQQQQS 146
+ G ++ P P L+SGQ +H Q +FP H+
Sbjct: 176 SINPNPGFFSDPPSSPPAKR----LNSGQP--GSQHLQW-----VFPFSDPGHESHD--- 221
Query: 147 SVVPFAKQQKVSSSTTGDDASIQLQQS-IFDQLFKTA-ELIEAG---NPVQAQGILARLN 201
PF K++ G+D + Q Q + I DQLF A EL G NPV AQGILARLN
Sbjct: 222 ---PFLTPPKIA----GEDQNDQDQSAVIIDQLFSAAAELTTNGGDNNPVLAQGILARLN 274
Query: 202 HQLSPIGN--------PFQRASFYMKEALQLMLHSNGNNLTAFSPI-SFIFKIGAYKSFS 252
H L+ + PF RA+ Y+ EAL +L + + + SP + IF+I AY++FS
Sbjct: 275 HNLNNNNDDTNNNPKPPFHRAASYITEALHSLLQDSSLSPPSLSPPQNLIFRIAAYRAFS 334
Query: 253 EISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIV---LRSNGKPSL 309
E SP LQF NFT NQ+++E+ E FDRIH++DFDIG+G QW+S +QE+ RS+ PSL
Sbjct: 335 ETSPFLQFVNFTANQTILESFEGFDRIHIVDFDIGYGGQWASLIQELAGKRNRSSSAPSL 394
Query: 310 KITAVVSPSS-CNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPS-CPLPGHFFDS 367
KITA SPS+ +E EL FT+ENL +A + + FE +LN+E L P+ PL
Sbjct: 395 KITAFASPSTVSDEFELRFTEENLRSFAGETGVSFEIELLNMEILLNPTYWPLSLFRSSE 454
Query: 368 NEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDR-MDVPLPTNVVHVLQC 426
EAI VN P+SS +S P+ L FLKQ+ P +VV D++CDR D P P V++ LQ
Sbjct: 455 KEAIAVNLPISSMVS--GYLPLILRFLKQISPNVVVCSDRSCDRNNDAPFPNGVINALQY 512
Query: 427 YSALLESLDAVNV-NLDVLQKIERHYIQPTINKIVLSHHNQRDKLPPWRNMFLQSGFSPF 485
Y++LLESLD+ N+ N + IER +QP+I K++ + + ++ PPWR++F Q GF+P
Sbjct: 513 YTSLLESLDSGNLNNAEAATSIERFCVQPSIQKLLTNRYRWMERSPPWRSLFGQCGFTPV 572
Query: 486 SFSNFTEAQAECLVQRAPVRGFQVERKPS---SLVLCWQRKELISVSTWRC 533
+ S E QAE L+QR P+RGF +E++ S SLVLCWQRKEL++VS W+C
Sbjct: 573 TLSQTAETQAEYLLQRNPMRGFHLEKRQSSSPSLVLCWQRKELVTVSAWKC 623
>At4g36710 SCARECROW-like protein
Length = 486
Score = 182 bits (462), Expect = 4e-46
Identities = 129/417 (30%), Positives = 214/417 (50%), Gaps = 17/417 (4%)
Query: 126 ETQLPLFPHHYLQHQQQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELI 185
++ LP FP +Q + S V P + Q D+ + L + + +
Sbjct: 76 DSNLPGFPDQ-IQPSDFESSSDVYP-GQNQTTGYGFNSLDSVDNGGFDFIEDLIRVVDCV 133
Query: 186 EAGNPVQAQGILARLNHQL-SPIGNPFQRASFYMKEAL-QLMLHSNGNNLTAFSPISFIF 243
E+ AQ +L+RLN +L SP G P QRA+FY KEAL + SN N + S +
Sbjct: 134 ESDELQLAQVVLSRLNQRLRSPAGRPLQRAAFYFKEALGSFLTGSNRNPIRLSSWSEIVQ 193
Query: 244 KIGAYKSFSEISPVLQFANFTCNQSLIEALERFDR---IHVIDFDIGFGVQWSSFMQEIV 300
+I A K +S ISP+ F++FT NQ+++++L +HV+DF+IGFG Q++S M+EI
Sbjct: 194 RIRAIKEYSGISPIPLFSHFTANQAILDSLSSQSSSPFVHVVDFEIGFGGQYASLMREIT 253
Query: 301 LRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPL 360
+S L++TAVV+ +E +ENL+Q+A ++ I F+ + +++ + S
Sbjct: 254 EKSVSGGFLRVTAVVAEECA--VETRLVKENLTQFAAEMKIRFQIEFVLMKTFEMLSFKA 311
Query: 361 PGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDV---PLP 417
+ ++ + +S + F ++ L+++ PK+VV +D
Sbjct: 312 IRFVEGERTVVLISPAIFRRLSGITDF---VNNLRRVSPKVVVFVDSEGWTEIAGSGSFR 368
Query: 418 TNVVHVLQCYSALLESLDAVNVNLDVLQKI-ERHYIQPTINKIVLSHHNQRDKLP-PWRN 475
V L+ Y+ +LESLDA D+++KI E ++P I+ V + ++R WR
Sbjct: 369 REFVSALEFYTMVLESLDAAAPPGDLVKKIVEAFVLRPKISAAVETAADRRHTGEMTWRE 428
Query: 476 MFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
F +G P S F + QAECL+++A VRGF V ++ LVLCW + L++ S WR
Sbjct: 429 AFCAAGMRPIQQSQFADFQAECLLEKAQVRGFHVAKRQGELVLCWHGRALVATSAWR 485
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 145 bits (365), Expect = 7e-35
Identities = 107/386 (27%), Positives = 190/386 (48%), Gaps = 25/386 (6%)
Query: 164 DDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQ 223
+D L+ + ++ A + ++ +P +A L ++ +S +G+P +R +FY EAL
