

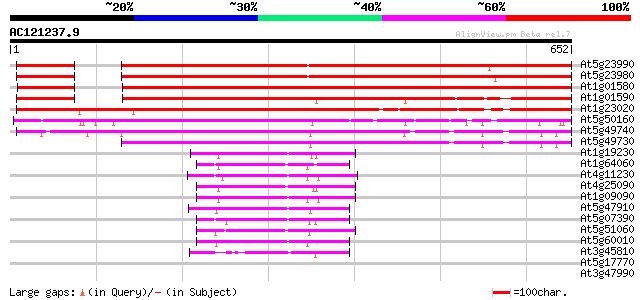
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.9 + phase: 0 /pseudo
(652 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g23990 FRO2 homolog 668 0.0
At5g23980 FRO2 homolog 660 0.0
At1g01580 hypothetical protein 605 e-173
At1g01590 hypothetical protein 552 e-157
At1g23020 putative superoxide-generating NADPH oxidase flavocyto... 551 e-157
At5g50160 FRO1 and FRO2-like protein 316 2e-86
At5g49740 FRO1-like protein; NADPH oxidase-like 295 4e-80
At5g49730 FRO2-like protein; NADPH oxidase-like 280 1e-75
At1g19230 hypothetical protein 100 2e-21
At1g64060 cytochrome b245 beta chain homolog RbohAp108 97 4e-20
At4g11230 respiratory burst oxidase homolog F - like protein 95 1e-19
At4g25090 respiratory burst oxidase - like protein 94 2e-19
At1g09090 respiratory burst oxidase protein B like 93 4e-19
At5g47910 respiratory burst oxidase protein 91 2e-18
At5g07390 respiratory burst oxidase protein A 90 3e-18
At5g51060 respiratory burst oxidase protein 88 2e-17
At5g60010 respiratory burst oxidase protein - like 79 8e-15
At3g45810 respiratory burst oxidase - like protein 72 1e-12
At5g17770 NADH-cytochrome b5 reductase 33 0.65
At3g47990 unknown protein 32 1.5
>At5g23990 FRO2 homolog
Length = 707
Score = 668 bits (1724), Expect = 0.0
Identities = 320/528 (60%), Positives = 408/528 (76%), Gaps = 7/528 (1%)
Query: 131 TRMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYV 190
TR S+ILPLVGLTSESSIKYHIWLG +S F H++ F+IYWA+ N+++E W+ TYV
Sbjct: 180 TRASTILPLVGLTSESSIKYHIWLGHVSNFCFLVHTVVFLIYWAMVNKLMETFAWNATYV 239
Query: 191 SNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPG 250
N+AG IA +I +A+W+TS+P RRK +E+FFYTHHLY LYI+F+AIH G C+I P
Sbjct: 240 PNLAGTIAMVIGIAIWVTSLPSFRRKKFEIFFYTHHLYGLYIVFYAIHVGDSWFCMILPN 299
Query: 251 VFLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKL 310
+FLF IDR+LRFLQS + +RL+SA++LP D LEL F+K L+Y PTS++F++VP +SKL
Sbjct: 300 IFLFFIDRYLRFLQSTKRSRLVSAKILPSDNLELTFAKTSGLHYTPTSILFLHVPSISKL 359
Query: 311 QWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFL 370
QWHPFT+TSS N+E + LSV I+ GSW+ KLY L SSS+D L +S EGPYGP++
Sbjct: 360 QWHPFTITSSSNLEKDTLSVVIRKQGSWTQKLYTHL-SSSIDSLEVSTEGPYGPNSFDVS 418
Query: 371 RHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPIS 430
RH+ +++V GGSGVTPFIS+IR+LIFQSQ + + P +LLVC FKNY DL LDL+ P
Sbjct: 419 RHDSLILVGGGSGVTPFISVIRELIFQSQNRSTKLPNVLLVCAFKNYHDLAFLDLIFPSD 478
Query: 431 GLKTRISQLQLQIEAYITREKQEP-SKDTQKQIQTIWFKTNLSDCPISAVLGPNNWLWLG 489
+ IS+L L+IEAYITRE ++P + D K +QT WFK D PIS VLGPNN+LWLG
Sbjct: 479 ISVSDISKLNLRIEAYITREDKKPETTDDHKLLQTKWFKPQPLDSPISPVLGPNNFLWLG 538
Query: 490 AIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICSSVVFLW 549
+I SSFVMFLL +GIVTRYYIYP+++N+G +YN+TY V+W MF C CI I SS++FLW
Sbjct: 539 VVILSSFVMFLLLIGIVTRYYIYPVDHNTGSIYNFTYRVLWVMFLGCVCIFISSSIIFLW 598
Query: 550 LKRLNKL----ENKHIMNVEVSTPARSPGSWIYGSERELESLPHQSLVQATNVHFGSRPD 605
K+ NK K + +VE TP SPGSW +G ERELES+P+QS+VQAT+VHFGS+P+
Sbjct: 599 RKKENKEGDKDSKKQVQSVEFQTPTSSPGSWFHGHERELESVPYQSIVQATSVHFGSKPN 658
Query: 606 LKKILFECEG-KDVGVMTCGPRKMRHEVARICASGLADNLHFESISFN 652
LKKILFE EG +DVGVM CGP+KMRHEVA+IC+SGLA NLHFE+ISFN
Sbjct: 659 LKKILFEAEGSEDVGVMVCGPKKMRHEVAKICSSGLAKNLHFEAISFN 706
Score = 75.1 bits (183), Expect = 1e-13
Identities = 30/67 (44%), Positives = 45/67 (66%)
Query: 9 LRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFTFPMMFIGA 68
++++ +V+FLGW+ VWI++ T ++ IW P L +TYF QG + + T PMMFI
Sbjct: 8 VKMLMVVLFLGWIFVWIMISTNRFQNIWTPKLAKYLKTTYFGPQGMNLVLLTVPMMFIAV 67
Query: 69 LSCIYLH 75
LSC+YLH
Sbjct: 68 LSCVYLH 74
>At5g23980 FRO2 homolog
Length = 699
Score = 660 bits (1704), Expect = 0.0
Identities = 319/528 (60%), Positives = 406/528 (76%), Gaps = 7/528 (1%)
Query: 131 TRMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYV 190
TR S+ILPLVGLTSESSIKYHIWLG +S F H++ F+IYWA+ N+++E W+ TYV
Sbjct: 172 TRASTILPLVGLTSESSIKYHIWLGHVSNFCFLVHTVVFLIYWAMINKLMETFAWNPTYV 231
