

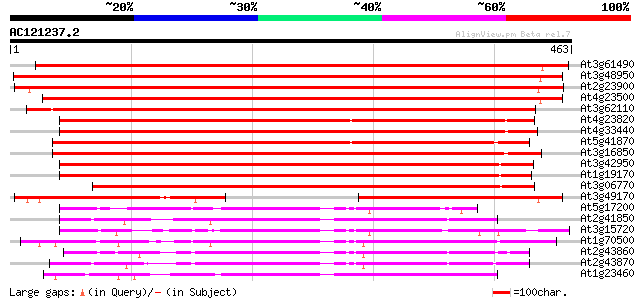
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.2 - phase: 0
(463 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g61490 unknown protein 649 0.0
At3g48950 endo-polygalacturonase - like protein 647 0.0
At2g23900 putative polygalacturonase 620 e-178
At4g23500 unknown protein (At4g23500) 610 e-175
At3g62110 unknown protein 436 e-122
At4g23820 polygalacturonase like protein 406 e-113
At4g33440 putative polygalacturonase 399 e-111
At5g41870 polygalacturonase-like protein 378 e-105
At3g16850 polygalacturonase like protein 373 e-103
At3g42950 polygalacturonase -like protein 358 5e-99
At1g19170 unknown protein 357 1e-98
At3g06770 unknown protein 328 4e-90
At3g49170 putative protein 211 7e-55
At5g17200 polygalacturonase-like protein 125 4e-29
At2g41850 polygalacturonase like protein 114 1e-25
At3g15720 polygalacturonase - like predicted GPI-anchored protein 112 3e-25
At1g70500 putative polygalacturonase 110 1e-24
At2g43860 putative polygalacturonase 103 3e-22
At2g43870 putative polygalacturonase 98 1e-20
At1g23460 putative polygalacturonase precursor 97 1e-20
>At3g61490 unknown protein
Length = 476
Score = 649 bits (1675), Expect = 0.0
Identities = 304/450 (67%), Positives = 373/450 (82%), Gaps = 10/450 (2%)
Query: 22 SIKLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPP 81
S F+Y AI CR HSA +T++G VGDGKTLNTKAF +A+ +LSQY+++GGAQL VP
Sbjct: 27 SQSFETFEYTAIICRSHSASITEYGGVGDGKTLNTKAFQSAVDHLSQYSSEGGAQLFVPA 86
Query: 82 GKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGT 141
GKWLTGSFNLTSHFTLFL KDA++L +QD +E+P L LPSYGRGRDA GRF+SLIFGT
Sbjct: 87 GKWLTGSFNLTSHFTLFLHKDAILLAAQDLNEYPILKALPSYGRGRDAAGGRFASLIFGT 146
Query: 142 NLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWF 201
NL+DVIITG+NGTIDGQGS+WW KFH +L TRPY+IE+M+SD IQISNLT ++SP+W
Sbjct: 147 NLSDVIITGNNGTIDGQGSFWWQKFHGGKLKYTRPYLIELMFSDTIQISNLTFLDSPSWN 206
Query: 202 VHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYG 261
+HP+YSSNII+ G+TI+APV SPNTDGINPDSCTN RIED +I+SGDDCIA+KSGWDEYG
Sbjct: 207 IHPVYSSNIIVKGVTIIAPVKSPNTDGINPDSCTNTRIEDCYIISGDDCIAVKSGWDEYG 266
Query: 262 IKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGR 321
I GMP++ ++IRRLTCISP SA +ALGSEMSGGI+DVR ED+TA TES +RIK+AVGR
Sbjct: 267 ISFGMPTKHLVIRRLTCISPYSAAIALGSEMSGGIEDVRAEDITAYQTESGVRIKTAVGR 326
Query: 322 GAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAG 381
GAFVK+I+VKGM+L+TMK+VFWMTG+Y H D+ +DP+ALP+I+GINYRD+ A+NV++AG
Sbjct: 327 GAFVKNIYVKGMNLHTMKWVFWMTGNYKAHADSHYDPHALPEITGINYRDIVAENVSMAG 386
Query: 382 KLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELLQEKEIE--- 438
+LEGIS DPFTGIC+SN TI M+A KK W C+D+ GVTS V PKPC+LL +E E
Sbjct: 387 RLEGISGDPFTGICISNATISMAAKHKKAIWMCSDVEGVTSGVDPKPCDLLDGQESETTK 446
Query: 439 -------CPFPTDKLAIENVQFKTCNFQSS 461
C FPTD L I+NV+ KTC++Q S
Sbjct: 447 KKMIDGGCDFPTDVLEIDNVELKTCSYQMS 476
>At3g48950 endo-polygalacturonase - like protein
Length = 469
Score = 647 bits (1669), Expect = 0.0
Identities = 304/459 (66%), Positives = 373/459 (81%), Gaps = 6/459 (1%)
Query: 4 VISVVLILGSLAECRVPSSIKLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFNAAI 63
++ + + L E R + + ++ A+NCRKHSA+LTDFGAVGDGKT NTKAF AI
Sbjct: 9 LVFLAIFLFPAIESRSHRNSVTSKIEFSALNCRKHSAILTDFGAVGDGKTSNTKAFRNAI 68
Query: 64 TNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPSY 123
+ LSQ A DGGAQL+VPPGKWLTGSFNLTSHFTLF+Q+ A IL SQDESEWP + LPSY
Sbjct: 69 SKLSQMATDGGAQLVVPPGKWLTGSFNLTSHFTLFIQRGATILASQDESEWPVIAPLPSY 128
Query: 124 GRGRDAPA-GRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIM 182
G+GRD GRF+SLI GTNLTDV+ITG+NGTI+GQG YWWDKF KKQ +TRPY+IEI+
Sbjct: 129 GKGRDGTGTGRFNSLISGTNLTDVVITGNNGTINGQGQYWWDKFKKKQFKITRPYLIEIL 188
Query: 183 YSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDN 242
+S IQISN+TLI+SP+W +HP+Y +++I+ +T+LAPV PNTDGINPDSCTN IED
Sbjct: 189 FSKNIQISNITLIDSPSWNIHPVYCNSVIVKSVTVLAPVTVPNTDGINPDSCTNTLIEDC 248
Query: 243 FIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRIE 302
+IVSGDDCIA+KSGWD+YGIK GMP+QQ+ IRRLTCISP SA VALGSEMSGGI+DVRIE
Sbjct: 249 YIVSGDDCIAVKSGWDQYGIKFGMPTQQLSIRRLTCISPKSAGVALGSEMSGGIKDVRIE 308
Query: 303 DVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALP 362