Sbjct: 207 EDDDFDLEPPLLKAIYDCARISDS-DPNEASKTLLQIRESVSELGDPTERVAFYFTEALS 265
Query: 224 LMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVID 283
L N + T+ S S I +YK+ ++ P +FA+ T NQ+++EA E+ ++IH++D
Sbjct: 266 NRLSPN-SPATSSSSSSTEDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHIVD 324
Query: 284 FDIGFGVQWSSFMQEIVLRSNGKPS-LKITAVVSPS--SCNEIELNFTQENLSQYAKDLN 340
F I G+QW + +Q + R++GKP+ ++++ + +PS E L T L +AK L+
Sbjct: 325 FGIVQGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKVLD 384
Query: 341 ILFEFNVLNIESLNLPSCPLPGHFF--DSNEAIGVNFPVSSF---ISNPSCFPVALHFLK 395
+ F+F + P L G F D +E + VNF + + P+ AL K
Sbjct: 385 LNFDF-----IPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAK 439
Query: 396 QLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQ--KIERHYIQ 453
L P++V + V V + LQ YSA+ ESL+ N+ D + ++ER
Sbjct: 440 SLNPRVVTLGEYEVSLNRVGFANRVKNALQFYSAVFESLEP-NLGRDSEERVRVERELFG 498
Query: 454 PTINKIVLS-----HHNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQ 508
I+ ++ H + ++ WR + +GF SN+ +QA+ L+
Sbjct: 499 RRISGLIGPEKTGIHRERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLY 558
Query: 509 --VERKPSSLVLCWQRKELISVSTWR 532
VE KP + L W L+++S+WR
Sbjct: 559 SIVESKPGFISLAWNDLPLLTLSSWR 584
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 135 bits (341), Expect = 4e-32
Identities = 102/372 (27%), Positives = 170/372 (45%), Gaps = 29/372 (7%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNN----L 233
L A+ + N + A+ + L +S G P QR YM E L L ++G++ L
Sbjct: 54 LVACAKAVSENNLLMARWCMGELRGMVSISGEPIQRLGAYMLEGLVARLAASGSSIYKSL 113
Query: 234 TAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWS 293
+ P S+ F Y E+ P +F + N ++ EA++ +RIH+IDF IG G QW
Sbjct: 114 QSREPESYEFLSYVYV-LHEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIGQGSQWI 172
Query: 294 SFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESL 353
+ +Q R G P+++IT V S L ++ L + AK ++ F FN ++
Sbjct: 173 ALIQAFAARPGGAPNIRITGVGDGS-----VLVTVKKRLEKLAKKFDVPFRFN-----AV 222
Query: 354 NLPSCPLPGHFFD--SNEAIGVNFPV------SSFISNPSCFPVALHFLKQLRPKIVVTL 405
+ PSC + D EA+GVNF +S + L +K L PK+V +
Sbjct: 223 SRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKVVTLV 282
Query: 406 DKNCDRMDVPLPTNVVHVLQCYSALLESLDA-VNVNLDVLQKIERHYIQPTINKIVLSHH 464
++ C+ P + L Y+A+ ES+D + N IE+H + + I+
Sbjct: 283 EQECNTNTSPFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNIIACEG 342
Query: 465 NQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCW 520
+R + L W++ F +GF P+ S+ A L+ R G+ +E + +L L W
Sbjct: 343 AERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALL-RDYSNGYAIEERDGALYLGW 401
Query: 521 QRKELISVSTWR 532
+ L+S W+
Sbjct: 402 MDRILVSSCAWK 413
>At3g50650 scarecrow-like 7 (SCL7)
Length = 542
Score = 127 bits (320), Expect = 1e-29
Identities = 104/367 (28%), Positives = 175/367 (47%), Gaps = 30/367 (8%)
Query: 187 AGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKIG 246
A P L R+ +S G+P QR +Y EAL H + ++ S S I
Sbjct: 185 ARKPETKPDTLIRIKESVSESGDPIQRVGYYFAEALS---HKETESPSSSSSSSLEDFIL 241
Query: 247 AYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGK 306
+YK+ ++ P +FA+ T NQ+++EA + + IH++DF I G+QWS+ +Q + RS+GK
Sbjct: 242 SYKTLNDACPYSKFAHLTANQAILEATNQSNNIHIVDFGIFQGIQWSALLQALATRSSGK 301
Query: 307 PS-LKITAVVSPSSCNE--IELNFTQENLSQYAKDLNILFEFN--VLNIESLNLPSCPLP 361
P+ ++I+ + +PS + L T L +A L++ FEF + I+ LN S +