Query: 191 SNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPG 250
N+AG IA +I +AMW+TS+P RRK +E+FFYTHHLY LYI+F+ IH G C+I P
Sbjct: 232 PNLAGTIAMVIGIAMWVTSLPSFRRKKFEIFFYTHHLYGLYIVFYVIHVGDSWFCMILPN 291
Query: 251 VFLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKL 310
+FLF IDR+LRFLQS + +RL+SAR+LP D LEL FSK P L+Y PTS++F++VP +SK+
Sbjct: 292 IFLFFIDRYLRFLQSTKRSRLVSARILPSDNLELTFSKTPGLHYTPTSILFLHVPSISKI 351
Query: 311 QWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLNISVEGPYGPHTAQFL 370
QWHPFT+TSS N+E + LSV I+ GSW+ KLY L SSS+D L +S EGPYGP++
Sbjct: 352 QWHPFTITSSSNLEKDTLSVVIRRQGSWTQKLYTHL-SSSIDSLEVSTEGPYGPNSFDVS 410
Query: 371 RHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPIS 430
RH +++VSGGSG+TPFIS+IR+LI QSQ + + P +LLVC FK+Y DL LDL+ P+
Sbjct: 411 RHNSLILVSGGSGITPFISVIRELISQSQNKSTKLPDVLLVCSFKHYHDLAFLDLIFPLD 470
Query: 431 GLKTRISQLQLQIEAYITREKQEP-SKDTQKQIQTIWFKTNLSDCPISAVLGPNNWLWLG 489
+ IS+L L+IEAYITRE ++P + D + +QT WFK D PIS VLGPNN+LWLG
Sbjct: 471 MSASDISRLNLRIEAYITREDKKPETTDDHRLLQTKWFKPQPLDSPISPVLGPNNFLWLG 530
Query: 490 AIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICSSVVFLW 549
+I SSFVMFLL +GIVTRYYIYP+++N+G +YN++Y +W MF +CI I SSVVFLW
Sbjct: 531 VVILSSFVMFLLLIGIVTRYYIYPVDHNTGSIYNFSYRGLWDMFLGSACIFISSSVVFLW 590
Query: 550 LKRLNKLENKHIMN----VEVSTPARSPGSWIYGSERELESLPHQSLVQATNVHFGSRPD 605
K+ NK +K N VE TP SPGSW +G ERELES+P+QS+VQAT+VHFGS+P+
Sbjct: 591 RKKQNKEGDKEFKNQVQSVEFQTPTSSPGSWFHGHERELESVPYQSIVQATSVHFGSKPN 650
Query: 606 LKKILFECEG-KDVGVMTCGPRKMRHEVARICASGLADNLHFESISFN 652
LKKIL E EG +DVGVM CGPRKMRHEVA+IC+SGLA NLHFE+ISFN
Sbjct: 651 LKKILLEAEGSEDVGVMVCGPRKMRHEVAKICSSGLAKNLHFEAISFN 698
Score = 78.6 bits (192), Expect = 1e-14
Identities = 31/67 (46%), Positives = 45/67 (66%)
Query: 9 LRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFTFPMMFIGA 68
++ + +V+FLGW+ VWI++ T ++K W P L N+TYF QG + + T PMMFI
Sbjct: 8 VKTLMVVLFLGWILVWIMISTNLFKSKWTPKLSKYLNTTYFGPQGTNLVLLTVPMMFIAV 67
Query: 69 LSCIYLH 75
LSC+YLH
Sbjct: 68 LSCVYLH 74
>At1g01580 hypothetical protein
Length = 725
Score = 605 bits (1559), Expect = e-173
Identities = 286/525 (54%), Positives = 391/525 (74%), Gaps = 4/525 (0%)
Query: 132 RMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYVS 191
R SS+LP +GLTSESSIKYHIWLG + M LF H + +IIYWA +++ +M+ W VS
Sbjct: 200 RGSSLLPAMGLTSESSIKYHIWLGHMVMALFTVHGLCYIIYWASMHEISQMIMWDTKGVS 259
Query: 192 NVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPGV 251
N+AGEIA L MW T+ P+IRR+ +EVFFYTH+LYI+++LFF +H G+ + PG
Sbjct: 260 NLAGEIALAAGLVMWATTYPKIRRRFFEVFFYTHYLYIVFMLFFVLHVGISFSFIALPGF 319
Query: 252 FLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQ 311
++FL+DR LRFLQSR++ RLL+AR+LP D +EL FSKN L Y+PTS++F+N+P +SKLQ
Sbjct: 320 YIFLVDRFLRFLQSRENVRLLAARILPSDTMELTFSKNSKLVYSPTSIMFVNIPSISKLQ 379
Query: 312 WHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSS-SLDHLNISVEGPYGPHTAQFL 370
WHPFT+TSS +E LS+ IK G WS KL+Q LSSS +D L +SVEGPYGP +A FL
Sbjct: 380 WHPFTITSSSKLEPEKLSIVIKKEGKWSTKLHQRLSSSDQIDRLAVSVEGPYGPASADFL 439
Query: 371 RHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPIS 430
RHE +VMV GGSG+TPFIS+IRDLI SQ++ + PK+ L+C FK ++++MLDL+LP+S
Sbjct: 440 RHEALVMVCGGSGITPFISVIRDLIATSQKETCKIPKITLICAFKKSSEISMLDLVLPLS 499
Query: 431 GLKTRI-SQLQLQIEAYITREKQEPSKDTQKQIQTIWFKTNLSDCPISAVLGPNNWLWLG 489
GL+T + S + ++IEA+ITR+ + +I+T+WFK +LSD IS++LGPN+WLWLG
Sbjct: 500 GLETELSSDINIKIEAFITRDNDAGDEAKAGKIKTLWFKPSLSDQSISSILGPNSWLWLG 559
Query: 490 AIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICSSVVFLW 549
AI+ SSF++F++ +GI+TRYYIYPI++N+ ++Y+ T + Y+ I S LW
Sbjct: 560 AILASSFLIFMIIIGIITRYYIYPIDHNTNKIYSLTSKTIIYILVISVSIMATCSAAMLW 619
Query: 550 -LKRLNKLENKHIMNVEVSTPARSP-GSWIYGSERELESLPHQSLVQATNVHFGSRPDLK 607
K+ K+E+K + NV+ +P SP SW Y S RE+ES P +SLVQ TN+HFG RP+LK
Sbjct: 620 NKKKYGKVESKQVQNVDRPSPTSSPTSSWGYNSLREIESTPQESLVQRTNLHFGERPNLK 679
Query: 608 KILFECEGKDVGVMTCGPRKMRHEVARICASGLADNLHFESISFN 652