DVT NTESAIRIK+AVGRGA+VKDI+ + + + TMKYVFWM+G+YG HPD GFDP ALP
Sbjct: 309 DVTLTNTESAIRIKTAVGRGAYVKDIYARRITMKTMKYVFWMSGNYGSHPDEGFDPKALP 368
Query: 363 KISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTS 422
+I+ INYRD+TA+NVT++ L+GI DPFTGIC+SNVTI ++A KK+ WNCTD++GVTS
Sbjct: 369 EITNINYRDMTAENVTMSASLDGIDKDPFTGICISNVTIALAAKAKKMQWNCTDVAGVTS 428
Query: 423 NVVPKPCELLQEKE-----IECPFPTDKLAIENVQFKTC 456
V P+PC LL EK+ ++C FP+D + IE+V K C
Sbjct: 429 RVTPEPCSLLPEKKAQAKNVDCAFPSDLIPIESVVLKKC 467
>At2g23900 putative polygalacturonase
Length = 466
Score = 620 bits (1599), Expect = e-178
Identities = 297/458 (64%), Positives = 363/458 (78%), Gaps = 5/458 (1%)
Query: 5 ISVVLILGSLA--ECRVPSSIKLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFNAA 62
I VVL + S + E R +S N +Y A+NCRKH+AVLT+FGAVGDGKT NTKAF A
Sbjct: 8 ILVVLAISSFSMMEARDLASKGKTNIEYMALNCRKHTAVLTEFGAVGDGKTSNTKAFKEA 67
Query: 63 ITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPS 122
IT L+ A DGG QLIVPPGKWLTGSFNLTSHFTLF+QK A IL SQDESE+P + LPS
Sbjct: 68 ITKLAPKAADGGVQLIVPPGKWLTGSFNLTSHFTLFIQKGATILASQDESEYPVVAPLPS 127
Query: 123 YGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLT-LTRPYMIEI 181
YG+GRDA F+SLI GTNLTDV+ITG+NGTI+GQG YWW K+ +TRPY IEI
Sbjct: 128 YGQGRDAAGPTFASLISGTNLTDVVITGNNGTINGQGKYWWVKYRSGGFKGITRPYTIEI 187
Query: 182 MYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIED 241
++S +QISN+T+I+SP W +HP+Y +N+I+ G+TILAP+DSPNTDGINPDSCTN IED
Sbjct: 188 IFSQNVQISNITIIDSPAWNIHPVYCNNVIVKGVTILAPIDSPNTDGINPDSCTNTLIED 247
Query: 242 NFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRI 301
++VSGDDCIA+KSGWD++GIKVGMP+QQ+ IRRLTCISPDSA +ALGSEMSGGI+DVRI
Sbjct: 248 CYVVSGDDCIAVKSGWDQFGIKVGMPTQQLSIRRLTCISPDSAGIALGSEMSGGIKDVRI 307
Query: 302 EDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNAL 361
ED+T + T+SAIRIK+AVGRG +VKDIF + + TMKYVFWM+G+Y HP +GFDP A+
Sbjct: 308 EDITLLQTQSAIRIKTAVGRGGYVKDIFARRFTMKTMKYVFWMSGAYNQHPASGFDPKAM 367
Query: 362 PKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVT 421
P I+ INYRD+TA NVT +L+G NDPFT IC+SN+ I+++A KKL WNCT ISGV+
Sbjct: 368 PVITNINYRDMTADNVTQPARLDGFKNDPFTKICMSNIKIDLAAEPKKLLWNCTSISGVS 427
Query: 422 SNVVPKPCELLQEK--EIECPFPTDKLAIENVQFKTCN 457
S V PKPC LL EK ++C FP DK+ IE+V C+
Sbjct: 428 SKVTPKPCSLLPEKGAPVDCAFPVDKIPIESVVLNKCS 465
>At4g23500 unknown protein (At4g23500)
Length = 495
Score = 610 bits (1574), Expect = e-175
Identities = 285/433 (65%), Positives = 352/433 (80%), Gaps = 4/433 (0%)
Query: 28 FDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTG 87
F+Y AI+CR +SA L +FGAVGDG T NT AF A++ LS++A+ GG+ L VP G+WLTG
Sbjct: 56 FEYSAISCRAYSASLDEFGAVGDGVTSNTAAFRDAVSQLSRFADYGGSLLFVPAGRWLTG 115
Query: 88 SFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVI 147
+FNLTSHFTLFL +DAVIL SQ+ES++ + LPSYGRGRD GRF SL+FG+NLTDV+
Sbjct: 116 NFNLTSHFTLFLHRDAVILASQEESDYEVIEPLPSYGRGRDTDGGRFISLLFGSNLTDVV 175
Query: 148 ITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYS 207
ITG+NGTIDGQG WW KF + +L TRPY+IEIM+SD IQISNLT +NSP+W +HP+YS
Sbjct: 176 ITGENGTIDGQGEPWWGKFKRGELKYTRPYLIEIMHSDGIQISNLTFLNSPSWHIHPVYS 235
Query: 208 SNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMP 267
SNI I GLTILAPV PNTDGINPDSCTN RIED +IVSGDDCIA+KSGWD+YGI GMP
Sbjct: 236 SNIYIQGLTILAPVTVPNTDGINPDSCTNTRIEDCYIVSGDDCIAVKSGWDQYGINYGMP 295
Query: 268 SQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKD 327
++Q++IRRLTCISPDSA++ALGSEMSGGI+DVR ED+ AIN+ES IRIK+A+GRG +VKD
Sbjct: 296 TKQLLIRRLTCISPDSAVIALGSEMSGGIEDVRAEDIVAINSESGIRIKTAIGRGGYVKD 355
Query: 328 IFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGIS 387
++V+GM + TMKYVFWMTGSYG HPD+ +DP ALP I INY+D+ A+NVT+ +L GIS
Sbjct: 356 VYVRGMTMMTMKYVFWMTGSYGSHPDDHYDPKALPVIQNINYQDMVAENVTMPAQLAGIS 415
Query: 388 NDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELLQEKE----IECPFPT 443
D FTGIC+SNVTI +S KK+ WNCTD+SG TS V P+PC+LL EK+ + C FP
Sbjct: 416 GDQFTGICISNVTITLSKKPKKVLWNCTDVSGYTSGVTPQPCQLLPEKQPGTVVPCNFPE 475
Query: 444 DKLAIENVQFKTC 456
D + I+ V+ + C
Sbjct: 476 DPIPIDEVKLQRC 488
>At3g62110 unknown protein
Length = 471
Score = 436 bits (1121), Expect = e-122
Identities = 208/421 (49%), Positives = 289/421 (68%), Gaps = 2/421 (0%)
