

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
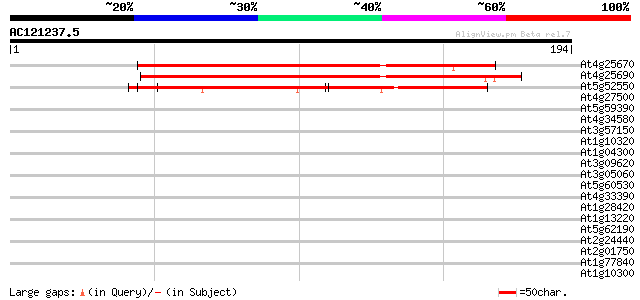
Query= AC121237.5 + phase: 2 /pseudo
(194 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g25670 unknown protein 130 3e-31
At4g25690 unknown protein 127 3e-30
At5g52550 unknown protein 126 6e-30
At4g27500 unknown protein 39 0.002
At5g59390 transcriptional regulator - like protein 36 0.015
At4g34580 putative protein 36 0.015
At3g57150 putative pseudouridine synthase (NAP57) 36 0.015
At1g10320 unknown protein 36 0.015
At1g04300 unknown protein 36 0.015
At3g09620 putative RNA helicase 35 0.019
At3g05060 putative SAR DNA-binding protein-1 35 0.019
At5g60530 late embryonic abundant protein - like 35 0.025
At4g33390 putative protein 35 0.025
At1g28420 hypothetical protein 35 0.025
At1g13220 putative nuclear matrix constituent protein 35 0.025
At5g62190 putative RNA helicase (MMI9.2) 35 0.033
At2g24440 unknown protein 34 0.043
At2g01750 putative myosin heavy chain-like protein 34 0.043
At1g77840 putative eukaryotic translation initiation factor 5 (E... 34 0.043
At1g10300 putative GTP-binding protein 34 0.043
>At4g25670 unknown protein
Length = 188
Score = 130 bits (328), Expect = 3e-31
Identities = 79/143 (55%), Positives = 89/143 (61%), Gaps = 21/143 (14%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
Q RK KKK KDE DR+KQAEKKKRRLEKALATSAAI +ELEKKKQK+ EEQQRLDEE
Sbjct: 10 QPVVRKAKKKHGKDEFDRIKQAEKKKRRLEKALATSAAIRAELEKKKQKRLEEQQRLDEE 69
Query: 105 GAAIAEAVALHVLLDEDSDDSYKVECKTWDDYNNNLDFFMSGKRACFP------------ 152
GAAIAEAVALHVLL EDSDDS +V K ++ +D F + P
Sbjct: 70 GAAIAEAVALHVLLGEDSDDSSRV--KFGEEKGFTMDLFRDERTNYVPRQSCASYAVQGI 127
Query: 153 -------NLDGSTWSVTSQNGKW 168
L S WS ++ G W
Sbjct: 128 GFVSNGYGLGDSNWSPFTRRGAW 150
>At4g25690 unknown protein
Length = 191
Score = 127 bits (320), Expect = 3e-30
Identities = 77/142 (54%), Positives = 93/142 (65%), Gaps = 12/142 (8%)
Query: 46 QHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEG 105
Q + KK KDE DR+KQAEKKKRRLEKALATSAAI +ELEKKKQK+ EEQQRLDEEG
Sbjct: 11 QPVTRKVKKHGKDEFDRIKQAEKKKRRLEKALATSAAIRAELEKKKQKRLEEQQRLDEEG 70
Query: 106 AAIAEAVALHVLLDEDSDDSYKVECKTWDDYNNNLDFFMSGKRACFPNLDGSTWSVTS-- 163
AAIAEAVALHVLL EDSDDS +V K ++ +D F + P ++++V
Sbjct: 71 AAIAEAVALHVLLGEDSDDSSRV--KFGEETGFGMDLFRDERTNYVPRQSCASYAVQGIG 128
Query: 164 --QNG------KWSISFRTVRK 177
NG WS+S++ K
Sbjct: 129 FVSNGYGLGDSNWSVSYKPFMK 150
>At5g52550 unknown protein
Length = 360
Score = 126 bits (317), Expect = 6e-30
Identities = 70/127 (55%), Positives = 89/127 (69%), Gaps = 7/127 (5%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