K+L + EG VGV+ CGP+KMR +VA IC+SGLA+NLHFESISF+
Sbjct: 680 KLLLDVEGSSVGVLVCGPKKMRQKVAEICSSGLAENLHFESISFS 724
Score = 69.7 bits (169), Expect = 5e-12
Identities = 29/67 (43%), Positives = 44/67 (65%), Gaps = 1/67 (1%)
Query: 10 RIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKF-NSTYFRGQGNYVFVFTFPMMFIGA 68
+ + +V+FLG + +WI++PT Y+ W P L+ KF STYF G +F++ FPMM +
Sbjct: 28 KFLMMVIFLGTIMLWIMMPTLTYRTKWLPHLRIKFGTSTYFGATGTTLFMYMFPMMVVAC 87
Query: 69 LSCIYLH 75
L C+YLH
Sbjct: 88 LGCVYLH 94
>At1g01590 hypothetical protein
Length = 704
Score = 552 bits (1422), Expect = e-157
Identities = 272/529 (51%), Positives = 378/529 (71%), Gaps = 23/529 (4%)
Query: 132 RMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMI-EMLEWSKTYV 190
R SS+L VGLTSESSIKYHIWLG L M++F +H + + IYW NQ++ +MLEW +T V
Sbjct: 190 RGSSLLAAVGLTSESSIKYHIWLGHLVMIIFTSHGLCYFIYWISKNQLVSKMLEWDRTAV 249
Query: 191 SNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPG 250
SN+AGEIA + L MW+T+ P+IRR+++EVFFY+H+LYI+++LFF H G+ + PG
Sbjct: 250 SNLAGEIALVAGLMMWVTTYPKIRRRLFEVFFYSHYLYIVFMLFFVFHVGISHALIPLPG 309
Query: 251 VFLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKL 310
++FL+DR LRFLQSR + +L+SAR+LPCD +ELNFSKNP L Y+PTS +F+N+P +SKL
Sbjct: 310 FYIFLVDRFLRFLQSRNNVKLVSARVLPCDTVELNFSKNPMLMYSPTSTMFVNIPSISKL 369
Query: 311 QWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSSLDHLN---ISVEGPYGPHTA 367
QWHPFT+ SS +E LSV IK+ G WS KLY LSSSS D +N +SVEGPYGP +
Sbjct: 370 QWHPFTIISSSKLEPETLSVMIKSQGKWSTKLYDMLSSSSSDQINRLAVSVEGPYGPSST 429
Query: 368 QFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLML 427
FLRHE +VMVSGGSG+TPFISI+RDL + S + + PK+ L+C FKN +DL+MLDL+L
Sbjct: 430 DFLRHESLVMVSGGSGITPFISIVRDLFYMSSTHKCKIPKMTLICAFKNSSDLSMLDLIL 489
Query: 428 PISGLKTRI-SQLQLQIEAYITREKQEPSKDT---QKQIQTIWFKTNLSDCPISAVLGPN 483
P SGL T + S + +QI+A++TRE++ K++ + I+T FK N+SD PIS +LGPN
Sbjct: 490 PTSGLTTDMASFVDIQIKAFVTREEKTSVKESTHNRNIIKTRHFKPNVSDQPISPILGPN 549
Query: 484 NWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICS 543
+WL L AI++SSF++F++ + I+TRY+I+PI+ NS E Y W Y + Y+ S + S
Sbjct: 550 SWLCLAAILSSSFMIFIVIIAIITRYHIHPIDQNS-EKYTWAYKSLIYLVSISITVVTTS 608
Query: 544 SVVFLWLKRLNKLENKHIMNVEVSTPARSPGSWIYGSERELESLPHQSLVQATNVHFGSR 603
+ LW K+ K K+ V+ +P +ES P Q + Q+T++H+G R
Sbjct: 609 TAAMLWNKK--KYYAKNDQYVDNLSPV------------IIESSPQQLISQSTDIHYGER 654
Query: 604 PDLKKILFECEGKDVGVMTCGPRKMRHEVARICASGLADNLHFESISFN 652
P+L K+L +G VG++ CGP+KMR +VA+IC+ G A+NLHFESISF+
Sbjct: 655 PNLNKLLVGLKGSSVGILVCGPKKMRQKVAKICSFGSAENLHFESISFS 703
Score = 59.3 bits (142), Expect = 6e-09
Identities = 24/68 (35%), Positives = 42/68 (61%), Gaps = 1/68 (1%)
Query: 9 LRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKF-NSTYFRGQGNYVFVFTFPMMFIG 67
++ + +V+ +G + +WI++PT YK+IW ++ K S Y+ G + V+ FPM+ +
Sbjct: 15 IKFLMMVILMGTIVIWIMMPTSTYKEIWLTSMRAKLGKSIYYGRPGVNLLVYMFPMILLA 74
Query: 68 ALSCIYLH 75
L CIYLH
Sbjct: 75 FLGCIYLH 82
>At1g23020 putative superoxide-generating NADPH oxidase
flavocytochrome
Length = 693
Score = 551 bits (1420), Expect = e-157
Identities = 297/698 (42%), Positives = 430/698 (61%), Gaps = 74/698 (10%)
Query: 9 LRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKF-NSTYFRGQGNYVFVFTFPMMFIG 67
++++ +V+ +G + +WI++PT YKKIW ++ K S YF G + V+ FPM+ +
Sbjct: 15 IKLLTMVILMGTVVIWIMMPTSTYKKIWLKSMRAKLGKSIYFGKPGVNLLVYMFPMILLA 74
Query: 68 ALSCIYLHAS*D-------------------------VHSWLCLQLELFQLWSLFLYSCL 102
+L IYLH V + L + ++ + + L
Sbjct: 75 SLGSIYLHLKKQTRVNQFNRMDRKKIDKFGALKRPMLVKAGLGIVTVTEVMFLMMFMALL 134
Query: 103 LHS*YGPFPITYIPVSITSACTNKMRKF-TRMSSILPLVGL------------------- 142
L S F T++ ++ S T+ + R+ SI +GL
Sbjct: 135 LWSLANYFYHTFVTITPQSLPTDGDNLWQARLDSIAVRLGLTGNICLGFLFYPVARGSSL 194
Query: 143 ------TSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEMLEWSKTYVSNVAGE 196
TSESS KYHIWLG L M LF +H + + IYW TNQ+ +MLEW +T +S++AGE
Sbjct: 195 LAAVGLTSESSTKYHIWLGNLVMTLFTSHGLCYCIYWISTNQVSQMLEWDRTGISHLAGE 254
