

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.18 - phase: 0
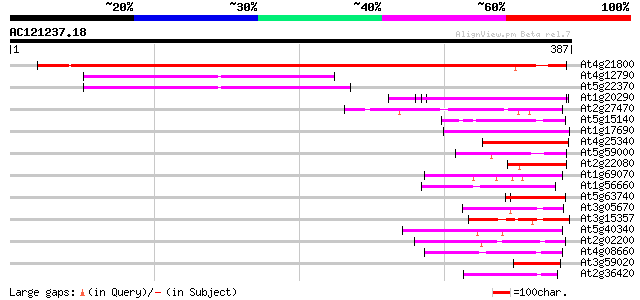
(387 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g21800 unknown protein 516 e-146
At4g12790 unknown protein 86 3e-17
At5g22370 putative protein 85 6e-17
At1g20290 hypothetical protein 48 8e-06
At2g27470 putative CCAAT-binding transcription factor subunit 46 4e-05
At5g15140 putative aldose 1-epimerase - like protein 45 5e-05
At1g17690 unknown protein 45 5e-05
At4g25340 unknown protein 45 9e-05
At5g59000 unknown protein 44 2e-04
At2g22080 En/Spm-like transposon protein 44 2e-04
At1g69070 hypothetical protein 44 2e-04
At1g56660 hypothetical protein 44 2e-04
At5g63740 unknown protein 43 3e-04
At3g05670 unknown protein 43 3e-04
At3g15357 unknown protein 42 4e-04
At5g40340 unknown protein 42 6e-04
At2g02200 putative protein on transposon FARE2.10 (cds2) 42 6e-04
At4g08660 putative MuDR-like transposon protein 42 7e-04
At3g59020 putative protein 42 7e-04
At2g36420 unknown protein 42 7e-04
>At4g21800 unknown protein
Length = 379
Score = 516 bits (1328), Expect = e-146
Identities = 259/371 (69%), Positives = 313/371 (83%), Gaps = 15/371 (4%)
Query: 20 KQKEELSESMKKLDIEGSSSGSPNFKRKPVIIIVVGMAGSGKTTLMHRLVTHTHLSNIRG 79
++ ++L +S+ KL + +SS S NFK+KP+IIIVVGMAGSGKT+ +HRLV HT S G
Sbjct: 14 EESQKLVDSLDKLRVSAASSSS-NFKKKPIIIIVVGMAGSGKTSFLHRLVCHTFDSKSHG 72
Query: 80 YVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQFNLGPNGGILTSLNLFATKFDEVISVIE 139
YV+NLDPAVM+LP+ +NIDIRDTVKYKEVMKQ+NLGPNGGILTSLNLFATKFDEV+SVIE
Sbjct: 73 YVVNLDPAVMSLPFGANIDIRDTVKYKEVMKQYNLGPNGGILTSLNLFATKFDEVVSVIE 132
Query: 140 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVAYVVDTPRAENPTTFMSNM 199
KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVV YVVDTPR+ +P TFMSNM
Sbjct: 133 KRADQLDYVLVDTPGQIEIFTWSASGAIITEAFASTFPTVVTYVVDTPRSSSPITFMSNM 192
Query: 200 LYACSILYKTRLPLILAFNKVDVAQHEFALEWMKDFEVFQAAASSDQSYTSNLTQSLSLA 259
LYACSILYKTRLPL+LAFNK DVA H+FALEWM+DFEVFQAA SD SYT+ L SLSL+
Sbjct: 193 LYACSILYKTRLPLVLAFNKTDVADHKFALEWMEDFEVFQAAIQSDNSYTATLANSLSLS 252
Query: 260 LDEFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKE 319
L EFY N+RS GVSA++GAG++GFFKA+EASAEEYMETYKADLD R+ +K+ LEE R+K
Sbjct: 253 LYEFYRNIRSVGVSAISGAGMDGFFKAIEASAEEYMETYKADLDMRKADKERLEEERKKH 312
Query: 320 NMDKLRREMEKSGGETVVLSTGLKNKEE------DEDEEDEEMDDDDNVDFGTYTEDEDA 373
M+KLR++ME S G TVVL+TGLK+++ +ED+ED +++D+++ D DA
Sbjct: 313 EMEKLRKDMESSQGGTVVLNTGLKDRDATEKMMLEEDDEDFQVEDEEDSD--------DA 364
Query: 374 IDEDEDEEVDK 384
IDED++++ K
Sbjct: 365 IDEDDEDDETK 375
>At4g12790 unknown protein
Length = 271
Score = 85.9 bits (211), Expect = 3e-17
Identities = 54/175 (30%), Positives = 88/175 (49%), Gaps = 3/175 (1%)
Query: 52 IVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQ 111
+V+G AGSGK+T L H +V+NLDPA Y +DIR+ + ++VM+
Sbjct: 6 LVIGPAGSGKSTYCSSLYEHCETIGRTMHVVNLDPAAEIFNYPVAMDIRELISLEDVMED 65
Query: 112 FNLGPNGGILTSLN-LFATKFDEVISVIEKRADQLDYVLVDTPGQIEIFTW-SASGAIIT 169
LGPNG ++ + L + D V +E D DY++ D PGQIE+FT +
Sbjct: 66 LKLGPNGALMYCMEYLEDSLHDWVDEELENYRDD-DYLIFDCPGQIELFTHVPVLKNFVE 124
Query: 170 EAFASTFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQ 224