+ A KKKQ ++EL+R+KQAE+KKRR+EK++ATSAAI +ELEKKK +K EEQ+RLDEE
Sbjct: 175 EDSAYAAKKKQEREELERIKQAERKKRRIEKSIATSAAIRAELEKKKLRKLEEQRRLDEE 234
Query: 105 GAAIAEAVALHVLLDEDSDDSYK------VECKTWDDYNNNLDFFMSGKRACFPNLDGST 158
GAAIAEAVALHVLL ED DDSY+ K W DY ++ F G FP+ S+
Sbjct: 235 GAAIAEAVALHVLLGEDCDDSYRNTLNQETGFKPW-DYTTKINLFSGGINRFFPHQRCSS 293
Query: 159 WSVTSQN 165
++V N
Sbjct: 294 YAVHDNN 300
Score = 87.8 bits (216), Expect = 3e-18
Identities = 45/66 (68%), Positives = 55/66 (83%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
+ A KKKQ +DEL+R+KQAE KK RLEK++ATSAAI++ELEKKK +K EEQ+RL EE
Sbjct: 72 EDSADAAKKKQERDELERIKQAENKKNRLEKSIATSAAIMAELEKKKLRKLEEQKRLAEE 131
Query: 105 GAAIAE 110
GAAIAE
Sbjct: 132 GAAIAE 137
Score = 73.9 bits (180), Expect = 5e-14
Identities = 43/69 (62%), Positives = 51/69 (73%), Gaps = 1/69 (1%)
Query: 42 K*GQQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQ-QR 100
K Q RK +KK+VK +D +KQAEKK RRLEKA+ATSAAI +ELEKKKQ KKE Q +
Sbjct: 9 KQAQPIVRKARKKKVKGVVDPIKQAEKKNRRLEKAIATSAAIRAELEKKKQMKKEGQLEA 68
Query: 101 LDEEGAAIA 109
DEE +A A
Sbjct: 69 ADEEDSADA 77
Score = 54.3 bits (129), Expect = 4e-08
Identities = 35/66 (53%), Positives = 45/66 (68%), Gaps = 7/66 (10%)
Query: 52 KKKQVKDELDRLKQ-----AEKKKRRLEKALATSAAIISELEKKKQKKKEEQ--QRLDEE 104
KK + +E RL + AEKKKRRLEKA+AT+AAI +ELEKKKQ KKE Q ++E+
Sbjct: 117 KKLRKLEEQKRLAEEGAAIAEKKKRRLEKAIATTAAIRAELEKKKQMKKEGQLDAAVEED 176
Query: 105 GAAIAE 110
A A+
Sbjct: 177 SAYAAK 182
>At4g27500 unknown protein
Length = 612
Score = 38.9 bits (89), Expect = 0.002
Identities = 17/53 (32%), Positives = 37/53 (69%)
Query: 48 ARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQR 100
A K+ + ++E+ + KQA ++K++L + A AAI ++ E +K++KKE++++
Sbjct: 468 AATAKEMRKQEEIAKAKQAMERKKKLAEKAAAKAAIRAQKEAEKKEKKEQEKK 520
>At5g59390 transcriptional regulator - like protein
Length = 561
Score = 35.8 bits (81), Expect = 0.015
Identities = 27/92 (29%), Positives = 53/92 (57%), Gaps = 9/92 (9%)
Query: 52 KKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGAAIAEA 111
KK+ +K +++ E+K L + ++ + +EKKKQ K+E +Q++DE + E+
Sbjct: 154 KKRDLKSISQIVEEDERKMVHLVENMSQT------IEKKKQSKQELEQKVDET-SRFLES 206
Query: 112 VALH-VLLDEDSDDSY-KVECKTWDDYNNNLD 141
+ LH VLL+++ + + K++ K + Y LD
Sbjct: 207 LELHNVLLNKNYQEGFQKMQMKMEELYQQVLD 238
>At4g34580 putative protein
Length = 560
Score = 35.8 bits (81), Expect = 0.015
Identities = 18/53 (33%), Positives = 35/53 (65%), Gaps = 4/53 (7%)
Query: 47 HARKTKKKQVKDELDRLKQAEKK----KRRLEKALATSAAIISELEKKKQKKK 95
H K+++++ L+R++ E++ K+ LE+AL + I++ +EKKK+KKK
Sbjct: 496 HVESEKEEKLQAALNRVQVLEQELTETKKALEEALVSQKEILAYIEKKKKKKK 548