Query: 197 IASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPGVFLFLI 256
IA + L MW T+ P IRR+ +EVFFYTH+LY++++LFF H G+ + PG ++F++
Sbjct: 255 IALVAGLLMWATTFPAIRRRFFEVFFYTHYLYMVFMLFFVFHVGISYALISFPGFYIFMV 314
Query: 257 DRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFT 316
DR LRFLQSR + +L+SAR+LPC+ +ELNFSKNP L Y+PTS++F+N+P +SKLQWHPFT
Sbjct: 315 DRFLRFLQSRNNVKLVSARVLPCETVELNFSKNPMLMYSPTSILFVNIPSISKLQWHPFT 374
Query: 317 VTSSCNMETNYLSVAIKNVGSWSNKLYQELSSSS-LDHLNISVEGPYGPHTAQFLRHEQI 375
+TSS +E LSV IK+ G WS+KL+ L+SS+ +DHL +SVEGPYGP + +LRH+ +
Sbjct: 375 ITSSSKLEPKKLSVMIKSQGKWSSKLHHMLASSNQIDHLAVSVEGPYGPASTDYLRHDSL 434
Query: 376 VMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPISGLKTR 435
VMVSGGSG+TPFISIIRDL++ S ++ PK+ L+C FKN +DL+ML+L+LP S T
Sbjct: 435 VMVSGGSGITPFISIIRDLLYVSSTNAYKTPKITLICAFKNSSDLSMLNLILPNS---TE 491
Query: 436 ISQ-LQLQIEAYITREKQEPSKDTQKQIQTIWFKTNLSDCPISAVLGPNNWLWLGAIITS 494
IS + +QI+A++TREK S I+T+ FK +SD PIS +LGPN+WLWL I++S
Sbjct: 492 ISSFIDIQIKAFVTREK--VSTCNMNIIKTLSFKPYVSDQPISPILGPNSWLWLATILSS 549
Query: 495 SFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICSSVVFLWLKRLN 554
SF++F++ + I++RY+IYPI+ +S E Y Y + Y+ + + S+V L K+
Sbjct: 550 SFMIFIIIIAIISRYHIYPIDQSSKE-YTSAYTSLIYLLAISISVVATSTVAMLCNKK-- 606
Query: 555 KLENKHIMNVEVSTPARSPGSWIYGSERELESLPHQSLVQATNVHFGSRPDLKKILFECE 614
+ + + A SP +ES P Q L + TN+H+G RP+L K+L +
Sbjct: 607 ----SYFKGLYQNVDALSP--------LMIESSPDQLLPEFTNIHYGERPNLNKLLVGLK 654
Query: 615 GKDVGVMTCGPRKMRHEVARICASGLADNLHFESISFN 652
G VGV+ CGPRKMR EVA+IC+ G A NL FESISFN
Sbjct: 655 GSSVGVLVCGPRKMREEVAKICSFGSAANLQFESISFN 692
>At5g50160 FRO1 and FRO2-like protein
Length = 728
Score = 316 bits (810), Expect = 2e-86
Identities = 223/740 (30%), Positives = 373/740 (50%), Gaps = 110/740 (14%)
Query: 5 TIIFLRIIFLVMFLGWLSVWILLPTKIYKKIWAPMLQNKFNSTYFRGQGNYVFVFTFPMM 64
T++ LR++ ++ +GW+S+WI+ PT I+ + W ++ T+F G VF+FP +
Sbjct: 6 TLLVLRLLMNLLLIGWISLWIIKPTTIWIQSWR-QAEDTARHTFFGYYGLNFAVFSFPPI 64
Query: 65 FIGALSCIYLHAS*DVH--------------------SWL----CLQLELFQLWSLFL-- 98
+ + IYL H S++ C ++ L+ LFL
Sbjct: 65 ALSIVGLIYLSLLPQHHHPTRGGRGAAITVSRPAIINSFIGIVSCFEILALLLFLLFLAW 124
Query: 99 --YSCLLHS*YGPFPITYIPVSI---------------TSACTNKMR-KFTRMSSILPLV 140
Y+ + + P+ + +++ AC + + R S+ L+
Sbjct: 125 NFYARVSNDFKKLMPVKTMNLNLWQLKYYRVATRFGLLAEACLSLLLFPVLRGLSMFRLL 184
Query: 141 GLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEML-EWSKTYVSNVAGEIAS 199
+ +S+KYH+W G + H + W IT+ + E + +W +T VAG I+
Sbjct: 185 NIEFAASVKYHVWFGTGLIFFSLVHGGSTLFIWTITHHIEEEIWKWQRTGRVYVAGLISL 244
Query: 200 LIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPGVFLFLIDRH 259
+ L MWITS+PQIRRK +EVF+YTHHLYI++++ F HAG + PG+FLF +D+
Sbjct: 245 VTGLLMWITSLPQIRRKNFEVFYYTHHLYIVFLVAFLFHAGDRHFYWVLPGMFLFGLDKI 304
Query: 260 LRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTS 319
LR +QSR + +LSA L C A+EL K+P L Y P+S +F+N+P VS+ QWHPF++ S
Sbjct: 305 LRIVQSRSESCILSANLFSCKAIELVLPKDPMLNYAPSSFIFLNIPLVSRFQWHPFSIIS 364
Query: 320 SCNMETNYLSVAIKNVGSWSNKLYQELSSSS-----LDHLNISVEGPYGPHTAQFLRHEQ 374
S +++ + LS+ +K G W+N +Y ++ ++ ++++ + VEGPYGP + FLR++
Sbjct: 365 SSSVDKHSLSIMMKCEGDWTNSVYNKIEEAANCENKINNIIVRVEGPYGPASVDFLRYDN 424
Query: 375 IVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLVCIFKNYADLTMLDLMLPISG-LK 433
+ +V+GG G+TPF+SI+++L S+ + P ++ LV + + DL ML LPI+ +
Sbjct: 425 LFLVAGGIGITPFLSILKEL--ASKNRLKSPKRVQLVFAVRTFQDLNML---LPIASIIF 479
Query: 434 TRISQLQLQIEAYITREKQEPSKDT--------QKQIQTIWFKTNLSDCPISAVLGPNNW 485
I L L+++ ++T+EK +PS T Q Q+Q+I T+ D + GP ++
Sbjct: 480 NPIYNLNLKLKVFVTQEK-KPSNGTTTLQEFLAQSQVQSIHLGTD-EDYSRFPIRGPESF 537
Query: 486 LWLGAIITSSFVMFLLFLGIVTRYYIYPIE--NNSGEVYNWTYGVM---------WYMFS 534
WL ++ + + FL FL ++ ++I P E N+SG + G M W
Sbjct: 538 RWLATLVLITVLTFLGFLIGLSHFFI-PSEHKNHSGVMKLAASGAMKTAKEKVPSWVP-- 594
Query: 535 FCSCICICSSVVFL----WLKRLNKLENKHIMNVEVSTPARSPGSWIYGSERELESLPHQ 590
I I S V+ + + + + KH P S I ER L
Sbjct: 595 --DLIIIVSYVIAISVGGFAATILQRRRKH-----KEAPRMSKEVVIKPEERNFTELKPI 647