F V Y++D+ + T F+S + + + + + LP + +K+D+ Q
Sbjct: 125 HLKQKNFNVCVVYLLDSQFITDVTKFISGCMSSLAAMIQLELPHVNILSKMDLLQ 179
>At5g22370 putative protein
Length = 291
Score = 85.1 bits (209), Expect = 6e-17
Identities = 51/186 (27%), Positives = 95/186 (50%), Gaps = 3/186 (1%)
Query: 52 IVVGMAGSGKTTLMHRLVTHTHLSNIRGYVMNLDPAVMTLPYASNIDIRDTVKYKEVMKQ 111
+V+G GSGKTT + + L + ++NLDPA LPY ++I + +K ++VM +
Sbjct: 6 VVIGPPGSGKTTYCNGMSQFLSLMGRKVAIVNLDPANDALPYECGVNIEELIKLEDVMSE 65
Query: 112 FNLGPNGGILTSLNLFATKFDEVISVIEKRADQLDYVLVDTPGQIEI-FTWSASGAIITE 170
+LGPNGG++ + D + S ++ Y+L D PGQ+E+ F ++ ++T+
Sbjct: 66 HSLGPNGGLVYCMEYLEKNIDWLESKLKPLLKD-HYILFDFPGQVELFFIHDSTKNVLTK 124
Query: 171 AFAS-TFPTVVAYVVDTPRAENPTTFMSNMLYACSILYKTRLPLILAFNKVDVAQHEFAL 229
S ++D+ +P ++S++L + S + LP + +K+D+ L
Sbjct: 125 LIKSLNLRLTAVQLIDSHLCCDPGNYVSSLLLSLSTMLHMELPHVNVLSKIDLIGSYGKL 184
Query: 230 EWMKDF 235
+ DF
Sbjct: 185 AFNLDF 190
>At1g20290 hypothetical protein
Length = 497
Score = 48.1 bits (113), Expect = 8e-06
Identities = 27/104 (25%), Positives = 54/104 (50%)
Query: 281 EGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLST 340
EG E EE E + + ++ EE++ EE +E ++ E E+ E
Sbjct: 357 EGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 416
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+ +EE+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 417 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 460
Score = 47.4 bits (111), Expect = 1e-05
Identities = 26/97 (26%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 386 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 445
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE +K
Sbjct: 446 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEK 482
Score = 47.4 bits (111), Expect = 1e-05
Identities = 26/98 (26%), Positives = 52/98 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 387 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 446
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDKF 385
+E+EE+EE ++++ + E+E+ +E+E+EE K+
Sbjct: 447 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKKY 484
Score = 47.0 bits (110), Expect = 2e-05
Identities = 26/100 (26%), Positives = 54/100 (54%)
Query: 285 KAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKN 344
+A E EE E + + ++ EE++ EE +E ++ E E+ E +
Sbjct: 362 QAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 421
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+EE+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 422 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 461
Score = 46.6 bits (109), Expect = 2e-05
Identities = 29/123 (23%), Positives = 61/123 (49%)
Query: 262 EFYSNLRSAGVSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENM 321
+F ++ G + A E + E EE E + + ++ EE++ EE +E
Sbjct: 347 KFTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 406
Query: 322 DKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
++ E E+ E + +EE+E+EE+EE ++++ + E+E+ +E+E+EE
Sbjct: 407 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 466
Query: 382 VDK 384
++
Sbjct: 467 EEE 469
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 382 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 441