Sbjct: 302 PTRIRISGIPAPSLGDSPGPSLIATGNRLRDFAAILDLNFEFYPVLTPIQLLNGSSFRV- 360
Query: 362 GHFFDSNEAIGVNFPVSSF---ISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPT 418
D +E + VNF + + + AL + L P+IV + V
Sbjct: 361 ----DPDEVLVVNFMLELYKLLDETATTVGTALRLARSLNPRIVTLGEYEVSLNRVEFAN 416
Query: 419 NVVHVLQCYSALLESLDAVNVNLDVLQ--KIERHYIQPTINKIVLSHHNQR--------- 467
V + L+ YSA+ ESL+ N++ D + ++ER I +V S +
Sbjct: 417 RVKNSLRFYSAVFESLEP-NLDRDSKERLRVERVLFGRRIMDLVRSDDDNNKPGTRFGLM 475
Query: 468 DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQ--VERKPSSLVLCWQRKEL 525
++ WR + ++GF P SN+ +QA+ L+ VE +P + L W L
Sbjct: 476 EEKEQWRVLMEKAGFEPVKPSNYAVSQAKLLLWNYNYSTLYSLVESEPGFISLAWNNVPL 535
Query: 526 ISVSTWR 532
++VS+WR
Sbjct: 536 LTVSSWR 542
>At3g54220 SCARECROW1
Length = 653
Score = 127 bits (318), Expect = 2e-29
Identities = 116/462 (25%), Positives = 193/462 (41%), Gaps = 52/462 (11%)
Query: 112 DSGQEVFARRHQQQETQLPLFPHHYLQHQQQQQQSSVVP--FAKQQKVSSST-------- 161
D + F +Q P P QHQQQQQQ P +Q++ +SST
Sbjct: 198 DPSPQTFEPLYQISNNPSP--PQQQQQHQQQQQQHKPPPPPIQQQERENSSTDAPPQPET 255
Query: 162 -TGDDASIQLQQS---------------------IFDQLFKTAELIEAGNPVQAQGILAR 199
T ++Q + + L + AE + A N +A +L
Sbjct: 256 VTATVPAVQTNTAEALRERKEEIKRQKQDEEGLHLLTLLLQCAEAVSADNLEEANKLLLE 315
Query: 200 LNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFK------IGAYKSFSE 253
++ +P G QR + Y EA+ L ++ + A P ++ + + A++ F+
Sbjct: 316 ISQLSTPYGTSAQRVAAYFSEAMSARLLNSCLGIYAALPSRWMPQTHSLKMVSAFQVFNG 375
Query: 254 ISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITA 313
ISP+++F++FT NQ++ EA E+ D +H+ID DI G+QW + R G P +++T
Sbjct: 376 ISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILASRPGGPPHVRLTG 435
Query: 314 VVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGV 373
+ + L T + LS +A L + FEF L + NL + L EA+ V
Sbjct: 436 LGTSMEA----LQATGKRLSDFADKLGLPFEFCPLAEKVGNLDTERLN---VRKREAVAV 488
Query: 374 NFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLES 433
++ S L L++L PK+V ++++ L V + YSAL +S
Sbjct: 489 HWLQHSLYDVTGSDAHTLWLLQRLAPKVVTVVEQDLSHAGSFL-GRFVEAIHYYSALFDS 547
Query: 434 LDA-VNVNLDVLQKIERHYIQPTINKIVLSHHNQRD---KLPPWRNMFLQSGFSPFSFSN 489
L A + +E+ + I ++ R K WR Q GF S +
Sbjct: 548 LGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFESWREKMQQCGFKGISLAG 607
Query: 490 FTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTW 531
QA L+ P G+ + +L L W+ L++ S W
Sbjct: 608 NAATQATLLLGMFPSDGYTLVDDNGTLKLGWKDLSLLTASAW 649
>At5g17490 RGA-like protein
Length = 523
Score = 122 bits (305), Expect = 6e-28
Identities = 101/380 (26%), Positives = 175/380 (45%), Gaps = 25/380 (6%)
Query: 164 DDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQ 223
++ ++L Q+ L AE ++ N A ++ R+ + + + Y EAL
Sbjct: 151 EETGVRLVQA----LVACAEAVQLENLSLADALVKRVGLLAASQAGAMGKVATYFAEALA 206
Query: 224 LMLHSNGNNLTAFSPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVID 283
++ + A P F+ +F + P L+FA+FT NQ+++EA+ +HVID
Sbjct: 207 RRIYRIHPSAAAIDPS---FEEILQMNFYDSCPYLKFAHFTANQAILEAVTTSRVVHVID 263
Query: 284 FDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEI-ELNFTQENLSQYAKDLNIL 342
+ G+QW + MQ + LR G PS ++T V +PS+ I EL + L+Q A+ + +
Sbjct: 264 LGLNQGMQWPALMQALALRPGGPPSFRLTGVGNPSNREGIQELGW---KLAQLAQAIGVE 320
Query: 343 FEFNVLNIESLNLPSCPLPGHFFDS---NEAIGVN--FPVSSFISNPSCFPVALHFLKQL 397
F+FN L E L+ L F++ +E + VN F + +S P L +K +
Sbjct: 321 FKFNGLTTERLS----DLEPDMFETRTESETLVVNSVFELHPVLSQPGSIEKLLATVKAV 376