>At3g57150 putative pseudouridine synthase (NAP57)
Length = 565
Score = 35.8 bits (81), Expect = 0.015
Identities = 19/70 (27%), Positives = 39/70 (55%)
Query: 41 VK*GQQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQR 100
VK ++ +K K+++ ++E K+ +KKK+ ++ + A +KKK+K K+ +
Sbjct: 473 VKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKKSKDTEAA 532
Query: 101 LDEEGAAIAE 110
+D E + AE
Sbjct: 533 VDAEDESAAE 542
Score = 35.8 bits (81), Expect = 0.015
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 9/98 (9%)
Query: 45 QQHARKTKKKQVKD-ELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDE 103
++ + +KKK+ KD E ++ ++A +K+ +K +I E+ K +KK++++ D
Sbjct: 470 EEKVKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKKSKDT 529
Query: 104 EGAAIAEAVALHVLLDEDSDDSYKVECKTWDDYNNNLD 141
E A AE DE + + + + K D N D
Sbjct: 530 EAAVDAE--------DESAAEKSEKKKKKKDKKKKNKD 559
Score = 35.4 bits (80), Expect = 0.019
Identities = 21/69 (30%), Positives = 39/69 (56%), Gaps = 8/69 (11%)
Query: 44 GQQHARKTKKKQVKDE-LDRLKQAEKKKRRLEKALATSAAIISE-------LEKKKQKKK 95
G + K KKK K+E ++ + + +K++ +K+ T AA+ +E EKKK+KK
Sbjct: 493 GSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKKSKDTEAAVDAEDESAAEKSEKKKKKKD 552
Query: 96 EEQQRLDEE 104
++++ D E
Sbjct: 553 KKKKNKDSE 561
Score = 32.0 bits (71), Expect = 0.21
Identities = 17/63 (26%), Positives = 39/63 (60%), Gaps = 1/63 (1%)
Query: 49 RKTKKKQVK-DELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGAA 107
+K+K K+V+ +E + ++ KKK++ +K SE ++KK+KK ++++ ++E +
Sbjct: 457 KKSKTKEVEGEEAEEKVKSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASP 516
Query: 108 IAE 110
+E
Sbjct: 517 KSE 519
Score = 27.7 bits (60), Expect = 4.0
Identities = 15/59 (25%), Positives = 30/59 (50%)
Query: 48 ARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGA 106
A + +K++ D D KK + ++ A + KKK+KK +E+++ +E G+
Sbjct: 436 AGEARKRKHDDSSDSPAPVTTKKSKTKEVEGEEAEEKVKSSKKKKKKDKEEEKEEEAGS 494
>At1g10320 unknown protein
Length = 757
Score = 35.8 bits (81), Expect = 0.015
Identities = 26/108 (24%), Positives = 49/108 (45%), Gaps = 2/108 (1%)
Query: 49 RKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGAAI 108
+K K+KQV+ E+ ++ E K + + A I E + ++++K E + +E A
Sbjct: 43 KKLKRKQVRKEIAAKEREEAKAKLNDPAEQERLKAIEEEDARRREK--ELKDFEESERAW 100
Query: 109 AEAVALHVLLDEDSDDSYKVECKTWDDYNNNLDFFMSGKRACFPNLDG 156
EA+ + +E+ + + E + W D SG C + DG
Sbjct: 101 REAMEIKRKKEEEEEAKREEEERRWKDLEELRKLEASGNDECGEDEDG 148
>At1g04300 unknown protein
Length = 1082
Score = 35.8 bits (81), Expect = 0.015
Identities = 23/66 (34%), Positives = 37/66 (55%), Gaps = 3/66 (4%)