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 442 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 478
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 379 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 438
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 439 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 475
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 381 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 440
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 441 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 477
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 380 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 439
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 440 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 476
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 374 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 433
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 434 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 470
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 377 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 436
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 437 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 473
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 376 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 435
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 436 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 472
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 385 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 444
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 445 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 481
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 384 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 443
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 444 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 480
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 378 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 437
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 438 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 474
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 383 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 442
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 443 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 479
Score = 45.8 bits (107), Expect = 4e-05
Identities = 25/97 (25%), Positives = 52/97 (52%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 375 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 434
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+E+EE+EE ++++ + E+E+ +E+E+EE ++
Sbjct: 435 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 471
Score = 29.3 bits (64), Expect = 3.8
Identities = 18/69 (26%), Positives = 33/69 (47%)
Query: 288 EASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEE 347
E EE E + + ++ EE++ EE +E ++ E E+ E + +EE
Sbjct: 417 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 476
Query: 348 DEDEEDEEM 356
+E+EE + M
Sbjct: 477 EEEEEKKYM 485
>At2g27470 putative CCAAT-binding transcription factor subunit
Length = 275
Score = 45.8 bits (107), Expect = 4e-05
Identities = 47/164 (28%), Positives = 73/164 (43%), Gaps = 24/164 (14%)
Query: 232 MKDFEVFQAAASSDQSYTSNLTQSLSLALDEFYSNL--RSAGVSAVTGAGIEGFFKAVEA 289
MK +VF+A D S + L +L++F + AG +A + K+
Sbjct: 70 MKADDVFKALEEMD---FSEFLEPLKSSLEDFKKKNAGKKAGAAAASYPAGGAALKSSSG 126
Query: 290 SAEEYMETYKADLDKRREEKQLLEENRRKENMDK-LRREMEKSGGETVVLSTGLKNKEED 348
+A + ET KR++E+ ++ RK +D+ +R E++ + G N EED
Sbjct: 127 TASKPKET-----KKRKQEEPSTQKGARKSKIDEETKRNDEETENDNTEEENG--NDEED 179
Query: 349 E---DEEDEEMD--------DDDNVDFGTYTEDEDAIDEDEDEE 381
E DEEDE D D++N D T D +E EDEE
Sbjct: 180 ENGNDEEDENDDENTEENGNDEENDDENTEENGNDEENEKEDEE 223
Score = 28.1 bits (61), Expect = 8.5
Identities = 18/82 (21%), Positives = 40/82 (47%), Gaps = 14/82 (17%)
Query: 303 DKRREEKQLLEENRRKE---NMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDD 359
D+ EE EEN +++ +M++ E E+SG N++ +E + +D
Sbjct: 205 DENTEENGNDEENEKEDEENSMEENGNESEESG-----------NEDHSMEENGSGVGED 253
Query: 360 DNVDFGTYTEDEDAIDEDEDEE 381
+ + G+ + + ++ DE++E
Sbjct: 254 NENEDGSVSGSGEEVESDEEDE 275
>At5g15140 putative aldose 1-epimerase - like protein
Length = 490
Score = 45.4 bits (106), Expect = 5e-05
Identities = 27/85 (31%), Positives = 51/85 (59%), Gaps = 7/85 (8%)
Query: 299 KADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
K + D ++++KQ EN+ K++ +K + +KSGG K+K++ +D +++ D
Sbjct: 74 KKEHDVQKKDKQ--HENKDKDD-EKKHVDKKKSGGHDKDDDDEKKHKDKKKDGHNDDDDS 130
Query: 359 DDNVDFGTYTEDEDAIDEDEDEEVD 383
DD+ D +D+D D+D+D+EVD
Sbjct: 131 DDDTD----DDDDDDDDDDDDDEVD 151
>At1g17690 unknown protein
Length = 754
Score = 45.4 bits (106), Expect = 5e-05
Identities = 26/88 (29%), Positives = 48/88 (54%), Gaps = 1/88 (1%)
Query: 300 ADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEED-EEMDD 358
AD++KRR+ ++ + + ++ D+ E E SG + + + G +K + +D+E + D
Sbjct: 78 ADMNKRRQREEEGKSDTEEDEDDEDEDEEENSGSDDLSSTDGEDDKSQGDDQETLGGLTD 137
Query: 359 DDNVDFGTYTEDEDAIDEDEDEEVDKFS 386
D D +E+ED D + DEEV + S
Sbjct: 138 DTQEDNDNQSEEEDPDDYETDEEVHELS 165
>At4g25340 unknown protein
Length = 477
Score = 44.7 bits (104), Expect = 9e-05
Identities = 21/60 (35%), Positives = 36/60 (60%), Gaps = 1/60 (1%)
Query: 327 EMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDA-IDEDEDEEVDKF 385
E +G ++ LS L+ ++DED+ + + DD D +D G ED+ D +EDE++D+F
Sbjct: 80 EFTVTGDRSIHLSGFLEYYQDDEDDYEHDEDDSDGIDVGESEEDDSCEYDSEEDEQLDEF 139
>At5g59000 unknown protein
Length = 231
Score = 43.5 bits (101), Expect = 2e-04
Identities = 26/80 (32%), Positives = 43/80 (53%), Gaps = 11/80 (13%)
Query: 308 EKQLLEENRRKENMDKLRREMEKS---GGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDF 364
EKQL +++ + RR+ +K G + + L+ +E EDE++EE DD D
Sbjct: 51 EKQLHSGGKKRRTRRRKRRKKKKKKKGGRDCRICHLPLETNKEAEDEDEEEEDDSD---- 106
Query: 365 GTYTEDEDAIDEDEDEEVDK 384
+DED DE+E+EE ++
Sbjct: 107 ----DDEDEEDEEEEEEEEE 122
>At2g22080 En/Spm-like transposon protein
Length = 140
Score = 43.5 bits (101), Expect = 2e-04
Identities = 22/46 (47%), Positives = 31/46 (66%), Gaps = 5/46 (10%)
Query: 344 NKEEDED-----EEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