Query: 15 AECRVPSSIKLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDGG 74
A C V S+ + +++ R HS +T+FGAVGDG TLNTKAF A+ L+ +++ GG
Sbjct: 19 AWCSVGESLHCEYSNLASLH-RPHSVSITEFGAVGDGVTLNTKAFQNALFYLNSFSDKGG 77
Query: 75 AQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRF 134
A+L VP G+WLTGSF+L SH TL+L K A ILGS WP + LPSYGRGR+ P R
Sbjct: 78 AKLFVPAGQWLTGSFDLISHLTLWLDKGATILGSTSSENWPVVDPLPSYGRGRELPGRRH 137
Query: 135 SSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTL 194
SLI+G NLTDV+ITG+NGTIDGQG+ WWD F +L TRP+++E+M S + ISNLT
Sbjct: 138 RSLIYGQNLTDVVITGENGTIDGQGTVWWDWFRNGELNYTRPHLVELMNSTGLIISNLTF 197
Query: 195 INSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIK 254
+NSP W +HP+Y ++++ LTILAP++SPNTDG++PDS TNV IED +IV+GDD ++IK
Sbjct: 198 LNSPFWNIHPVYCRDVVVKNLTILAPLESPNTDGVDPDSSTNVCIEDCYIVTGDDLVSIK 257
Query: 255 SGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIR 314
SGWDEYGI PS +I I RLT + S+ +A+GSEMSGG+ ++ I+D+ N+ + IR
Sbjct: 258 SGWDEYGISYARPSSKIKINRLTGQTTSSSGIAIGSEMSGGVSEIYIKDLHLFNSNTGIR 317
Query: 315 IKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTA 374
IK++ GRG +V+++ + + L+ +K TG YG+HPD +DP ALP I I + +V
Sbjct: 318 IKTSAGRGGYVRNVHILNVKLDNVKKAIRFTGKYGEHPDEKYDPKALPAIEKITFENVNG 377
Query: 375 KNVTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPK-PCELLQ 433
+ +AG LEGI D F IC NVT+ + + KK PW C+++ G + V P+ C+ L+
Sbjct: 378 DGIGVAGLLEGIEGDVFKNICFLNVTLRVKKNSKKSPWECSNVRGYSQWVSPEITCDSLK 437
Query: 434 E 434
E
Sbjct: 438 E 438
>At4g23820 polygalacturonase like protein
Length = 444
Score = 406 bits (1044), Expect = e-113
Identities = 199/392 (50%), Positives = 263/392 (66%), Gaps = 2/392 (0%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQK 101
+TDFG VGDG+T+NTKAF AAI + GG L +PPG +LT SFNLTSH TL+L K
Sbjct: 43 ITDFGGVGDGRTVNTKAFRAAIYRIQHLKRRGGTLLYIPPGVYLTESFNLTSHMTLYLAK 102
Query: 102 DAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSY 161
AVI QD WP + LPSYGRGR+ P GR+ S I G L DV+ITG NGTIDGQG
Sbjct: 103 GAVIRAVQDTWNWPLIDPLPSYGRGRELPGGRYMSFIHGDGLRDVVITGQNGTIDGQGEV 162
Query: 162 WWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPV 221
WW+ + + L TRP +IE S +I ISN+ NSP W +HP+Y SN++I+ +TILAP
Sbjct: 163 WWNMWRSRTLKYTRPNLIEFKDSKEIIISNVIFQNSPFWNIHPVYCSNVVIHHVTILAPQ 222
Query: 222 DSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISP 281
DSPNTDGI+PDS NV IED++I +GDD +AIKSGWD+YGI G PS I IRR+T SP
Sbjct: 223 DSPNTDGIDPDSSYNVCIEDSYISTGDDLVAIKSGWDQYGIAYGRPSSNITIRRITGSSP 282
Query: 282 DSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYV 341
A +A+GSE SGGI+++ E +T N + IK+ +GRG ++K+I + + ++T KY
Sbjct: 283 -FAGIAIGSETSGGIKNIIAEHITLSNMGVGVNIKTNIGRGGYIKNIKISDVYVDTAKYG 341
Query: 342 FWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTI 401
+ G GDHPD ++PNALP + GI+ ++V NV AG ++G+ PFTGIC+S + +
Sbjct: 342 IKIAGDTGDHPDENYNPNALPVVKGIHIKNVWGVNVRNAGSIQGLKGSPFTGICLSEINL 401
Query: 402 EMSAHKKKLPWNCTDISGVTSNVVPKPCELLQ 433
S + K W C+D+SG + V P PC L+
Sbjct: 402 HGSLNSYK-TWKCSDVSGTSLKVSPWPCSELR 432
>At4g33440 putative polygalacturonase
Length = 475
Score = 399 bits (1024), Expect = e-111
Identities = 197/395 (49%), Positives = 270/395 (67%), Gaps = 3/395 (0%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQK 101
+TDFG VGDGKT NT AF A+ +L +A +GGAQL VP G WL+GSFNLTS+FTLFL++
Sbjct: 74 ITDFGGVGDGKTSNTAAFRRAVRHLEGFAAEGGAQLNVPEGTWLSGSFNLTSNFTLFLER 133
Query: 102 DAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSY 161
A+ILGS+D EWP + LPSYGRGR+ P GR SLI G NLT+V+ITG+NGTIDGQG
Sbjct: 134 GALILGSKDLDEWPIIEPLPSYGRGRERPGGRHISLIHGDNLTNVVITGENGTIDGQGKM 193
Query: 162 WWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPV 221
WW+ + + L TR ++IE+ S I ISNLTL+NSP W +HP+Y SN++I +TILAP+
Sbjct: 194 WWELWWNRTLVHTRGHLIELKNSHNILISNLTLLNSPFWTIHPVYCSNVVIRNMTILAPM 253
Query: 222 DSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISP 281
++PNTDGI+PDS TNV IED +I SGDD +A+KSGWD+YG+ V PS I+IRR++ +
Sbjct: 254 NAPNTDGIDPDSSTNVCIEDCYIESGDDLVAVKSGWDQYGMAVARPSSNIVIRRISGTTR 313
Query: 282 DSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYV 341
+ V +GSEMSGGI ++ +ED+ ++ + +RIK+ GRG ++ +I + L +K