Query: 46 QHARKTKKKQVKDELDRLKQAEKKKRRLEK-ALATSAAIISELEKKKQKKKEEQQRLDEE 104
+ +K+KKKQ K + R K K KR+ EK + AT A + E + + Q +EE+ + E+
Sbjct: 450 EREKKSKKKQAKQK--RNKNKGKDKRKEEKVSFATHAKDLEENQNQNQNDEEEKDSVTEK 507
Query: 105 GAAIAE 110
+ AE
Sbjct: 508 AQSSAE 513
>At3g09620 putative RNA helicase
Length = 989
Score = 35.4 bits (80), Expect = 0.019
Identities = 30/119 (25%), Positives = 59/119 (49%), Gaps = 21/119 (17%)
Query: 49 RKTKKKQVKDELDRL-KQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGAA 107
+KT+ +QV+DE +L ++ EK++RR++ E ++ K++ E+ +++ +G
Sbjct: 143 KKTRDEQVEDEQKQLAEEVEKRRRRVQ-------------EWQELKRQNEEAQIESKGPE 189
Query: 108 IAEAVALHVLLDEDSDDSYKVECKTWDDYNNNLDFFMSGKRACFPNLDGSTWSVTSQNG 166
+A LD +SDD K + + D + L+ +G A + T S+NG
Sbjct: 190 TGKAWT----LDGESDDEVKSDSEMDVDRDTKLE---NGGDAKMVASENETAVTVSENG 241
>At3g05060 putative SAR DNA-binding protein-1
Length = 533
Score = 35.4 bits (80), Expect = 0.019
Identities = 17/55 (30%), Positives = 38/55 (68%), Gaps = 1/55 (1%)
Query: 50 KTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
K KKK+V++E ++ +KK++ +KA A + A++ +++K+K K++++ +EE
Sbjct: 460 KKKKKKVEEEKPEEEEPSEKKKK-KKAEAETEAVVEVAKEEKKKNKKKRKHEEEE 513
Score = 27.3 bits (59), Expect = 5.2
Identities = 16/60 (26%), Positives = 32/60 (52%), Gaps = 7/60 (11%)
Query: 45 QQHARKTKKKQVKDELDRL-------KQAEKKKRRLEKALATSAAIISELEKKKQKKKEE 97
++ + K KKK+ + E + + K+ KKKR+ E+ T + +K+K+KK ++
Sbjct: 474 EEPSEKKKKKKAEAETEAVVEVAKEEKKKNKKKRKHEEEETTETPAKKKDKKEKKKKSKD 533
>At5g60530 late embryonic abundant protein - like
Length = 439
Score = 35.0 bits (79), Expect = 0.025
Identities = 17/59 (28%), Positives = 36/59 (60%)
Query: 49 RKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGAA 107
++ K K+ K++ D+ ++ ++KK +LEK E ++K++K KE++ + + E AA
Sbjct: 78 KEKKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEKERKEKERKAKEKKDKEESEAAA 136
Score = 28.9 bits (63), Expect = 1.8
Identities = 14/60 (23%), Positives = 36/60 (59%), Gaps = 5/60 (8%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
++ +K K+K KD+ ++ K+ +++K + +K E + K +K+K++++R ++E
Sbjct: 61 KEQEKKDKEKAAKDKKEKEKKDKEEKEKKDKERKEK-----EKKDKLEKEKKDKERKEKE 115
Score = 28.9 bits (63), Expect = 1.8
Identities = 17/74 (22%), Positives = 44/74 (58%), Gaps = 1/74 (1%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
++ A+ K+K+ KD+ ++ K+ +++K + +K E ++K++K+KE + + +++
Sbjct: 69 EKAAKDKKEKEKKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEKERKEKERKAK-EKK 127
Query: 105 GAAIAEAVALHVLL 118
+EA A + +L
Sbjct: 128 DKEESEAAARYRIL 141
>At4g33390 putative protein
Length = 779
Score = 35.0 bits (79), Expect = 0.025