Query: 398 RPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTIN 457
+P +V +++ + L YS+L +SL+ V + + Y+ I
Sbjct: 377 KPGLVTVVEQEANHNGDVFLDRFNEALHYYSSLFDSLEDGVVIPSQDRVMSEVYLGRQIL 436
Query: 458 KIVLSHHNQR----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ-RAPVRGFQVERK 512
+V + + R + L WR +GF P + + QA L+ G++VE
Sbjct: 437 NLVATEGSDRIERHETLAQWRKRMGSAGFDPVNLGSDAFKQASLLLALSGGGDGYRVEEN 496
Query: 513 PSSLVLCWQRKELISVSTWR 532
SL+L WQ K LI+ S W+
Sbjct: 497 DGSLMLAWQTKPLIAASAWK 516
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 121 bits (304), Expect = 8e-28
Identities = 88/370 (23%), Positives = 171/370 (45%), Gaps = 17/370 (4%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAF- 236
L A+ + + + A ++ +L +S G P QR Y+ E L L S+G+++
Sbjct: 123 LVSCAKAMSENDLMMAHSMMEKLRQMVSVSGEPIQRLGAYLLEGLVAQLASSGSSIYKAL 182
Query: 237 --SPISFIFKIGAYKSFS-EISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWS 293
P ++ +Y E+ P +F + N ++ EA++ +R+H+IDF IG G QW
Sbjct: 183 NRCPEPASTELLSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQWV 242
Query: 294 SFMQEIVLRSNGKPSLKITAV--VSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIE 351
+ +Q R G P ++IT + ++ + L+ L++ AK N+ FEFN +++
Sbjct: 243 TLIQAFAARPGGPPRIRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVSVS 302
Query: 352 SLNLPSCPL---PGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKN 408
+ L PG N A ++ +S + L +K L PK+V +++
Sbjct: 303 VSEVKPKNLGVRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVEQE 362
Query: 409 CDRMDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKI--ERHYIQPTINKIVLSHHNQ 466
+ + + Y+A+ ES+D V + D Q+I E+H + + I+
Sbjct: 363 SNTNTAAFFPRFMETMNYYAAMFESID-VTLPRDHKQRINVEQHCLARDVVNIIACEGAD 421
Query: 467 R----DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQR 522
R + L WR+ F +GF+P+ S + + L++ + +++E + +L L W
Sbjct: 422 RVERHELLGKWRSRFGMAGFTPYPLSPLVNSTIKSLLRNYSDK-YRLEERDGALYLGWMH 480
Query: 523 KELISVSTWR 532
++L++ W+
Sbjct: 481 RDLVASCAWK 490
>At3g03450 RGA1-like protein
Length = 547
Score = 120 bits (302), Expect = 1e-27
Identities = 93/366 (25%), Positives = 164/366 (44%), Gaps = 15/366 (4%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLT-AF 236
L AE I N A ++ R+ + + Y +AL ++ + T
Sbjct: 184 LVACAEAIHQENLNLADALVKRVGTLAGSQAGAMGKVATYFAQALARRIYRDYTAETDVC 243
Query: 237 SPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFM 296
+ ++ F+ F E P L+FA+FT NQ+++EA+ R+HVID + G+QW + M
Sbjct: 244 AAVNPSFEEVLEMHFYESCPYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALM 303
Query: 297 QEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLP 356
Q + LR G PS ++T + P + N L L+Q+A+++ + FEF L ESL+
Sbjct: 304 QALALRPGGPPSFRLTGIGPPQTENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLS-- 361
Query: 357 SCPLPGHFFDS---NEAIGVN--FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDR 411
L F++ +E + VN F + ++ L+ +K ++P IV +++ +
Sbjct: 362 --DLEPEMFETRPESETLVVNSVFELHRLLARSGSIEKLLNTVKAIKPSIVTVVEQEANH 419
Query: 412 MDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQR---- 467
+ L YS+L +SL+ + + Y+ I +V + + R
Sbjct: 420 NGIVFLDRFNEALHYYSSLFDSLEDSYSLPSQDRVMSEVYLGRQILNVVAAEGSDRVERH 479
Query: 468 DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ-RAPVRGFQVERKPSSLVLCWQRKELI 526
+ WR +GF P + QA L+ A G++VE L++ WQ + LI
Sbjct: 480 ETAAQWRIRMKSAGFDPIHLGSSAFKQASMLLSLYATGDGYRVEENDGCLMIGWQTRPLI 539
Query: 527 SVSTWR 532
+ S W+
Sbjct: 540 TTSAWK 545