Sbjct: 314 TCSGVGIGSEMSGGIFNITVEDIHVWDSAAGLRIKTDKGRGGYISNITFNNVLLEKVKVP 373
Query: 342 FWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTI 401
+ DH D+ +DP ALP++ GI +V + N A L G+ F +C+ NVT+
Sbjct: 374 IRFSSGSNDHSDDKWDPKALPRVKGIYISNVVSLNSRKAPMLLGVEGTSFQDVCLRNVTL 433
Query: 402 EMSAHKKKLPWNCTDISGVTSNVVPKPC-ELLQEK 435
+K W C D+SG S+V P C +LLQ+K
Sbjct: 434 LGLPKTEK--WKCKDVSGYASDVFPLSCPQLLQKK 466
>At5g41870 polygalacturonase-like protein
Length = 449
Score = 378 bits (970), Expect = e-105
Identities = 193/395 (48%), Positives = 258/395 (64%), Gaps = 4/395 (1%)
Query: 36 RKHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDG-GAQLIVPPGKWLTGSFNLTSH 94
R ++DFGAVGDGKTLNTKAFN+AI + N G L VP G +LT SFNLTSH
Sbjct: 41 RNEMLSISDFGAVGDGKTLNTKAFNSAIDRIRNSNNSNEGTLLYVPRGVYLTQSFNLTSH 100
Query: 95 FTLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGT 154
TL+L AVI QD +WP LPSYGRGR+ P R+ S I G L DV+ITG NGT
Sbjct: 101 MTLYLADGAVIKAVQDTEKWPLTDPLPSYGRGREHPGRRYISFIHGDGLNDVVITGRNGT 160
Query: 155 IDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIING 214
IDGQG WW+ + L TRP +IE S I +S++ L NSP W +HP+Y SN++++
Sbjct: 161 IDGQGEPWWNMWRHGTLKFTRPGLIEFNNSTNILVSHVVLQNSPFWTLHPVYCSNVVVHH 220
Query: 215 LTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIR 274
+TILAP DS NTDGI+PDS +NV IED++I +GDD +A+KSGWDEYGI PS+ I IR
Sbjct: 221 VTILAPTDSYNTDGIDPDSSSNVCIEDSYISTGDDLVAVKSGWDEYGIAYNRPSRDITIR 280
Query: 275 RLTCISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMD 334
R+T SP A +A+GSE SGGIQ+V +E++T N+ I IK+ +GRG ++ I + G+
Sbjct: 281 RITGSSP-FAGIAIGSETSGGIQNVTVENITLYNSGIGIHIKTNIGRGGSIQGITISGVY 339
Query: 335 LNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGI 394
L ++ ++G GDHPD+ F+ +ALP + GI ++V V AG ++G+ + PFT +
Sbjct: 340 LEKVRTGIKISGDTGDHPDDKFNTSALPIVRGITIKNVWGIKVERAGMVQGLKDSPFTNL 399
Query: 395 CVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPC 429
C SNVT ++ K+ W C+D+ G V P PC
Sbjct: 400 CFSNVT--LTGTKRSPIWKCSDVVGAADKVNPTPC 432
>At3g16850 polygalacturonase like protein
Length = 455
Score = 373 bits (958), Expect = e-103
Identities = 184/404 (45%), Positives = 256/404 (62%), Gaps = 3/404 (0%)
Query: 36 RKHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHF 95
R HS + +FGAVGDGKTLNT AF A+ L +A+ GGAQL VPPGKWLTGSFNLTSH
Sbjct: 36 RPHSVSILEFGAVGDGKTLNTIAFQNAVFYLKSFADKGGAQLYVPPGKWLTGSFNLTSHL 95
Query: 96 TLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTI 155
TLFL+K A IL S D S W + LPSYGRG + P R+ SLI G NL DV+ITG+NGT
Sbjct: 96 TLFLEKGATILASPDPSHWDVVSPLPSYGRGIELPGKRYRSLINGDNLIDVVITGENGTF 155
Query: 156 DGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGL 215
DGQG+ WW+ L +RP++IE + S I ISNLT +N+P +HP+Y S I I +
Sbjct: 156 DGQGAAWWEWLESGSLNYSRPHIIEFVSSKHILISNLTFLNAPAINIHPVYCSQIHIRKV 215
Query: 216 TILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRR 275
I VDSP+ G+ PDS NV IED+ I G D +++KSGWD+YGI G P+ + IR
Sbjct: 216 LIETSVDSPHVLGVAPDSSDNVCIEDSTINVGHDAVSLKSGWDQYGIHYGRPTTAVHIRN 275
Query: 276 LTCISPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDL 335
L SP A ++ GSEMSGG+ DV +E + ++ + ++ GRG ++++I + +DL
Sbjct: 276 LRLKSPTGAGISFGSEMSGGVSDVTVERLNIHSSLIGVAFRTTRGRGGYIRNITISDVDL 335
Query: 336 NTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGIC 395
++ G G HPD+ FD +ALP ++ I R+ T ++ +AG L GI PFT IC
Sbjct: 336 TSVDTAIVANGHTGSHPDDKFDRDALPVVTHIVMRNFTGVDIGVAGNLTGIGESPFTSIC 395
Query: 396 VSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPCELLQEKEIEC 439
++++ ++ + + W C+++SG + +V P+PC+ L C
Sbjct: 396 LADIHLQTRSEES---WICSNVSGFSDDVSPEPCQELMSSPSSC 436
>At3g42950 polygalacturonase -like protein
Length = 484
Score = 358 bits (918), Expect = 5e-99
Identities = 177/393 (45%), Positives = 257/393 (65%), Gaps = 3/393 (0%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQK 101
LTDFGAVGDG T+NT+AF AI +S+ A GG QL VPPG+WLT FNLTS+ TLFL +
Sbjct: 68 LTDFGAVGDGVTINTEAFEKAIYKISKLAKKGGGQLNVPPGRWLTAPFNLTSYMTLFLSE 127
Query: 102 DAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSY 161
+A IL QDE W L LPSYG GR+ R+ S I G NL DV++TG+NG+I+GQG
Sbjct: 128 NAEILALQDEKYWSLLPPLPSYGYGREHHGPRYGSFIHGQNLRDVVVTGNNGSINGQGQT 187
Query: 162 WWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPV 221