N +ED+D +EDEE D+D+N D G +DEDA E+E+EE D+
Sbjct: 81 NDDEDDDNHENDDEDEEEDEDENDDGGEEDDDEDAEVEEEEEEEDE 126
Score = 35.8 bits (81), Expect = 0.041
Identities = 17/39 (43%), Positives = 27/39 (68%), Gaps = 3/39 (7%)
Query: 343 KNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+++EEDEDE D+ ++DD+ D E+E +EDED+E
Sbjct: 94 EDEEEDEDENDDGGEEDDDEDAEVEEEEE---EEDEDDE 129
Score = 32.0 bits (71), Expect = 0.59
Identities = 15/42 (35%), Positives = 23/42 (54%)
Query: 342 LKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
L + D D+E E D +D+ D + D++ +EDEDE D
Sbjct: 64 LNGEAGDNDDEPEGDDGNDDEDDDNHENDDEDEEEDEDENDD 105
>At1g69070 hypothetical protein
Length = 901
Score = 43.5 bits (101), Expect = 2e-04
Identities = 33/109 (30%), Positives = 56/109 (51%), Gaps = 14/109 (12%)
Query: 287 VEASAEEYMETYKADLDKRREEKQLLEENRRK--ENMDKLRREMEKSGGE------TVVL 338
+ A E +T + K RE+ + LEE R+K + ++L E+ GGE TV+
Sbjct: 265 IRARPSERTKTPEEIAQKEREKLEALEEERKKRMQETEELSDGDEEIGGEESTKRLTVIS 324
Query: 339 STGLKNK---EEDEDEE---DEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
L + EED+ + D+ ++ +DNVD EDED+ E+E+++
Sbjct: 325 GDDLGDSFSVEEDKPKRGWIDDVLEREDNVDNSESDEDEDSESEEEEDD 373
Score = 32.3 bits (72), Expect = 0.45
Identities = 19/73 (26%), Positives = 42/73 (57%), Gaps = 9/73 (12%)
Query: 292 EEYMETYKADLDKRREEKQLLEENRRKENMDKLRR------EMEKSGGETVVLSTGLKNK 345
E+ ++ ++D D+ E ++ +++ + D+ +R + E+S E L L+++
Sbjct: 351 EDNVDNSESDEDEDSESEEEEDDDGESDGGDEKQRKGHHLEDWEQSDDE---LGAELEDE 407
Query: 346 EEDEDEEDEEMDD 358
EED+DEED++ +D
Sbjct: 408 EEDDDEEDDDEED 420
>At1g56660 hypothetical protein
Length = 522
Score = 43.5 bits (101), Expect = 2e-04
Identities = 24/92 (26%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Query: 285 KAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKN 344
K + EE+ + +K +K+ EE + +E ++K+N ++E ++SG E +
Sbjct: 102 KESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKN----KKEKDESGPEEKNKKADKEK 157
Query: 345 KEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDE 376
K ED +E EE++++D +DE +E
Sbjct: 158 KHEDVSQEKEELEEEDGKKNKKKEKDESGTEE 189
Score = 37.4 bits (85), Expect = 0.014
Identities = 25/97 (25%), Positives = 46/97 (46%), Gaps = 14/97 (14%)
Query: 299 KADLDKRREEK-----QLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEED 353
K DL+K EEK + +E + K++ ++E ++S E + KE+DE E
Sbjct: 221 KGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEK 280
Query: 354 EEM---------DDDDNVDFGTYTEDEDAIDEDEDEE 381
E+ + + D G T++ DA +++ D+E
Sbjct: 281 EDKKLKGKKGKGEKPEKEDEGKKTKEHDATEQEMDDE 317
Score = 33.5 bits (75), Expect = 0.20
Identities = 20/73 (27%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Query: 304 KRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVD 363
K++ +K+ ++ K N DK + + +K GE L + K+++ DE D+EM + D+
Sbjct: 191 KKKPKKEKKQKEESKSNEDK-KVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKK 249
Query: 364 FGTYTEDEDAIDE 376
+DE +E
Sbjct: 250 NKKKEKDESCAEE 262
Score = 30.4 bits (67), Expect = 1.7
Identities = 26/93 (27%), Positives = 38/93 (39%), Gaps = 6/93 (6%)
Query: 296 ETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDE---- 351
E D+ E + EE+ +E E G+ KNK++D E