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 120 bits (302), Expect = 1e-27
Identities = 96/363 (26%), Positives = 161/363 (43%), Gaps = 14/363 (3%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFS 237
L AE I+ N A+ ++ ++ ++ + Y EAL ++ +
Sbjct: 225 LMACAEAIQQNNLTLAEALVKQIGCLAVSQAGAMRKVATYFAEALARRIYRLS---PPQN 281
Query: 238 PISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQ 297
I F E P L+FA+FT NQ+++EA E R+HVIDF + G+QW + MQ
Sbjct: 282 QIDHCLSDTLQMHFYETCPYLKFAHFTANQAILEAFEGKKRVHVIDFSMNQGLQWPALMQ 341
Query: 298 EIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESL-NLP 356
+ LR G P+ ++T + P+ N L+ L+Q A+ +++ FE+ SL +L
Sbjct: 342 ALALREGGPPTFRLTGIGPPAPDNSDHLHEVGCKLAQLAEAIHVEFEYRGFVANSLADLD 401
Query: 357 SCPLPGHFFDSNEAIGVN--FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDV 414
+ L D+ EA+ VN F + + P L +KQ++P I +++ +
Sbjct: 402 ASMLELRPSDT-EAVAVNSVFELHKLLGRPGGIEKVLGVVKQIKPVIFTVVEQESNHNGP 460
Query: 415 PLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQR----DKL 470
L YS L +SL+ V + D + + Y+ I +V R + L
Sbjct: 461 VFLDRFTESLHYYSTLFDSLEGVPNSQD--KVMSEVYLGKQICNLVACEGPDRVERHETL 518
Query: 471 PPWRNMFLQSGFSPFSFSNFTEAQAECLVQ-RAPVRGFQVERKPSSLVLCWQRKELISVS 529
W N F SG +P + QA L+ +G++VE L+L W + LI+ S
Sbjct: 519 SQWGNRFGSSGLAPAHLGSNAFKQASMLLSVFNSGQGYRVEESNGCLMLGWHTRPLITTS 578
Query: 530 TWR 532
W+
Sbjct: 579 AWK 581
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 117 bits (293), Expect = 2e-26
Identities = 92/366 (25%), Positives = 159/366 (43%), Gaps = 26/366 (7%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHS-NGNNLTAF 236
L AE ++ N A ++ + S ++ + Y E L ++ + A
Sbjct: 156 LLACAEAVQQNNLKLADALVKHVGLLASSQAGAMRKVATYFAEGLARRIYRIYPRDDVAL 215
Query: 237 SPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFM 296
S S +I Y+S P L+FA+FT NQ+++E +++HVID + G+QW + +
Sbjct: 216 SSFSDTLQIHFYES----CPYLKFAHFTANQAILEVFATAEKVHVIDLGLNHGLQWPALI 271
Query: 297 QEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLP 356
Q + LR NG P ++T + + ++ L Q A + + FEF + + +L+
Sbjct: 272 QALALRPNGPPDFRLTGI----GYSLTDIQEVGWKLGQLASTIGVNFEFKSIALNNLS-- 325
Query: 357 SCPLPGHFFDSN---EAIGVN--FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDR 411
L D E++ VN F + +++P L +K +RP I+ +++ +
Sbjct: 326 --DLKPEMLDIRPGLESVAVNSVFELHRLLAHPGSIDKFLSTIKSIRPDIMTVVEQEANH 383
Query: 412 MDVPLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQR---- 467
L YS+L +SL+ V+ ++ ++ I +V R
Sbjct: 384 NGTVFLDRFTESLHYYSSLFDSLEGPPSQDRVMSEL---FLGRQILNLVACEGEDRVERH 440
Query: 468 DKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ-RAPVRGFQVERKPSSLVLCWQRKELI 526
+ L WRN F GF P S + QA L+ A G+ VE L+L WQ + LI
Sbjct: 441 ETLNQWRNRFGLGGFKPVSIGSNAYKQASMLLALYAGADGYNVEENEGCLLLGWQTRPLI 500
Query: 527 SVSTWR 532
+ S WR
Sbjct: 501 ATSAWR 506
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 115 bits (287), Expect = 8e-26
Identities = 97/410 (23%), Positives = 173/410 (41%), Gaps = 25/410 (6%)
Query: 138 QHQQQQQQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGIL 197
QHQ ++SS S++ + Q + L A + G +A ++
Sbjct: 194 QHQDSPKESS--------SADSNSHVSSKEVVSQATPKQILISCARALSEGKLEEALSMV 245
Query: 198 ARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGN----NLTAFSPISFIFKIGAYKSFSE 253
L +S G+P QR + YM E L + ++G L P S ++ A + E
Sbjct: 246 NELRQIVSIQGDPSQRIAAYMVEGLAARMAASGKFIYRALKCKEPPSDE-RLAAMQVLFE 304
Query: 254 ISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITA 313