WW K+ +K L TR +++IM+S I +N+TL +SP W +HP N+ I +TILAPV
Sbjct: 188 WWKKYRQKLLNHTRGPLVQIMWSSDIVFANITLRDSPFWTLHPYDCKNVTITNMTILAPV 247
Query: 222 -DSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCIS 280
++PNTDGI+PDSC ++ IE+++I GDD IAIKSGWD+YG G PS+ I+IR L S
Sbjct: 248 FEAPNTDGIDPDSCEDMLIENSYISVGDDGIAIKSGWDQYGTTYGKPSKNILIRNLIIRS 307
Query: 281 PDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKY 340
SA +++GSEMSGG+ ++ +E++ ++ +RIK+A GRG +V+DI + + L+ ++
Sbjct: 308 MVSAGISIGSEMSGGVSNITVENILIWSSRRGVRIKTAPGRGGYVRDITFRNVTLDELRV 367
Query: 341 VFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVT 400
+ Y +HPD GF+P A P + INY + + V + +++G P + +++
Sbjct: 368 GIVIKTDYNEHPDGGFNPQAFPILENINYTGIYGQGVRVPVRIQGSKEIPVKNVTFRDMS 427
Query: 401 IEMSAHKKKLPWNCTDISG-VTSNVVPKPCELL 432
+ ++ +KKK + C + G V + P PC+ L
Sbjct: 428 VGIT-YKKKHIFQCAYVEGRVIGTIFPAPCDNL 459
>At1g19170 unknown protein
Length = 506
Score = 357 bits (915), Expect = 1e-98
Identities = 179/392 (45%), Positives = 258/392 (65%), Gaps = 4/392 (1%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYA-NDGGAQLIVPPGKWLTGSFNLTSHFTLFLQ 100
L DFG VGDG TLNT+AF A+ ++S+ + GG QL VPPG+WLT FNLTSH TLFL
Sbjct: 89 LKDFGGVGDGFTLNTEAFERAVISISKLGGSSGGGQLNVPPGRWLTAPFNLTSHMTLFLA 148
Query: 101 KDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGS 160
+D+ ILG +DE WP + LPSYG GR+ P R+ SLI G NL D++ITG NGTI+GQG
Sbjct: 149 EDSEILGVEDEKYWPLMPPLPSYGYGRERPGPRYGSLIHGQNLKDIVITGHNGTINGQGQ 208
Query: 161 YWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAP 220
WW K ++ L TR +++IM+S I I+N+T+ +SP W +HP N+ I +TILAP
Sbjct: 209 SWWKKHQRRLLNYTRGPLVQIMWSSDIVIANITMRDSPFWTLHPYDCKNVTIRNVTILAP 268
Query: 221 V-DSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCI 279
V +PNTDGI+PDSC ++ IED +I +GDD IAIKSGWD++GI G PS I+IR L
Sbjct: 269 VTGAPNTDGIDPDSCEDMVIEDCYISTGDDAIAIKSGWDQFGIAYGRPSTNILIRNLVVR 328
Query: 280 SPDSAMVALGSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMK 339
S SA V++GSEMSGGI +V IE++ N+ IRIK+A GRG ++++I K + L+ ++
Sbjct: 329 SVISAGVSIGSEMSGGISNVTIENLLIWNSRRGIRIKTAPGRGGYIRNITYKNLTLDNVR 388
Query: 340 YVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNV 399
+ Y +H D+ +D A P +SG ++ + + V + ++ G P + ++
Sbjct: 389 VGIVIKTDYNEHADDNYDRKAYPILSGFSFAGIHGQGVRVPVRIHGSEQIPVRNVTFRDM 448
Query: 400 TIEMSAHKKKLPWNCTDISG-VTSNVVPKPCE 430
++ ++ +KKK + C+ + G V ++ P+PCE
Sbjct: 449 SVGLT-YKKKHIFQCSFVKGRVFGSIFPRPCE 479
>At3g06770 unknown protein
Length = 377
Score = 328 bits (841), Expect = 4e-90
Identities = 160/365 (43%), Positives = 235/365 (63%), Gaps = 1/365 (0%)
Query: 69 YANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLVLPSYGRGRD 128
+A+ GGAQL VPPG WLTGSF+LTSH TLFL+ AVI+ SQD S W + LPSYGRG D
Sbjct: 3 FADKGGAQLYVPPGHWLTGSFSLTSHLTLFLENGAVIVASQDPSHWEVVDPLPSYGRGID 62
Query: 129 APAGRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQ 188
P R+ SLI G L DV++TGDNGTIDGQG WWD+F L RP++IE + S+ +
Sbjct: 63 LPGKRYKSLINGNKLHDVVVTGDNGTIDGQGLVWWDRFTSHSLKYNRPHLIEFLSSENVI 122
Query: 189 ISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGD 248
+SNLT +N+P + ++ IYSS++ I+ + + SP T GI PDS V I+++ I G
Sbjct: 123 VSNLTFLNAPAYSIYSIYSSHVYIHKILAHSSPKSPYTIGIVPDSSDYVCIQNSTINVGY 182
Query: 249 DCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGSEMSGGIQDVRIEDVTAIN 308
D I++KSGWDEYGI P++ + IR + + ++ GSEMSGGI DV +++
Sbjct: 183 DAISLKSGWDEYGIAYSRPTENVHIRNVYLRGASGSSISFGSEMSGGISDVVVDNAHIHY 242
Query: 309 TESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGIN 368
+ + I ++ GRG ++K+I + +D+ + GS+G HPD+ +D NALP +S I
Sbjct: 243 SLTGIAFRTTKGRGGYIKEIDISNIDMLRIGTAIVANGSFGSHPDDKYDVNALPLVSHIR 302
Query: 369 YRDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKP 428
+++ +N+ IAGKL GI PF+ + +SNV++ MS+ + W C+ + G + +V+P+P
Sbjct: 303 LSNISGENIGIAGKLFGIKESPFSSVTLSNVSLSMSS-GSSVSWQCSYVYGSSESVIPEP 361
Query: 429 CELLQ 433
C L+
Sbjct: 362 CPELK 366
>At3g49170 putative protein
Length = 1113
Score = 211 bits (537), Expect = 7e-55
Identities = 101/172 (58%), Positives = 126/172 (72%), Gaps = 4/172 (2%)
Query: 289 GSEMSGGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSY 348
G + DV I +VT INT+SAIRIK+A+GRG ++KDIF + + TMKYVFWMTGSY