Identities = 24/100 (24%), Positives = 48/100 (48%), Gaps = 5/100 (5%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
+ H + + KQ ++EL RLKQ + L+ L ++A++ +L+K+ KE + +E
Sbjct: 372 ETHRWEKELKQAEEELQRLKQHLVSTKELQVKLEFASALLLDLKKELADHKESSKVKEET 431
Query: 105 GAAIAEAVALHVLLDEDSDDSYKVEC---KTWDDYNNNLD 141
+ + + L E + D K K ++ N N++
Sbjct: 432 SETV--VTNIEISLQEKTTDIQKAVASAKKELEEVNANVE 469
>At1g28420 hypothetical protein
Length = 1703
Score = 35.0 bits (79), Expect = 0.025
Identities = 26/84 (30%), Positives = 44/84 (51%), Gaps = 6/84 (7%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
Q+ + +KK+ KDE+ R K A ++K +EKA A A K+ E++Q E
Sbjct: 442 QRENERAEKKKQKDEIRREKDAIRRKLAIEKATARRIA------KESMDLIEDEQLELME 495
Query: 105 GAAIAEAVALHVLLDEDSDDSYKV 128
AAI++ + + LD D+ + +V
Sbjct: 496 LAAISKGLPSVLQLDHDTLQNLEV 519
Score = 26.6 bits (57), Expect = 8.9
Identities = 13/70 (18%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
+++ R+ +K++ + +R+K+ E+ +R + + + + E ++ +KK+++ + E
Sbjct: 402 ERNERERRKEEERLMRERIKEEERLQREQRREVERREKFL-QRENERAEKKKQKDEIRRE 460
Query: 105 GAAIAEAVAL 114
AI +A+
Sbjct: 461 KDAIRRKLAI 470
>At1g13220 putative nuclear matrix constituent protein
Length = 1128
Score = 35.0 bits (79), Expect = 0.025
Identities = 26/81 (32%), Positives = 40/81 (49%), Gaps = 3/81 (3%)
Query: 46 QHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQ--RLDE 103
Q A++ K++ L L E+++ L KAL + ELEK ++ +EE RL
Sbjct: 125 QEAQEILKREQSSHLYALTTVEQREENLRKALGLEKQCVQELEKALREIQEENSKIRLSS 184
Query: 104 EGAAIAEAVALHVLLDEDSDD 124
E A + EA AL ++ S D
Sbjct: 185 E-AKLVEANALVASVNGRSSD 204
Score = 27.3 bits (59), Expect = 5.2
Identities = 21/75 (28%), Positives = 34/75 (45%), Gaps = 8/75 (10%)
Query: 52 KKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKK------KQKKKEEQ--QRLDE 103
K++ + ++ L Q E+K +EK L + E +K K K+ EE +RL+E
Sbjct: 275 KEESITEQKRNLNQREEKVNEIEKKLKLKEKELEEWNRKVDLSMSKSKETEEDITKRLEE 334
Query: 104 EGAAIAEAVALHVLL 118
EA L + L
Sbjct: 335 LTTKEKEAHTLQITL 349
>At5g62190 putative RNA helicase (MMI9.2)
Length = 671
Score = 34.7 bits (78), Expect = 0.033
Identities = 20/60 (33%), Positives = 33/60 (54%), Gaps = 4/60 (6%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
++ +K KKK D + + KK + E+ L S + E EKKK KKK+++++ EE
Sbjct: 9 KKEEKKMKKKMALDT----PELDSKKGKKEQKLKLSDSDEEESEKKKSKKKDKKRKASEE 64
>At2g24440 unknown protein
Length = 183
Score = 34.3 bits (77), Expect = 0.043
Identities = 26/96 (27%), Positives = 46/96 (47%), Gaps = 15/96 (15%)
Query: 51 TKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEEGAAIAE 110
TK+ +L +++ EKKKR+ KA AA KKK KK+E ++++E +