Sbjct: 390 EVMSRDIKLEEPEAEKKEEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDTKEPKMT 449
Query: 352 EDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
EDEE DD+ D E A +E +D++V K
Sbjct: 450 EDEEEKKDDSKD--VKIEGSKAKEEKKDKDVKK 480
Score = 28.5 bits (62), Expect = 6.5
Identities = 22/93 (23%), Positives = 42/93 (44%), Gaps = 12/93 (12%)
Query: 299 KADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDD 358
K + DK ++++ +++E KE DK + GET K K+ ++ E+D + D
Sbjct: 327 KKNKDKAKKKETVIDEVCEKETKDK-----DDDEGETKQKKNKKKEKKSEKGEKDVKEDK 381
Query: 359 DDNVDFGTYT-------EDEDAIDEDEDEEVDK 384
T E+ +A ++ED+ +K
Sbjct: 382 KKENPLETEVMSRDIKLEEPEAEKKEEDDTEEK 414
>At5g63740 unknown protein
Length = 226
Score = 43.1 bits (100), Expect = 3e-04
Identities = 17/38 (44%), Positives = 28/38 (72%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
E+++D++D++ DDDD+ D ED+D D+DEDE+ D
Sbjct: 87 EDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDEDDD 124
Score = 43.1 bits (100), Expect = 3e-04
Identities = 20/42 (47%), Positives = 31/42 (73%), Gaps = 1/42 (2%)
Query: 343 KNKEEDEDEEDEE-MDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
++++EDEDE+D++ DDDD+ D +DED DED+DE+ D
Sbjct: 81 EDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDED 122
Score = 41.2 bits (95), Expect = 0.001
Identities = 19/38 (50%), Positives = 28/38 (73%), Gaps = 4/38 (10%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+EDEDE+++E DDDD+ D +D+DA D D+DE+ D
Sbjct: 80 DEDEDEDEDEDDDDDDDD----DDDDDADDADDDEDDD 113
Score = 40.8 bits (94), Expect = 0.001
Identities = 19/42 (45%), Positives = 30/42 (71%), Gaps = 1/42 (2%)
Query: 344 NKEEDEDE-EDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVDK 384
+++EDEDE ED++ DDDD+ D D+D D+DED++ D+
Sbjct: 80 DEDEDEDEDEDDDDDDDDDDDDDADDADDDEDDDDEDDDEDE 121
Score = 35.0 bits (79), Expect = 0.070
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 4/35 (11%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDE 380
++D D+ D++ DDDD D EDED D+D+DE
Sbjct: 100 DDDADDADDDEDDDDEDD----DEDEDDDDDDDDE 130
Score = 34.7 bits (78), Expect = 0.091
Identities = 15/38 (39%), Positives = 25/38 (65%), Gaps = 6/38 (15%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+ D+DE+D++ DDD++ D D+D D++ DEE D
Sbjct: 105 DADDDEDDDDEDDDEDED------DDDDDDDENDEECD 136
Score = 34.7 bits (78), Expect = 0.091
Identities = 15/38 (39%), Positives = 27/38 (70%), Gaps = 4/38 (10%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
++DED++DE+ D+D++ D +D+D DE+ D+E D
Sbjct: 107 DDDEDDDDEDDDEDEDDD----DDDDDENDEECDDEYD 140
Score = 33.5 bits (75), Expect = 0.20
Identities = 12/37 (32%), Positives = 25/37 (67%)
Query: 347 EDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+ +++EDE+ D D++ D +D+D D+D+D++ D
Sbjct: 68 DGDEDEDEDADADEDEDEDEDEDDDDDDDDDDDDDAD 104
Score = 33.1 bits (74), Expect = 0.27
Identities = 17/38 (44%), Positives = 26/38 (67%), Gaps = 6/38 (15%)
Query: 346 EEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+EDEDE D + D+D++ D EDED D+D+D++ D
Sbjct: 70 DEDEDE-DADADEDEDED-----EDEDDDDDDDDDDDD 101
Score = 32.3 bits (72), Expect = 0.45
Identities = 27/75 (36%), Positives = 35/75 (46%), Gaps = 8/75 (10%)
Query: 318 KENMDKLRREMEKSGGETVVLST-GLKNKEEDE------DEEDEEMDDDDNVDFGTYTED 370