Query: 591 SLVQATNVHFGSRPDLKKILFECE-----GKDVGVMTCGPRKMRHEVARICAS-----GL 640
+ + +H G RP L++I+ E E VGV+ CGP ++ VA +C G+
Sbjct: 648 PITEEHEIHIGERPKLEEIMSEFEKNLRGWSSVGVLVCGPESVKEAVASMCRQWPQCFGV 707
Query: 641 AD--------NLHFESISFN 652
D NL+F S++FN
Sbjct: 708 EDLRRSRMKMNLNFHSLNFN 727
>At5g49740 FRO1-like protein; NADPH oxidase-like
Length = 739
Score = 295 bits (756), Expect = 4e-80
Identities = 202/723 (27%), Positives = 357/723 (48%), Gaps = 98/723 (13%)
Query: 9 LRIIFLVMFLGWLSVWILLPTKIYKKI---WAPMLQNKFNSTYFRGQGNYVFVFTFPMMF 65
L+++ V+F+ W+ ++ P + +I W + N T F G+ +F+ P++
Sbjct: 35 LKVVMSVIFVTWVVFLMMYPGSLGDQILTNWRAISSN----TLFGLTGSMFLIFSGPILV 90
Query: 66 IGALSCIYLHAS*DVHSWLCLQL-------------------------ELFQLWSLFLYS 100
I L+ +YL S + + +L +F LW+++ Y+
Sbjct: 91 IAILASLYLIISGEETVFTKFRLWTFPVLVDGPFGVVSAAEFLGIMVFSVFFLWAIYAYT 150
Query: 101 CLLHS*YGPFPITYIPVSITSACTNKMR--------------KFTRMSSILPLVGLTSES 146
+ F + SI +R +R S +L L+ + E
Sbjct: 151 LRNLNVLDYFHVLPNNRSIFLLELTGLRFGMIGLLCMVFLFLPISRGSILLRLIDIPFEH 210
Query: 147 SIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEML-EWSKTYVSNVAGEIASLIALAM 205
+ +YH+WLG ++M F+ H + +++ W I Q++E+L EW T ++ + G I+ + L M
Sbjct: 211 ATRYHVWLGHITMTFFSLHGLCYVVGWTIQGQLLELLFEWKATGIAVLPGVISLVAGLLM 270
Query: 206 WITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPGVFLFLIDRHLRFLQS 265
W+TS+ +R+ +E+FFYTH LYI++++F A+H G ++A G+FLF++DR LRF QS
Sbjct: 271 WVTSLHTVRKNYFELFFYTHQLYIVFVVFLALHVGDYLFSIVAGGIFLFILDRFLRFYQS 330
Query: 266 RQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMET 325
R+ ++SA+ LPC LEL SK P++ YN S +F+ V ++S LQWHPF+V+SS
Sbjct: 331 RRTVDVISAKSLPCGTLELVLSKPPNMRYNALSFIFLQVKELSWLQWHPFSVSSSPLDGN 390
Query: 326 NYLSVAIKNVGSWSNKLYQELSS-------------SSLDHLNISVEGPYGPHTAQFLRH 372
++++V IK +G W+ KL +LS+ S + VEGPYG + L +
Sbjct: 391 HHVAVLIKVLGGWTAKLRDQLSTLYEAENQDQLISPESYPKITTCVEGPYGHESPYHLAY 450
Query: 373 EQIVMVSGGSGVTPFISIIRDLIFQSQQ-QEFQPPKLLLVCIFKNYADLTMLDLMLPISG 431
E +V+V+GG G+TPF +I+ D++ + + ++ P K+L+V KN +L++L + S
Sbjct: 451 ENLVLVAGGIGITPFFAILSDILHRKRDGKDCLPGKVLVVWAIKNSDELSLLSAIDIPSI 510
Query: 432 LKTRISQLQLQIEAYITREKQEPSKD------TQKQIQTIWFKTNLSDCPISAVLGPNNW 485
+L L+I Y+TR+ + +D ++T W + C +S ++G +
Sbjct: 511 CHFFSKKLNLEIHIYVTRQSEPCLEDGMVHKVVHPSVKTPW----TNGCSMSVLVGTGDN 566
Query: 486 LWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCSCICICSSV 545
+W G + S + F+ + +V +YI N + W Y + ++ + + I +
Sbjct: 567 IWSGLYLIISTIGFIAMITLVDIFYI-----NKYNITTWWYKGLLFVVCMVASVLIFGGL 621
Query: 546 VFL----WLKRLNKLENKHIMNVEVS-TPARSPGSWIYGSERELESLPHQSLVQ-ATNVH 599
V + W + ++E V+++ +P S EL+ L + VQ T +
Sbjct: 622 VVVFWHRWEHKTGEVEANGNDKVDLNGEETHNP------SAAELKGLAIEEDVQNYTTIR 675
Query: 600 FGSRPDLKKILFECEGK----DVGVMTCGPRKMRHEVAR------ICASGLADNLHFESI 649
+G+RP ++I GK DVGV+ CGP ++ VA+ I S HF S
Sbjct: 676 YGTRPAFREIFESLNGKWGSVDVGVIVCGPATLQTTVAKEIRSHSIWRSANHPLFHFNSH 735
Query: 650 SFN 652
SF+
Sbjct: 736 SFD 738
>At5g49730 FRO2-like protein; NADPH oxidase-like
Length = 738
Score = 280 bits (717), Expect = 1e-75
Identities = 171/555 (30%), Positives = 296/555 (52%), Gaps = 44/555 (7%)
Query: 131 TRMSSILPLVGLTSESSIKYHIWLGQLSMVLFAAHSIGFIIYWAITNQMIEML-EWSKTY 189
+R S +L L+ + E + +YH+WLG ++M F+ H + +++ W I Q++E++ W+
Sbjct: 194 SRGSILLRLIDIPFEHATRYHVWLGHITMAFFSLHGLCYVVGWIIQGQLLELIFSWNAIG 253
Query: 190 VSNVAGEIASLIALAMWITSIPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAP 249
++ + G I+ + L MW+TS+ +R+ +E+FFYTH LYI++I+F A+H G ++A
Sbjct: 254 IAILPGVISLVAGLLMWVTSLHTVRKNYFELFFYTHQLYIVFIVFLALHVGDYMFSIVAG 313
Query: 250 GVFLFLIDRHLRFLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSK 309
G+FLF++DR LRF QSR+ ++SA+ LPC LEL SK P++ YN S +F+ V ++S
Sbjct: 314 GIFLFILDRFLRFCQSRRTVDVISAKSLPCGTLELVLSKPPNMRYNALSFIFLQVRELSW 373
Query: 310 LQWHPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQELSS-------------SSLDHLNI 356
LQWHPF+V+SS ++++V IK +G W+ KL +LS+ S +
Sbjct: 374 LQWHPFSVSSSPLDGNHHVAVLIKVLGGWTAKLRDQLSNLYEAENQDQLISPQSYPKITT 433