Sbjct: 32 TKRDGSSSKLMKIESPEKKKRKTTKAKNVGAA------KKKVKKEEVAVKIEKEEEEDDD 85
Query: 111 AVALHVLLDEDSDDSYKV------ECKTWDDYNNNL 140
A ++D D K+ +CK++ + N +
Sbjct: 86 AAEKE---EDDDSDKKKIVIEHCKQCKSFKERANEV 118
>At2g01750 putative myosin heavy chain-like protein
Length = 629
Score = 34.3 bits (77), Expect = 0.043
Identities = 36/137 (26%), Positives = 63/137 (45%), Gaps = 11/137 (8%)
Query: 52 KKKQVKDELDRLKQAEKKK-----RRLEKAL---ATSAAIISELEKKKQKKKEEQQRLDE 103
++ ++ D+L R K AE +K R LE+A+ T+A + + ++K Q+ EE++ LD
Sbjct: 253 EENRILDKLHRQKVAEVEKFTQTVRELEEAVLAGGTAANAVRDYQRKFQEMNEERRILDR 312
Query: 104 E--GAAIAEAVALHVLLDEDSDDSYKV-ECKTWDDYNNNLDFFMSGKRACFPNLDGSTWS 160
E A ++ + V+ +E D S KV K W + L M R D + S
Sbjct: 313 ELARAKVSASRVATVVANEWKDGSDKVMPVKQWLEERRFLQGEMQQLRDKLAIADRAAKS 372
Query: 161 VTSQNGKWSISFRTVRK 177
K+ + R + +
Sbjct: 373 EAQLKEKFQLRLRVLEE 389
>At1g77840 putative eukaryotic translation initiation factor 5
(EIF-5) sp|P48724; similar to ESTs emb|F19992,
gb|N96933, emb|Z33699, emb|F19991
Length = 437
Score = 34.3 bits (77), Expect = 0.043
Identities = 27/88 (30%), Positives = 36/88 (40%), Gaps = 4/88 (4%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
Q+ K KK K E +RLK+ E K A A L K K + DE+
Sbjct: 149 QKKVSKDKKAMRKAEKERLKEGELADEEQRKLKAKKKA----LSNGKDSKTSKNHSSDED 204
Query: 105 GAAIAEAVALHVLLDEDSDDSYKVECKT 132
+ + AL V DED DD + + T
Sbjct: 205 ISPKHDENALEVDEDEDDDDGVEWQTDT 232
>At1g10300 putative GTP-binding protein
Length = 687
Score = 34.3 bits (77), Expect = 0.043
Identities = 31/102 (30%), Positives = 49/102 (47%), Gaps = 12/102 (11%)
Query: 45 QQHARKTKKKQVKDELDRLKQAEKKKRRLEKALATSAAIISELEKKKQKKKEEQQRLDEE 104
Q+ A K K K++ D L+R A K R ++ L I ++ +K+K+K E+ DE
Sbjct: 374 QRVAAKMKSKKIGDHLNRFHVAMPKSRDDQERLPC----IPQVAGEKEKRKTEKDLEDEN 429
Query: 105 GAAIAEAVAL---HVLLDED-SDDSYKVECKTWDDYNNNLDF 142
G A + +L ++L E+ DD C D +N DF
Sbjct: 430 GGAGVYSASLKKNYILAKEEWKDDIIPEIC----DCHNVADF 467
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,192,367
Number of Sequences: 26719
Number of extensions: 187955
Number of successful extensions: 3065
Number of sequences better than 10.0: 261
Number of HSP's better than 10.0 without gapping: 119
Number of HSP's successfully gapped in prelim test: 145
Number of HSP's that attempted gapping in prelim test: 2172
Number of HSP's gapped (non-prelim): 755
length of query: 194
length of database: 11,318,596
effective HSP length: 94
effective length of query: 100
effective length of database: 8,807,010
effective search space: 880701000
effective search space used: 880701000
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC121237.5