KE M+K R + + ST G+ EDE D ++ D D G ED
Sbjct: 15 KEEMEKRIRTVVIDSNLVLSNSTCGICRTAEDEYIKVYDDHNNDGEGDGDGDGDGDEDED 74
Query: 371 EDA-IDEDEDEEVDK 384
EDA DEDEDE+ D+
Sbjct: 75 EDADADEDEDEDEDE 89
Score = 28.9 bits (63), Expect = 5.0
Identities = 14/37 (37%), Positives = 23/37 (61%), Gaps = 3/37 (8%)
Query: 347 EDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
+ + +EDE+ D D + D EDED D+D+D++ D
Sbjct: 66 DGDGDEDEDEDADADED---EDEDEDEDDDDDDDDDD 99
>At3g05670 unknown protein
Length = 883
Score = 42.7 bits (99), Expect = 3e-04
Identities = 28/74 (37%), Positives = 41/74 (54%), Gaps = 7/74 (9%)
Query: 313 EENRRKENMDKLRREMEKSGGETVVLSTGLKN---KEEDEDEEDEEMDDDDNVDFGTYTE 369
EE R EN+D + G + + + KE+D D EDE+ DDD + DF T
Sbjct: 179 EEERDVENVDSNSLHDGEDGKMALEEQDNVSHETEKEDDGDYEDEDEDDDGDEDF---TA 235
Query: 370 DED-AIDEDEDEEV 382
DED ++DE+E+EE+
Sbjct: 236 DEDVSLDEEEEEEI 249
>At3g15357 unknown protein
Length = 143
Score = 42.4 bits (98), Expect = 4e-04
Identities = 28/72 (38%), Positives = 45/72 (61%), Gaps = 11/72 (15%)
Query: 317 RKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDD--DNVDFGTYTEDEDAI 374
+KE + + +ME + G+ VV NK+ D EED + D+D D+VDF +DED+
Sbjct: 77 QKEAVSERVGKMEAAHGKRVV-----DNKDADS-EEDTDFDEDEIDDVDF---EDDEDSD 127
Query: 375 DEDEDEEVDKFS 386
++DE+EE DK++
Sbjct: 128 EDDEEEEDDKYT 139
>At5g40340 unknown protein
Length = 1008
Score = 42.0 bits (97), Expect = 6e-04
Identities = 32/119 (26%), Positives = 61/119 (50%), Gaps = 9/119 (7%)
Query: 272 VSAVTGAGIEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENM-----DKLRR 326
+ V G G + K A E +T K D + ++ + +K ++ +K +
Sbjct: 3 IEVVLGIGEDAGPKPCSAEIESAEKTLKDDGVVQENGVRVSDNGEKKSDVVVDVDEKNEK 62
Query: 327 EMEKSGG-ETVVL---STGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+ +SG E V+ S+ LK KE+ E+EE+EE ++++ + G E+E+ +E+E+EE
Sbjct: 63 NLNESGVIEDCVMNGVSSLLKLKEDVEEEEEEEEEEEEEEEDGEDEEEEEEEEEEEEEE 121
Score = 29.6 bits (65), Expect = 2.9
Identities = 13/47 (27%), Positives = 28/47 (58%), Gaps = 3/47 (6%)
Query: 341 GLKNKEEDEDEEDEEMDDDDNVDF---GTYTEDEDAIDEDEDEEVDK 384
GL++K D+D++DEE + +D + + + E+ A+D +E + +
Sbjct: 316 GLEDKNNDDDDDDEEKNVNDGLQWRAKRSRVEEVAALDHEESSSLQR 362
>At2g02200 putative protein on transposon FARE2.10 (cds2)
Length = 586
Score = 42.0 bits (97), Expect = 6e-04
Identities = 29/113 (25%), Positives = 57/113 (49%), Gaps = 15/113 (13%)
Query: 280 IEGFFKAVEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKL---------RREMEK 330
+EG+ A E K + +K+ +E++ EE ++E ++K+ ++E+ K
Sbjct: 188 LEGYLGIERVPASYTREEQKEEDEKKEQEEEKQEEEGKEEKLEKIEYRGDEGTEKQEIPK 247
Query: 331 SGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEEVD 383
G E + G + K+E+E +E+EE + D GT ++ I + DEE++
Sbjct: 248 QGDEEM---EGEEEKQEEEGKEEEEEKVEYRGDEGT---EKQEIPKQGDEEME 294
Score = 33.1 bits (74), Expect = 0.27
Identities = 31/111 (27%), Positives = 51/111 (45%), Gaps = 16/111 (14%)
Query: 287 VEASAEEYMETYKADLDKRREEKQL-----LEENRRKENMDKLRREMEKSGGETVVLSTG 341
+E E+ E K + +++ E + +EE ++EN + EME+ G+ +
Sbjct: 293 MEGEEEKQKEEGKEEEEEKVEYRDHHSTCNVEETEKQENPKQGDEEMEREEGKEEKVLKE 352
Query: 342 LKNKEEDEDEEDEEM-------DDDDNVDFGTY-TEDEDAIDEDEDEEVDK 384
+E DE +E E+ DD+DN GT TE E ++E EV K