Sbjct: 940 GLVYGSNLTDVVITEVTLINTQSAIRIKTAIGRGGYIKDIFARRFTMKTMKYVFWMTGSY 999
Query: 349 GDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLEGISNDPFTGICVSNVTIEMSAHKK 408
HP GFDP ALP+IS I+YRD+TA+NVTI+ KLE I N PFTG+C+S+VTI +S K
Sbjct: 1000 KLHPVGGFDPKALPEISNIHYRDMTAENVTISAKLERIKNGPFTGLCMSSVTIALSPDPK 1059
Query: 409 KLPWNCTDISGVTSNVVPKPCELLQEK----EIECPFPTDKLAIENVQFKTC 456
KL WNCTD+SGVTS V P+PC LL +K + +C FPTDK+ IE+V C
Sbjct: 1060 KLQWNCTDVSGVTSRVTPEPCSLLPDKRTTMDSDCDFPTDKIPIESVVLNKC 1111
Score = 166 bits (421), Expect = 2e-41
Identities = 97/189 (51%), Positives = 122/189 (64%), Gaps = 21/189 (11%)
Query: 5 ISVVLILGS--LAECRVPSSI---KLNNFDYPAINCRKHSAVLTDFGAVGDGKTLNTKAF 59
I VVL + + E R+ S++ + +Y A+NCRKHSAVLT+FGA+GDGK NTKAF
Sbjct: 810 ILVVLAISGFPMMESRIQSNLGSTSNKSIEYLALNCRKHSAVLTNFGAIGDGKPSNTKAF 869
Query: 60 NAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAVILGSQDESEWPQLLV 119
AI+ L+ A DGG QLIVPPG WLTGSFNLTSHFTLF+Q+ A IL SQDESE+P +
Sbjct: 870 REAISKLTPLAADGGVQLIVPPGSWLTGSFNLTSHFTLFIQQGATILASQDESEYPMIPP 929
Query: 120 LPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDN----------GTIDGQGSYWWDKFHKK 169
LPSYG DA RF+ L++G+NLTDV+IT T G+G Y D F ++
Sbjct: 930 LPSYG---DA---RFTGLVYGSNLTDVVITEVTLINTQSAIRIKTAIGRGGYIKDIFARR 983
Query: 170 QLTLTRPYM 178
T Y+
Sbjct: 984 FTMKTMKYV 992
>At5g17200 polygalacturonase-like protein
Length = 421
Score = 125 bits (315), Expect = 4e-29
Identities = 105/351 (29%), Positives = 171/351 (47%), Gaps = 46/351 (13%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQK 101
+T FGA+GDGKT +TKAF A + + ++ +++VP GK F+ K
Sbjct: 41 VTSFGAIGDGKTDDTKAFLKAWEAVCKGGHN--RKILVPQGK-------------TFMLK 85
Query: 102 DAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSY 161
+G S + G AGR+++ I ++ +++TG GTIDG+GS
Sbjct: 86 PLTFIGPCKSSTISLSIRGNLVAPGYTWYAGRYTTWISFDSINGLVVTG-GGTIDGRGSL 144
Query: 162 WWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPV 221
WW + RP + + ++ISNL +NSP V S NI + GL + AP
Sbjct: 145 WWGNVNN------RPCAMHFNNCNGLRISNLRHLNSPRNHVGLSCSQNIEVRGLRMTAPG 198
Query: 222 DSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISP 281
DSPNTDGI+ +C V I D+ I +GDDCIAI SG S I I + C P
Sbjct: 199 DSPNTDGIDISNCIGVHIHDSVIATGDDCIAINSG-----------SSHINITGIFC-GP 246
Query: 282 DSAMVALGSEMSGG----IQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNT 337
+++GS G +++VR+++ T T++ +RIK+ + + I + +++
Sbjct: 247 GHG-ISVGSLGVTGDFETVEEVRVKNCTFTKTQNGVRIKTYQNGSGYARKISFEDINMVA 305
Query: 338 MKYVFWMTGSYGDHPDNGFDPNALPKISGINYRD--VTAKNVTIAGKLEGI 386
+ + +Y + NG + K S +Y++ +TAK T +G +G+
Sbjct: 306 SENPIIIDQTYHNGGTNG----GISK-SSSSYQNCHLTAKQRTPSGNGKGV 351
>At2g41850 polygalacturonase like protein
Length = 433
Score = 114 bits (285), Expect = 1e-25
Identities = 96/371 (25%), Positives = 169/371 (44%), Gaps = 41/371 (11%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGK-WLTGSFNLTS--HFTLF 98
++DFGA GDGKT +T+AF A +++G L+VP G +L S LT + L
Sbjct: 70 VSDFGAKGDGKTDDTQAFVNAWKKAC--SSNGAVNLLVPKGNTYLLKSIQLTGPCNSILT 127
Query: 99 LQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITG-DNGTIDG 157
+Q + SQ S++ + S I + ++ + G D G +DG
Sbjct: 128 VQIFGTLSASQKRSDYKDI-----------------SKWIMFDGVNNLSVDGGDTGVVDG 170
Query: 158 QGSYWWD---KFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIING 214
G WW K +K + P + S + + NL + N+ + SN+ ++
Sbjct: 171 NGETWWQNSCKRNKAKPCTKAPTALTFYNSKSLIVKNLKVRNAQQIQISIEKCSNVQVSN 230
Query: 215 LTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIR 274
+ + AP DSPNTDGI+ + N+R+ ++ I +GDDCI+I+SG SQ + I
Sbjct: 231 VVVTAPADSPNTDGIHITNTQNIRVSESIIGTGDDCISIESG-----------SQNVQIN 279
Query: 275 RLTCISPDSAMV-ALGSEMSGG-IQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKG 332
+TC + +LG + S + V ++ T++ +RIK+ G +I +
Sbjct: 280 DITCGPGHGISIGSLGDDNSKAFVSGVTVDGAKLSGTDNGVRIKTYQGGSGTASNIIFQN 339
Query: 333 MDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKNVT-IAGKLEGISNDPF 391
+ ++ +K + Y D + +A+ ++ + YRD++ + + A N P
Sbjct: 340 IQMDNVKNPIIIDQDYCDKSKCTTEKSAV-QVKNVVYRDISGTSASENAITFNCSKNYPC 398
Query: 392 TGICVSNVTIE 402
GI + V I+
Sbjct: 399 QGIVLDRVNIK 409
>At3g15720 polygalacturonase - like predicted GPI-anchored protein
Length = 456
Score = 112 bits (281), Expect = 3e-25