+ P +F N +++EA++ + +H+IDFDI G Q+ + ++ I +P L++T
Sbjct: 305 VCPCFKFGFLAANGAILEAIKGEEEVHIIDFDINQGNQYMTLIRSIAELPGKRPRLRLTG 364
Query: 314 VVSPSSCNEI--ELNFTQENLSQYAKDLNILFEFNVL----NIESLNLPSCPLPGHFFDS 367
+ P S L L Q A+D + F+F + +I S + C PG
Sbjct: 365 IDDPESVQRSIGGLRIIGLRLEQLAEDNGVSFKFKAMPSKTSIVSPSTLGCK-PGETLIV 423
Query: 368 NEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCY 427
N A ++ ++ + LH +K L PK+V ++++ + P + + Y
Sbjct: 424 NFAFQLHHMPDESVTTVNQRDELLHMVKSLNPKLVTVVEQDVNTNTSPFFPRFIEAYEYY 483
Query: 428 SALLESLD-AVNVNLDVLQKIERHYIQPTINKIVLSHHNQR----DKLPPWRNMFLQSGF 482
SA+ ESLD + +ER + I IV +R + WR + +GF
Sbjct: 484 SAVFESLDMTLPRESQERMNVERQCLARDIVNIVACEGEERIERYEAAGKWRARMMMAGF 543
Query: 483 SPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
+P S + L+++ ++++ + L CW+ K LI S WR
Sbjct: 544 NPKPMSAKVTNNIQNLIKQQYCNKYKLKEEMGELHFCWEEKSLIVASAWR 593
>At1g14920 signal response protein (GAI)
Length = 533
Score = 114 bits (286), Expect = 1e-25
Identities = 92/363 (25%), Positives = 159/363 (43%), Gaps = 14/363 (3%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNH-QLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAF 236
L AE ++ N A+ ++ ++ +S IG ++ + Y EAL ++ +
Sbjct: 173 LLACAEAVQKENLTVAEALVKQIGFLAVSQIG-AMRKVATYFAEALARRIYRLSPSQ--- 228
Query: 237 SPISFIFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFM 296
SPI F E P L+FA+FT NQ+++EA + R+HVIDF + G+QW + M
Sbjct: 229 SPIDHSLSDTLQMHFYETCPYLKFAHFTANQAILEAFQGKKRVHVIDFSMSQGLQWPALM 288
Query: 297 QEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLP 356
Q + LR G P ++T + P+ N L+ L+ A+ +++ FE+ +L
Sbjct: 289 QALALRPGGPPVFRLTGIGPPAPDNFDYLHEVGCKLAHLAEAIHVEFEYRGFVANTLADL 348
Query: 357 SCPLPGHFFDSNEAIGVN--FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDV 414
+ E++ VN F + + P L + Q++P+I +++ +
Sbjct: 349 DASMLELRPSEIESVAVNSVFELHKLLGRPGAIDKVLGVVNQIKPEIFTVVEQESNHNSP 408
Query: 415 PLPTNVVHVLQCYSALLESLDAVNVNLDVLQKIERHYIQPTINKIVLSHHNQR----DKL 470
L YS L +SL+ V D + + Y+ I +V R + L
Sbjct: 409 IFLDRFTESLHYYSTLFDSLEGVPSGQD--KVMSEVYLGKQICNVVACDGPDRVERHETL 466
Query: 471 PPWRNMFLQSGFSPFSFSNFTEAQAECLVQR-APVRGFQVERKPSSLVLCWQRKELISVS 529
WRN F +GF+ + QA L+ G++VE L+L W + LI+ S
Sbjct: 467 SQWRNRFGSAGFAAAHIGSNAFKQASMLLALFNGGEGYRVEESDGCLMLGWHTRPLIATS 526
Query: 530 TWR 532
W+
Sbjct: 527 AWK 529
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 112 bits (279), Expect = 7e-25
Identities = 98/417 (23%), Positives = 179/417 (42%), Gaps = 53/417 (12%)
Query: 144 QQSSVVPFAKQQKVSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQ 203
QQ VV A + + + GD + L++ A+ +E + ++++L
Sbjct: 206 QQHGVVSSAMYRSMEMISRGDLKGV---------LYECAKAVENYDLEMTDWLISQLQQM 256
Query: 204 LSPIGNPFQRASFYMKEALQLMLHSNGNNL-------TAFSPISFIFKIGAYKSFSEISP 256
+S G P QR YM E L L S+G+++ P + Y E P
Sbjct: 257 VSVSGEPVQRLGAYMLEGLVARLASSGSSIYKALRCKDPTGPELLTYMHILY----EACP 312
Query: 257 VLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVS 316
+F + N ++ EA++ +H+IDF I G QW S ++ + R G P+++IT +
Sbjct: 313 YFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWVSLIRALGARPGGPPNVRITGIDD 372
Query: 317 PSS--CNEIELNFTQENLSQYAKDLNILFEFN-------VLNIESLNLPSCPLPGHFFDS 367
P S + L + L + A+ + FEF+ + IE L + +
Sbjct: 373 PRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALCCTEVEIEKLGV----------RN 422
Query: 368 NEAIGVNFPVSSFISNPSCFPVALH------FLKQLRPKIVVTLDKNCDRMDVPLPTNVV 421