Sbjct: 353 ENVEEHDEHDETEDQEAYVILSDDEDN---GTAPTEKESESQKEETTEVAK 400
Score = 32.3 bits (72), Expect = 0.45
Identities = 22/89 (24%), Positives = 41/89 (45%), Gaps = 4/89 (4%)
Query: 297 TYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEM 356
T K ++ K+ +E+ EE ++KE + E + + K+E+ + DEEM
Sbjct: 280 TEKQEIPKQGDEEMEGEEEKQKEEGKEEEEEKVEYRDHHSTCNVEETEKQENPKQGDEEM 339
Query: 357 DDDDNVDFGTY----TEDEDAIDEDEDEE 381
+ ++ + E+ D DE ED+E
Sbjct: 340 EREEGKEEKVLKEENVEEHDEHDETEDQE 368
>At4g08660 putative MuDR-like transposon protein
Length = 714
Score = 41.6 bits (96), Expect = 7e-04
Identities = 30/95 (31%), Positives = 47/95 (48%), Gaps = 7/95 (7%)
Query: 287 VEASAEEYMETYKADLDKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKE 346
VE +AE+ + + D + R K+ +E + R E+ K+ G E V
Sbjct: 106 VEFTAEDSSDISRKDGSRGRGSKRPIESDSRAESTFKV----PGGGFEGVCYDYQDDEVG 161
Query: 347 EDEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDEE 381
+DE +DEE DD+ VD D++A DE+ED+E
Sbjct: 162 DDESGDDEEWDDESGVD---EASDKEAGDEEEDDE 193
>At3g59020 putative protein
Length = 1112
Score = 41.6 bits (96), Expect = 7e-04
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 348 DEDEEDEEMDDDDNVDFGTYTEDEDAIDEDEDE 380
+E+EEDE+ DDDD +F T EDED DE+ DE
Sbjct: 972 EEEEEDEDGDDDDMDEFQTDDEDEDGDDENPDE 1004
Score = 29.3 bits (64), Expect = 3.8
Identities = 24/101 (23%), Positives = 47/101 (45%), Gaps = 25/101 (24%)
Query: 293 EYMETYKADL-DKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDE 351
E + YK L + + E++ +E+ ++MD+ + + E E+ +DE
Sbjct: 956 ELLVAYKDQLAEAAKAEEEEEDEDGDDDDMDEFQTDDED---------------EDGDDE 1000
Query: 352 EDEEMDDD-------DNVDFGTYTEDEDAIDED--EDEEVD 383
+E D DF +Y++D+D D+D +DEE++
Sbjct: 1001 NPDETDGSTLRKLAAQAKDFRSYSDDDDFSDDDFSDDEELE 1041
>At2g36420 unknown protein
Length = 439
Score = 41.6 bits (96), Expect = 7e-04
Identities = 22/65 (33%), Positives = 38/65 (57%), Gaps = 2/65 (3%)
Query: 314 ENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNVDFGTYTEDEDA 373
+ ++ + KLRR + +G + V L + +E++E+EE EE ++DDN+ Y DE+
Sbjct: 287 QRAKRRLLKKLRRFEKLAGLDPVELEGKMSEEEDEEEEEYEESEEDDNIRI--YDSDEEY 344
Query: 374 IDEDE 378
D DE
Sbjct: 345 EDVDE 349
Score = 32.3 bits (72), Expect = 0.45
Identities = 19/60 (31%), Positives = 32/60 (52%), Gaps = 2/60 (3%)
Query: 303 DKRREEKQLLEENRRKENMDKLRREMEKSGGETVVLSTGLKNKEEDEDEEDEEMDDDDNV 362
D+ +E + LE+ R +E DK + E+ +V+ L+ +EEDED E D +N+
Sbjct: 222 DEDSDETESLEKVRGQEEEDKEEEDKEQCSPVSVL--DPLEEEEEDEDHHQHEPDPPNNL 279
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,854,388
Number of Sequences: 26719
Number of extensions: 420985
Number of successful extensions: 10086
Number of sequences better than 10.0: 586
Number of HSP's better than 10.0 without gapping: 276
Number of HSP's successfully gapped in prelim test: 327
Number of HSP's that attempted gapping in prelim test: 5161
Number of HSP's gapped (non-prelim): 2496
length of query: 387
length of database: 11,318,596
effective HSP length: 101
effective length of query: 286
effective length of database: 8,619,977
effective search space: 2465313422
effective search space used: 2465313422
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC121237.18