Identities = 125/442 (28%), Positives = 190/442 (42%), Gaps = 76/442 (17%)
Query: 42 LTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLT-------GSFNLTSH 94
+T FGAVGDG T +++AF A + DG Q +VP G GS T
Sbjct: 26 VTQFGAVGDGVTDDSQAFLKAWEAVCSGTGDG--QFVVPAGMTFMLQPLKFQGSCKSTPV 83
Query: 95 FTLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGT 154
F L K LV PS G + G I T++ ++I GD G
Sbjct: 84 FVQMLGK----------------LVAPSKGNWK----GDKDQWILFTDIEGLVIEGD-GE 122
Query: 155 IDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIING 214
I+GQGS WW+ HK +RP ++ + +++S LT ++SP +H + + I+
Sbjct: 123 INGQGSSWWE--HKG----SRPTALKFRSCNNLRLSGLTHLDSPMAHIHISECNYVTISS 176
Query: 215 LTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIR 274
L I AP SPNTDGI+ + +NV I+D I +GDDCIAI SG + I I
Sbjct: 177 LRINAPESSPNTDGIDVGASSNVVIQDCIIATGDDCIAINSG-----------TSNIHIS 225
Query: 275 RLTCISPDSAMVALGSEMSGG----IQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFV 330
+ C P +++GS G +++V +++ T + RIK+ G + + I
Sbjct: 226 GIDC-GPGHG-ISIGSLGKDGETATVENVCVQNCNFRGTMNGARIKTWQGGSGYARMITF 283
Query: 331 KGMDLNTMKYVFWMTGSY-GDHPDNGFDPNALPKISGINYRDVTAKNVTIAGKLE-GI-- 386
G+ L+ ++ + Y G DN D K S + V N K E G+
Sbjct: 284 NGITLDNVENPIIIDQFYNGGDSDNAKD----RKSSAVEVSKVVFSNFIGTSKSEYGVDF 339
Query: 387 ---SNDPFTGICVSNVTIE--MSAHKKKLPWNCTDISGVTSNVVPKPCELLQEKEIEC-P 440
P T I + ++ IE S + C ++ G ++ VP +EC
Sbjct: 340 RCSERVPCTEIFLRDMKIETASSGSGQVAQGQCLNVRGASTIAVP---------GLECLE 390
Query: 441 FPTDKLAIENVQFKTCNFQSSV 462
TD + + +TC SV
Sbjct: 391 LSTDMFSSAQLLEQTCMSAQSV 412
>At1g70500 putative polygalacturonase
Length = 468
Score = 110 bits (276), Expect = 1e-24
Identities = 116/459 (25%), Positives = 201/459 (43%), Gaps = 51/459 (11%)
Query: 10 ILGSLAECRVPSSI---KLNNFDYPAINCR---KHSAVLTDFGAVGDGKTLNTKAFNAAI 63
ILG L VP + FD+ + + K+ + F A GDG + +T+AF A
Sbjct: 31 ILGKLENLDVPEDDIEDDVTFFDFSSFTSQYSGKNLVNVDSFNASGDGVSDDTQAFIRAW 90
Query: 64 TNLSQYANDGGAQLIVPPGK-WLTGS--FNLTSHFTLFLQKDAVILGSQDESEWPQLLVL 120
T N + L+VP G+ +L + F+ L +Q D I+ + S+W
Sbjct: 91 TMACSAPN---SVLLVPQGRSYLVNATKFDGPCQEKLIIQIDGTIIAPDEPSQWD----- 142
Query: 121 PSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQGSYWWD---KFHKKQLTLTRPY 177
P + R + + + L V+ G NG IDG G+ WW K +K + P
Sbjct: 143 PKFPR----------NWLQFSKLQGVVFQG-NGVIDGSGTKWWAASCKKNKSNPCVGAPT 191
Query: 178 MIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNV 237
+ I S + + LT+ NS + S+ + I+ + + +P DSPNTDGI+ + T+V
Sbjct: 192 ALTIYSSSNVYVRGLTIRNSQQMHLIIQRSTTVRISRVMVTSPGDSPNTDGIHITASTDV 251
Query: 238 RIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISPDSAMVALGS----EMS 293
++D+ I +GDDC++I +G S +I ++R+ C P +++GS
Sbjct: 252 VVQDSKISTGDDCVSIVNG-----------SAKIKMKRIYC-GPGHG-ISIGSLGQGHSK 298
Query: 294 GGIQDVRIEDVTAINTESAIRIKSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPD 353
G + V +E NT + +RIK+ G +VK + + + + + + Y D P
Sbjct: 299 GTVTAVVLETAFLKNTTNGLRIKTWQGGNGYVKGVRFENVVMQDVANPIIIDQFYCDSPS 358
Query: 354 NGFDPNALPKISGINYRDVTAKNVTI-AGKLEGISNDPFTGICVSNVTIEMSAHKKKLPW 412
+ + IS I YR++T + A + P + I ++N+ +E + K+
Sbjct: 359 TCQNQTSAVHISEIMYRNITGTTKSSKAINFKCSDAVPCSHIVLNNINLE--GNDGKVEA 416
Query: 413 NCTDISGVTSNVVPKPCELLQEKEIECPFPTDKLAIENV 451
C G VV + L + + TD+ E +
Sbjct: 417 YCNSAEGFGYGVVHPSADCLYSHDDKSLNQTDQYLAETI 455
>At2g43860 putative polygalacturonase
Length = 405
Score = 103 bits (256), Expect = 3e-22
Identities = 112/395 (28%), Positives = 184/395 (46%), Gaps = 45/395 (11%)
Query: 45 FGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTSHFTLFLQKDAV 104
+GA DG +TKAF AA AN +IVP G++L G NL H Q
Sbjct: 38 YGAKPDGSKDSTKAFLAAWDVACASANP--TTIIVPKGRFLVG--NLVFHGNECKQAPIS 93
Query: 105 I--LGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGTNLTDVIITGDNGTIDGQG-SY 161
I GS E +++ + I+ ++TDV I G G +D QG S
Sbjct: 94 IRIAGSIVAPEDFRIIASSKHW-------------IWFEDVTDVSIYG--GILDAQGTSL 138
Query: 162 WWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWFVHPIYSSNIIINGLTILAPV 221
W K + T + S+ I+IS LT INS + + S+N+ I+G+ + A
Sbjct: 139 WKCKNNGGHNCPTGAKSLVFSGSNNIKISGLTSINSQKFHIVIDNSNNVNIDGVKVSADE 198
Query: 222 DSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYGIKVGMPSQQIIIRRLTCISP 281
+SPNTDGI+ +S +V I ++ I +GDDCI+I G S + I+ + C P