EA+ VNFP+ V H +K L P +V +++ + P V
Sbjct: 423 GEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQEANTNTAPFLPRFV 482
Query: 422 HVLQCYSALLESLDAVNVNLDVLQKI--ERHYIQPTINKIV----LSHHNQRDKLPPWRN 475
+ Y A+ ES+D V + D ++I E+H + + ++ + + + L WR+
Sbjct: 483 ETMNHYLAVFESID-VKLARDHKERINVEQHCLAREVVNLIACEGVEREERHEPLGKWRS 541
Query: 476 MFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRKELISVSTWR 532
F +GF P+ S++ A + L++ + + +E + +L L W+ + LI+ WR
Sbjct: 542 RFHMAGFKPYPLSSYVNATIKGLLESYSEK-YTLEERDGALYLGWKNQPLITSCAWR 597
>At5g41920 SCARECROW gene regulator-like protein
Length = 405
Score = 108 bits (269), Expect = 1e-23
Identities = 96/369 (26%), Positives = 168/369 (45%), Gaps = 26/369 (7%)
Query: 178 LFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASFYMKEALQLMLHSNGNNLTAFS 237
L + AE + + +A +L+ ++ SP G+ +R Y +ALQ + S+ + A S
Sbjct: 44 LLQCAEYVATDHLREASTLLSEISEICSPFGSSPERVVAYFAQALQTRVISSYLS-GACS 102
Query: 238 PISF----------IFKIGAYKSFSEISPVLQFANFTCNQSLIEALERFDRIHVIDFDIG 287
P+S IF A ++++ +SP+++F++FT NQ++ +AL+ D +H+ID D+
Sbjct: 103 PLSEKPLTVVQSQKIFS--ALQTYNSVSPLIKFSHFTANQAIFQALDGEDSVHIIDLDVM 160
Query: 288 FGVQWSSFMQEIVLRSNGKPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNV 347
G+QW + + R S++IT S S L T L+ +A LN+ FEF+
Sbjct: 161 QGLQWPALFHILASRPRKLRSIRITGFGSSSDL----LASTGRRLADFASSLNLPFEFHP 216
Query: 348 LN--IESLNLPSCPLPGHFFDSNEAIGVNFPVSSFISNPSCFPVALHFLKQLRPKIVVTL 405
+ I +L PS EA+ V++ L L++L+P ++ +
Sbjct: 217 IEGIIGNLIDPSQLAT----RQGEAVVVHWMQHRLYDVTGNNLETLEILRRLKPNLITVV 272
Query: 406 DKNCDRMD-VPLPTNVVHVLQCYSALLESL-DAVNVNLDVLQKIERHYIQPTINKIVLSH 463
++ D V L YSAL ++L D + +E+ + I IV +H
Sbjct: 273 EQELSYDDGGSFLGRFVEALHYYSALFDALGDGLGEESGERFTVEQIVLGTEIRNIV-AH 331
Query: 464 HNQRDKLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQRAPVRGFQVERKPSSLVLCWQRK 523
R K W+ + GF P S QA L+ P G+ + + +L L W+
Sbjct: 332 GGGRRKRMKWKEELSRVGFRPVSLRGNPATQAGLLLGMLPWNGYTLVEENGTLRLGWKDL 391
Query: 524 ELISVSTWR 532
L++ S W+
Sbjct: 392 SLLTASAWK 400
>At1g55580 unknown protein
Length = 445
Score = 102 bits (255), Expect = 4e-22
Identities = 101/426 (23%), Positives = 179/426 (41%), Gaps = 58/426 (13%)
Query: 157 VSSSTTGDDASIQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQLSPIGNPFQRASF 216
++SS+ AS L++ LF A + N AQ +L+ L+ SP G+ +R
Sbjct: 28 IASSSAATSASHHLRRL----LFTAANFVSQSNFTAAQNLLSILSLNSSPHGDSTERLVH 83
Query: 217 YMKEALQLMLHSNGNNLTAFSPISFI-------------------------------FKI 245
+AL + ++ + TA + ++ F+
Sbjct: 84 LFTKALSVRINRQQQDQTAETVATWTTNEMTMSNSTVFTSSVCKEQFLFRTKNNNSDFES 143
Query: 246 GAYKSFSEISPVLQFANFTCNQSLIEALERFDR--IHVIDFDIGFGVQWSSFMQEIVLRS 303
Y ++++P ++F + T NQ++++A E D +H++D DI G+QW MQ + RS
Sbjct: 144 CYYLWLNQLTPFIRFGHLTANQAILDATETNDNGALHILDLDISQGLQWPPLMQALAERS 203
Query: 304 NG----KPSLKITAVVSPSSCNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCP 359
+ PSL+IT + LN T + L+++A L + F+F+ L I +L
Sbjct: 204 SNPSSPPPSLRITG----CGRDVTGLNRTGDRLTRFADSLGLQFQFHTLVIVEEDLAGLL 259
Query: 360 LPGHFFD----SNEAIGVN---FPVSSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRM 412
L E I VN F F + L +K L +IV ++ +
Sbjct: 260 LQIRLLALSAVQGETIAVNCVHFLHKIFNDDGDMIGHFLSAIKSLNSRIVTMAEREANHG 319
Query: 413 DVPLPTNVVHVLQCYSALLESLDA-VNVNLDVLQKIERHYIQPTINKIVLSHHNQRD--- 468
D + Y A+ +SL+A + N +E+ + I +V + +R
Sbjct: 320 DHSFLNRFSEAVDHYMAIFDSLEATLPPNSRERLTLEQRWFGKEILDVVAAEETERKQRH 379
Query: 469 -KLPPWRNMFLQSGFSPFSFSNFTEAQAECLVQ-RAPVRGFQVERKPSSLVLCWQRKELI 526