Query: 357 SVEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQ-QEFQPPKLLLVCIFK 415
VEGPYG + L +E +V+V+GG G+TPF +I+ D++ + + + P K+L+V K
Sbjct: 434 CVEGPYGHESPYHLAYENLVLVAGGIGITPFFAILSDILHRKRDGKACLPSKVLVVWAIK 493
Query: 416 NYADLTMLDLMLPISGLKTRISQLQLQIEAYITREKQEPSKD--TQKQIQTIWFKTNLSD 473
N +L++L + S +L L+I YITR+ + +D K + +
Sbjct: 494 NSDELSLLSAIDIPSICPFFSKKLNLEIHIYITRQSEPRLEDGMVHKVVHPSVKLPRTNG 553
Query: 474 CPISAVLGPNNWLWLGAIITSSFVMFLLFLGIVTRYYIYPIENNSGEVYNWTYGVMWYMF 533
C +S ++G + +W G + S + F+ + ++ +YI + W Y + ++
Sbjct: 554 CSMSVLVGTGDNIWSGLYLIISTIGFISMITLLDIFYI-----KRYNITTWWYKGLLFVG 608
Query: 534 SFCSCICICSSVVFL----WLKRLNKLENKHIMNVEVS-TPARSPGSWIYGSERELESLP 588
+ + I +V + W + ++E V+++ +P S EL+ L
Sbjct: 609 CMVASVLIFGGLVVVFWHRWEHKTGEVEANGNDKVDLNGEETHNP------SAAELKGLA 662
Query: 589 HQSLVQ-ATNVHFGSRPDLKKILFECEGK----DVGVMTCGPRKMRHEVAR------ICA 637
+ VQ T + +G+RP ++I GK DVGV+ CGP ++ VA+ I
Sbjct: 663 IEEDVQNYTTIRYGTRPAFREIFESLNGKWGSVDVGVIVCGPATLQTTVAKEIRSHSIWR 722
Query: 638 SGLADNLHFESISFN 652
S HF S SF+
Sbjct: 723 SANHPLFHFNSHSFD 737
>At1g19230 hypothetical protein
Length = 926
Score = 100 bits (250), Expect = 2e-21
Identities = 59/223 (26%), Positives = 112/223 (49%), Gaps = 33/223 (14%)
Query: 211 PQIRRKMYEVFFYTHHLYILYILFFAIHAGV----------GSMCVIAPGVFLFLIDRHL 260
P R + F+YTHHL ++ + +H + I+ + L++ +R L
Sbjct: 536 PLDRLTGFNAFWYTHHLLVVVYIMLIVHGTFLFFADKWYQKTTWMYISVPLVLYVAERSL 595
Query: 261 RFLQSRQHA-RLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTS 319
R +S+ ++ ++L +LP + L L SK P Y +F+ P +S+ +WHPF++TS
Sbjct: 596 RACRSKHYSVKILKVSMLPGEVLSLIMSKPPGFKYKSGQYIFLQCPTISRFEWHPFSITS 655
Query: 320 SCNMETNYLSVAIKNVGSWSNKLYQELSSS-----------------SLDHLN---ISVE 359
+ + LSV I+ +G W+ +L + L+ ++D +N + V+
Sbjct: 656 APG--DDQLSVHIRTLGDWTEELRRVLTVGKDLSTCVIGRSKFSAYCNIDMINRPKLLVD 713
Query: 360 GPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQE 402
GPYG + ++ ++++ G G TPFISI++DL+ S+ ++
Sbjct: 714 GPYGAPAQDYRSYDVLLLIGLGIGATPFISILKDLLNNSRDEQ 756
>At1g64060 cytochrome b245 beta chain homolog RbohAp108
Length = 944
Score = 96.7 bits (239), Expect = 4e-20
Identities = 59/210 (28%), Positives = 105/210 (49%), Gaps = 37/210 (17%)
Query: 218 YEVFFYTHHLYILYILFFAIHAGV-----------GSMCVIAPGVFLFLIDRHLRFLQSR 266
+ F+Y+HHL+++ + +H G+ + +A V L+ +R LR+ +S
Sbjct: 565 FNAFWYSHHLFVIVYILLILH-GIFLYFAKPWYVRTTWMYLAVPVLLYGGERTLRYFRSG 623
Query: 267 QHA-RLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMET 325
++ RLL + P + L L SK Y +F+ P VS +WHPF++TS+ E
Sbjct: 624 SYSVRLLKVAIYPGNVLTLQMSKPTQFRYKSGQYMFVQCPAVSPFEWHPFSITSA--PED 681
Query: 326 NYLSVAIKNVGSWSNKLYQ--------------------ELSSSSLDHLNISVEGPYGPH 365
+Y+S+ I+ +G W+ +L + E + SL L ++GPYG
Sbjct: 682 DYISIHIRQLGDWTQELKRVFSEVCEPPVGGKSGLLRADETTKKSLPKL--LIDGPYGAP 739
Query: 366 TAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
+ +++ +++V G G TPFISI++DL+
Sbjct: 740 AQDYRKYDVLLLVGLGIGATPFISILKDLL 769
>At4g11230 respiratory burst oxidase homolog F - like protein
Length = 942
Score = 94.7 bits (234), Expect = 1e-19
Identities = 66/230 (28%), Positives = 109/230 (46%), Gaps = 35/230 (15%)
Query: 207 ITSIPQIRRKM--YEVFFYTHHLYILYILFFAIHAGVGSMC-----------VIAPGVFL 253
+T +P+ K+ Y F+Y+HHL + + IH GV +A V L
Sbjct: 540 LTKLPKPFDKLTGYNAFWYSHHLLLTVYVLLVIH-GVSLYLEHKWYRKTVWMYLAVPVLL 598
Query: 254 FLIDRHLRFLQSRQHA-RLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQW 312
++ +R RF +SR + + + P + + L SK S Y VF+ P VSK +W
Sbjct: 599 YVGERIFRFFRSRLYTVEICKVVIYPGNVVVLRMSKPTSFDYKSGQYVFVQCPSVSKFEW 658
Query: 313 HPFTVTSSCNMETNYLSVAIKNVGSWSNKLYQ---------ELSSSSLDHLNIS------ 357
HPF++TSS +YLS+ I+ G W+ + + E S L ++
Sbjct: 659 HPFSITSSPG--DDYLSIHIRQRGDWTEGIKKAFSVVCHAPEAGKSGLLRADVPNQRSFP 716
Query: 358 ---VEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQ 404
++GPYG +++ +++V G G TPF+SI+RDL+ +Q+ Q
Sbjct: 717 ELLIDGPYGAPAQDHWKYDVVLLVGLGIGATPFVSILRDLLNNIIKQQEQ 766
>At4g25090 respiratory burst oxidase - like protein