Sbjct: 199 NSPNTDGIHVESSHSVHITNSRIGTGDDCISIGPG-----------STNVFIQTIRC-GP 246
Query: 282 DSAMVALGS----EMSGGIQDVRIEDVTAINTESAIRIKS-AVGRGAFVKDIFVKGMDLN 336
+++GS E G+ +V + +V + T + +RIK+ +F ++I + +++
Sbjct: 247 GHG-ISIGSLGRAEEEQGVDNVTVSNVDFMGTNNGVRIKTWGKDSNSFARNIVFQHINMK 305
Query: 337 TMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDV-TAKNVTIAGKLEGISNDPFTGIC 395
+K + Y H + + K+S + Y D+ N +A L+ P TGI
Sbjct: 306 MVKNPIIIDQHYCLHKPCPKQESGV-KVSNVRYEDIHGTSNTEVAVLLDCSKEKPCTGIV 364
Query: 396 VSNVTIEMSAHKKKLPWNCTDISGVTSNVVP-KPC 429
+ +V + +S H+ +C + +G ++VVP PC
Sbjct: 365 MDDVNL-VSVHRPAQA-SCDNANGSANDVVPFTPC 397
>At2g43870 putative polygalacturonase
Length = 384
Score = 97.8 bits (242), Expect = 1e-20
Identities = 105/414 (25%), Positives = 176/414 (42%), Gaps = 63/414 (15%)
Query: 34 NCRKHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGKWLTGSFNLTS 93
+C S + FGA DGKT TKAF A A+ ++VP G++L S
Sbjct: 15 SCSAQSYNVLSFGAKPDGKTDATKAFMAVWQTAC--ASSRPVTIVVPKGRFLLRSVTFDG 72
Query: 94 HF------------TLFLQKDAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSLIFGT 141
TL D ++G++D W IF
Sbjct: 73 SKCKPKPVTFRIDGTLVAPADYRVIGNEDY--W-----------------------IFFQ 107
Query: 142 NLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINSPTWF 201
+L + + G G +D +G+ WD + + I S + +S LT +NS +
Sbjct: 108 HLDGITVYG--GVLDARGASLWDCKKSGKNCPSGATTIGFQSSSNVVVSGLTSLNSQMFH 165
Query: 202 VHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGWDEYG 261
V +N+ + G+ +LA +SPNTDGI+ S ++V I + I +GDDC++I G
Sbjct: 166 VVINGCNNVKLQGVKVLAAGNSPNTDGIHVQSSSSVSIFNTKISTGDDCVSIGPG----- 220
Query: 262 IKVGMPSQQIIIRRLTCISPDSAMVALGS----EMSGGIQDVRIEDVTAINTESAIRIKS 317
+ + I + C P +++GS + G+Q+V ++ VT T++ +RIKS
Sbjct: 221 ------TNGLWIENVAC-GPGHG-ISIGSLGKDSVESGVQNVTVKTVTFTGTDNGVRIKS 272
Query: 318 -AVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAKN 376
A F K+I + +N ++ + +Y D + + KIS + + D+ +
Sbjct: 273 WARPSSGFAKNIRFQHCVMNNVENPIIIDQNYCPDHDCPRQVSGI-KISDVLFVDIHGTS 331
Query: 377 VTIAG-KLEGISNDPFTGICVSNVTIEMSAHKKKLPWNCTDISGVTSNVVPKPC 429
T G KL+ S P TGI + +V +++ K CT G+ + C
Sbjct: 332 ATEVGVKLDCSSKKPCTGIRLEDV--KLTYQNKPAASACTHAGGIEAGFFQPNC 383
>At1g23460 putative polygalacturonase precursor
Length = 457
Score = 97.4 bits (241), Expect = 1e-20
Identities = 97/388 (25%), Positives = 168/388 (43%), Gaps = 48/388 (12%)
Query: 29 DYPAINCR---KHSAVLTDFGAVGDGKTLNTKAFNAAITNLSQYANDGGAQLIVPPGK-W 84
D+P+ R K+ + FGA GDG + +T+AF +A S+ + + +VP G+ +
Sbjct: 52 DWPSFTSRHSGKNLVNVDTFGAAGDGVSDDTQAFVSA---WSKACSTSKSVFLVPEGRRY 108
Query: 85 LTGS--FNLTSHFTLFLQK-----DAVILGSQDESEWPQLLVLPSYGRGRDAPAGRFSSL 137
L + FN L +Q D I+ + S W +F +
Sbjct: 109 LVNATKFNGPCEQKLIIQVKLSSIDGTIVAPDEPSNWDS----------------KFQRI 152
Query: 138 IFGTNLTDVIITGDNGTIDGQGSYWWDKFHKKQLTLTRPYMIEIMYSDQIQISNLTLINS 197
+ ++ G IDG GS WW KK + + I S +++S LT+ NS
Sbjct: 153 WLEFSKLKGVVFQGKGVIDGSGSKWWAASCKKN----KSNALTIESSSGVKVSGLTIQNS 208
Query: 198 PTWFVHPIYSSNIIINGLTILAPVDSPNTDGINPDSCTNVRIEDNFIVSGDDCIAIKSGW 257
S ++ ++ + + +P DSPNTDGI+ TNV ++D I +GDDC++I +
Sbjct: 209 QQMNFIIARSDSVRVSKVMVSSPGDSPNTDGIHITGSTNVILQDCKIGTGDDCVSIVNA- 267
Query: 258 DEYGIKVGMPSQQIIIRRLTCISPDSAMV-ALGSEMSGGIQDVRIEDVTAI-NTESAIRI 315
S I ++ + C + +LG + + GI + D + T + +RI
Sbjct: 268 ----------SSNIKMKNIYCGPGHGISIGSLGKDNTTGIVTQVVLDTALLRETTNGLRI 317
Query: 316 KSAVGRGAFVKDIFVKGMDLNTMKYVFWMTGSYGDHPDNGFDPNALPKISGINYRDVTAK 375
K+ G +V+ I +++ + + Y D P + + KIS I YR++T
Sbjct: 318 KTYQGGSGYVQGIRFTNVEMQDVANPILIDQFYCDSPTTCQNQTSAVKISQIMYRNITGT 377
Query: 376 NVTI-AGKLEGISNDPFTGICVSNVTIE 402
+ A K P + I ++NV +E
Sbjct: 378 TKSAKAIKFACSDTVPCSHIVLNNVNLE 405
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,867,816
Number of Sequences: 26719
Number of extensions: 488183
Number of successful extensions: 1113
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 938
Number of HSP's gapped (non-prelim): 85
length of query: 463
length of database: 11,318,596
effective HSP length: 103
effective length of query: 360
effective length of database: 8,566,539
effective search space: 3083954040
effective search space used: 3083954040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC121237.2