+ W M + GF +F +QA+ L++ P G+ ++ +SL L WQ + L
Sbjct: 380 RRFEIWEEMMKRFGFVNVPIGSFALSQAKLLLRLHYPSEGYNLQFLNNSLFLGWQNRPLF 439
Query: 527 SVSTWR 532
SVS+W+
Sbjct: 440 SVSSWK 445
>At1g63100 transcription factor SCARECROW, putative
Length = 658
Score = 100 bits (248), Expect = 3e-21
Identities = 105/422 (24%), Positives = 179/422 (41%), Gaps = 52/422 (12%)
Query: 150 PFAKQQKVSSSTTGDDAS------IQLQQSIFDQLFKTAELIEAGNPVQAQGILARLNHQ 203
P++ + TTG+ + +Q + + L + I + N +AR
Sbjct: 244 PYSHRGATEERTTGNINNNNNRNDLQRDFELVNLLTGCLDAIRSRNIAAINHFIARTGDL 303
Query: 204 LSPIGN-PFQRASFYMKEALQLMLHSNGNNLTAFSPISFIFKI------GAYKSFSEISP 256
SP G P R Y EAL L + ++ +P + A + ++++P
Sbjct: 304 ASPRGRTPMTRLIAYYIEALALRVARMWPHIFHIAPPREFDRTVEDESGNALRFLNQVTP 363
Query: 257 VLQFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVS 316
+ +F +FT N+ L+ A E +R+H+IDFDI G+QW SF Q + R N ++IT +
Sbjct: 364 IPKFIHFTANEMLLRAFEGKERVHIIDFDIKQGLQWPSFFQSLASRINPPHHVRITGIGE 423
Query: 317 PSSCNEIELNFTQENLSQYAKDLNILFEFN--VLNIESLNLPSCPLPGHFFDSNEAIGVN 374
+++ELN T + L +A+ +N+ FEF+ V +E + L + E++ VN
Sbjct: 424 ----SKLELNETGDRLHGFAEAMNLQFEFHPVVDRLEDVRLWMLHV-----KEGESVAVN 474
Query: 375 FPV----SSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSAL 430
+ + + + L ++ P +V ++ + L T V + L+ YSA+
Sbjct: 475 CVMQMHKTLYDGTGAAIRDFLGLIRSTNPIALVLAEQEAEHNSEQLETRVCNSLKYYSAM 534
Query: 431 LESLDAVNVNL--DVLQ--KIERHYIQPTINKIVL---SHHNQRD-KLPPWRNMFLQSGF 482
DA++ NL D L K+E I IV SH +R WR M Q GF
Sbjct: 535 ---FDAIHTNLATDSLMRVKVEEMLFGREIRNIVACEGSHRQERHVGFRHWRRMLEQLGF 591
Query: 483 SPFSFSNFTEAQAECLVQRAPVRG---FQVER----------KPSSLVLCWQRKELISVS 529
S Q++ L++ F VER + + L W + L ++S
Sbjct: 592 RSLGVSEREVLQSKMLLRMYGSDNEGFFNVERSDEDNGGEGGRGGGVTLRWSEQPLYTIS 651
Query: 530 TW 531
W
Sbjct: 652 AW 653
>At4g17230 scarecrow-like 13 (SCL13)
Length = 287
Score = 90.1 bits (222), Expect = 3e-18
Identities = 78/279 (27%), Positives = 125/279 (43%), Gaps = 21/279 (7%)
Query: 204 LSPIGNPFQRASFYMKEALQLMLHSNGNN----LTAFSPISFIFKIGAYKS-FSEISPVL 258
+S G+P QR YM E L+ L +G+N L P ++ +Y S EI P
Sbjct: 2 VSVSGSPIQRLGTYMAEGLRARLEGSGSNIYKSLKCNEPTGR--ELMSYMSVLYEICPYW 59
Query: 259 QFANFTCNQSLIEALERFDRIHVIDFDIGFGVQWSSFMQEIVLRSNGKPSLKITAVVSPS 318
+FA T N ++EA+ R+H+IDF I G Q+ +QE+ G P L++T V
Sbjct: 60 KFAYTTANVEILEAIAGETRVHIIDFQIAQGSQYMFLIQELAKHPGGPPLLRVTGVDDSQ 119
Query: 319 S--CNEIELNFTQENLSQYAKDLNILFEFNVLNIESLNLPSCPLPGHFFDSNEAIGVNFP 376
S L+ E L+ A+ + FEF+ + + L + A+ VNFP
Sbjct: 120 STYARGGGLSLVGERLATLAQSCGVPFEFHDAIMSGCKVQREHLG---LEPGFAVVVNFP 176
Query: 377 V------SSFISNPSCFPVALHFLKQLRPKIVVTLDKNCDRMDVPLPTNVVHVLQCYSAL 430
+S + LH +K L PK+V +++ + P + V L Y+A+
Sbjct: 177 YVLHHMPDESVSVENHRDRLLHLIKSLSPKLVTLVEQESNTNTSPFLSRFVETLDYYTAM 236
Query: 431 LESLDAVNVNLDVLQKI--ERHYIQPTINKIVLSHHNQR 467
ES+DA D Q+I E+H + I ++ ++R
Sbjct: 237 FESIDAARPR-DDKQRISAEQHCVARDIVNMIACEESER 274
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,073,400
Number of Sequences: 26719
Number of extensions: 526339
Number of successful extensions: 1793
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1502
Number of HSP's gapped (non-prelim): 135
length of query: 533
length of database: 11,318,596
effective HSP length: 104
effective length of query: 429
effective length of database: 8,539,820
effective search space: 3663582780
effective search space used: 3663582780
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC121238.1