Length = 863
Score = 94.0 bits (232), Expect = 2e-19
Identities = 62/224 (27%), Positives = 103/224 (45%), Gaps = 41/224 (18%)
Query: 218 YEVFFYTHHLYILYILFFAIHAGV----------GSMCVIAPGVFLFLIDRHLR-FLQSR 266
+ F+YTHHL+++ + +H + +A V L+ +R +R F S
Sbjct: 484 FNAFWYTHHLFVIVYILLVLHGYYIYLNKEWYKKTTWMYLAVPVALYAYERLIRAFRSSI 543
Query: 267 QHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMETN 326
+ ++L P L L SK + Y +F+N P VS +WHPF++TS+ + +
Sbjct: 544 RTVKVLKMAAYPGKVLTLQMSKPTNFKYMSGQYMFVNCPAVSPFEWHPFSITST--PQDD 601
Query: 327 YLSVAIKNVGSWSNKLYQELSSSSL-------------------DHLN---------ISV 358
YLSV IK +G W+ + S S D LN I +
Sbjct: 602 YLSVHIKALGDWTEAIQGVFSESREKIVEKHHVIFQVSKPPPVGDMLNGANSPRFPKIMI 661
Query: 359 EGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQE 402
+GPYG + ++E ++++ G G TP ISII+D+I ++ +E
Sbjct: 662 DGPYGAPAQDYKKYEVVLLIGLGIGATPMISIIKDIINNTETKE 705
>At1g09090 respiratory burst oxidase protein B like
Length = 843
Score = 93.2 bits (230), Expect = 4e-19
Identities = 60/216 (27%), Positives = 103/216 (46%), Gaps = 34/216 (15%)
Query: 218 YEVFFYTHHLYILYILFFAIHAGV----------GSMCVIAPGVFLFLIDRHLR-FLQSR 266
+ F+Y+HHL+++ + +H + +A V L+ +R +R F
Sbjct: 477 FNAFWYSHHLFVIVYVLLIVHGYFVYLSKEWYHKTTWMYLAVPVLLYAFERLIRAFRPGA 536
Query: 267 QHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMETN 326
+ ++L + P + L L SK Y ++IN VS LQWHPF++TS+ +
Sbjct: 537 KAVKVLKVAVYPGNVLSLYMSKPKGFKYTSGQYIYINCSDVSPLQWHPFSITSASG--DD 594
Query: 327 YLSVAIKNVGSWSNKLYQ------ELSSSSLDHLNIS---------------VEGPYGPH 365
YLSV I+ +G W+++L +L S+S L I+ ++GPYG
Sbjct: 595 YLSVHIRTLGDWTSQLKSLYSKVCQLPSTSQSGLFIADIGQANNITRFPRLLIDGPYGAP 654
Query: 366 TAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQ 401
+ ++ +++V G G TP ISIIRD++ + Q
Sbjct: 655 AQDYRNYDVLLLVGLGIGATPLISIIRDVLNNIKNQ 690
>At5g47910 respiratory burst oxidase protein
Length = 921
Score = 91.3 bits (225), Expect = 2e-18
Identities = 56/219 (25%), Positives = 102/219 (46%), Gaps = 34/219 (15%)
Query: 209 SIPQIRRKM--YEVFFYTHHLYILYILFFAIHA----------GVGSMCVIAPGVFLFLI 256
++P +K+ + F+YTHHL+I+ +H + +A + L+
Sbjct: 543 NLPNFLKKLTGFNAFWYTHHLFIIVYALLIVHGIKLYLTKIWYQKTTWMYLAVPILLYAS 602
Query: 257 DRHLR-FLQSRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPF 315
+R LR F S + +++ + P + L L+ +K Y + +N VS +WHPF
Sbjct: 603 ERLLRAFRSSIKPVKMIKVAVYPGNVLSLHMTKPQGFKYKSGQFMLVNCRAVSPFEWHPF 662
Query: 316 TVTSSCNMETNYLSVAIKNVGSWSNKLYQELSS-------------------SSLDHLNI 356
++TS+ +YLSV I+ +G W+ KL S +L +
Sbjct: 663 SITSAPG--DDYLSVHIRTLGDWTRKLRTVFSEVCKPPTAGKSGLLRADGGDGNLPFPKV 720
Query: 357 SVEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
++GPYG + +++ +++V G G TP ISI++D+I
Sbjct: 721 LIDGPYGAPAQDYKKYDVVLLVGLGIGATPMISILKDII 759
>At5g07390 respiratory burst oxidase protein A
Length = 902
Score = 90.1 bits (222), Expect = 3e-18
Identities = 60/212 (28%), Positives = 101/212 (47%), Gaps = 37/212 (17%)
Query: 218 YEVFFYTHHLYILYILFFAIHAGVGSMCVIAPG------------VFLFLIDRHLR-FLQ 264
+ F+Y+HHL+++ +H G +I P V L+L +R +R F
Sbjct: 522 FNAFWYSHHLFVIVYSLLVVH-GFYVYLIIEPWYKKTTWMYLMVPVVLYLCERLIRAFRS 580
Query: 265 SRQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNME 324
S + +L +LP + L L+ S+ + Y +++N VS L+WHPF++TS+
Sbjct: 581 SVEAVSVLKVAVLPGNVLSLHLSRPSNFRYKSGQYMYLNCSAVSTLEWHPFSITSAPG-- 638
Query: 325 TNYLSVAIKNVGSWSNKLYQELS-----------------SSSLDHL----NISVEGPYG 363
+YLSV I+ +G W+ +L S S D++ I ++GPYG
Sbjct: 639 DDYLSVHIRVLGDWTKQLRSLFSEVCKPRPPDEHRLNRADSKHWDYIPDFPRILIDGPYG 698
Query: 364 PHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
+ + E +++V G G TP ISI+ D+I
Sbjct: 699 APAQDYKKFEVVLLVGLGIGATPMISIVSDII 730
>At5g51060 respiratory burst oxidase protein
Length = 905
Score = 87.8 bits (216), Expect = 2e-17
Identities = 58/218 (26%), Positives = 105/218 (47%), Gaps = 36/218 (16%)
Query: 218 YEVFFYTHHLYIL-YILFFAI----------HAGVGSMCVIAPGVFLFLIDRHLR-FLQS 265
+ F+YTHHL+++ YIL A H M ++ P V L+ +R +R F S
Sbjct: 531 FNAFWYTHHLFVIVYILLVAHGYYLYLTRDWHNKTTWMYLVVP-VVLYACERLIRAFRSS 589
Query: 266 RQHARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMET 325
+ + + P + L ++ S+ + Y +F+N VS +WHPF++TS+ +
Sbjct: 590 IKAVTIRKVAVYPGNVLAIHLSRPQNFKYKSGQYMFVNCAAVSPFEWHPFSITSA--PQD 647
Query: 326 NYLSVAIKNVGSWSNKLYQELS---------------------SSSLDHLNISVEGPYGP 364
+YLSV I+ +G W+ L S +++ D + ++GPYG
Sbjct: 648 DYLSVHIRVLGDWTRALKGVFSEVCKPPPAGVSGLLRADMLHGANNPDFPKVLIDGPYGA 707
Query: 365 HTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQE 402
+ ++E +++V G G TP ISI++D++ + +E
Sbjct: 708 PAQDYKKYEVVLLVGLGIGATPMISIVKDIVNNIKAKE 745
>At5g60010 respiratory burst oxidase protein - like
Length = 839
Score = 79.0 bits (193), Expect = 8e-15
Identities = 55/222 (24%), Positives = 92/222 (40%), Gaps = 46/222 (20%)
Query: 218 YEVFFYTHHLYILYILFFAIHA-----------GVGSMCVIAPGVFLFLIDRHLRFLQSR 266
+ F+Y HHL +L + IH M + P +F R LQ
Sbjct: 494 FNAFWYAHHLLVLAYILLIIHGYYLIIEKPWYQKTTWMYLAVPMLFYASERLFSRLLQEH 553
Query: 267 QH-ARLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMET 325
H ++ A + + L L +K P Y +F+ P +SK +WHPF++TS+
Sbjct: 554 SHRVNVIKAIVYSGNVLALYVTKPPGFKYKSGMYMFVKCPDLSKFEWHPFSITSAPG--D 611
Query: 326 NYLSVAIKNVGSWSNKLYQ--------------------------------ELSSSSLDH 353
+YLSV I+ +G W+ +L + S +
Sbjct: 612 DYLSVHIRALGDWTTELRSRFAKTCEPTQAAAKPKPNSLMRMETRAAGVNPHIEESQVLF 671
Query: 354 LNISVEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
I ++GPYG + + + +++V G G TPFISI++D++
Sbjct: 672 PKIFIKGPYGAPAQNYQKFDILLLVGLGIGATPFISILKDML 713
>At3g45810 respiratory burst oxidase - like protein
Length = 835
Score = 71.6 bits (174), Expect = 1e-12
Identities = 51/217 (23%), Positives = 92/217 (41%), Gaps = 54/217 (24%)
Query: 210 IPQIRRKMYEVFFYTHHLYILYILFFAIHAGVGSMCVIAPGVFLFLIDRHLRFLQSRQHA 269
IP R + F+Y HHL ++ IH ++ +I++ + Q
Sbjct: 497 IPFNRLAGFNSFWYAHHLLVIAYALLIIHG------------YILIIEKP--WYQK---- 538
Query: 270 RLLSARLLPCDALELNFSKNPSLYYNPTSLVFINVPKVSKLQWHPFTVTSSCNMETNYLS 329
+A + + L L +K Y +F+ P +SK +WHPF++TS+ E YLS
Sbjct: 539 ---TAIVYSGNVLALYMTKPQGFKYKSGMYMFVKCPDISKFEWHPFSITSAPGDE--YLS 593
Query: 330 VAIKNVGSWSNKLYQELSSSSLDHL-------------------------------NISV 358
V I+ +G W+++L + + H I +
Sbjct: 594 VHIRALGDWTSELRNRFAETCEPHQKSKPSPNDLIRMETRARGANPHVEESQALFPRIFI 653
Query: 359 EGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLI 395
+GPYG + + + ++++ G G TPFISI++D++
Sbjct: 654 KGPYGAPAQSYQKFDILLLIGLGIGATPFISILKDML 690
>At5g17770 NADH-cytochrome b5 reductase
Length = 281
Score = 32.7 bits (73), Expect = 0.65
Identities = 26/85 (30%), Positives = 43/85 (50%), Gaps = 13/85 (15%)
Query: 352 DHLNISVEGPYGPHTAQFLRHEQIVMVSGGSGVTPFISIIRDLIFQSQQQEFQPPKLLLV 411
DHL +V+GP G Q + M++GGSG+TP + R ++ + K+ L+
Sbjct: 135 DHL--AVKGPKGRFKYQPGQFRAFGMLAGGSGITPMFQVARAIL----ENPTDKTKVHLI 188
Query: 412 CIFKNYADLTMLDLML--PISGLKT 434
YA++T D++L + GL T
Sbjct: 189 -----YANVTYDDILLKEELEGLTT 208
>At3g47990 unknown protein
Length = 341
Score = 31.6 bits (70), Expect = 1.5
Identities = 22/92 (23%), Positives = 39/92 (41%), Gaps = 11/92 (11%)
Query: 484 NWLWLGAIITSSFVMFLLF------LGIVTRYYIYPIENNSGEVYNWTYGVMWYMFSFCS 537
N ++ G ++ S + LL+ I T+++ E W + ++W MFS+C
Sbjct: 61 NAMFCGRVVVLSVLSLLLYPFLWAWTVIGTQWFTKSKTCLPEEGQKWGF-LIWLMFSYCG 119
Query: 538 CICICSSVVFLWLKRLNKLENKHIMNVEVSTP 569
+CI V WL R H++ + P
Sbjct: 120 LLCIAFICVGKWLTR----RQVHLLRAQQGIP 147
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.140 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,631,181
Number of Sequences: 26719
Number of extensions: 605223
Number of successful extensions: 1907
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1794
Number of HSP's gapped (non-prelim): 48
length of query: 652
length of database: 11,318,596
effective HSP length: 106
effective length of query: 546
effective length of database: 8,486,382
effective search space: 4633564572
effective search space used: 4633564572
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC121237.9


