

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121234.11 - phase: 0 /pseudo
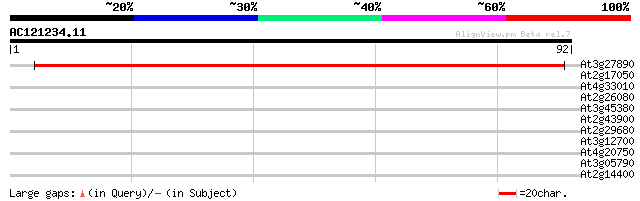
(92 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g27890 NADPH:quinone oxidoreductase (NQR) 128 4e-31
At2g17050 putative disease resistance protein 27 1.8
At4g33010 P-Protein - like protein 26 3.0
At2g26080 putative glycine dehydrogenase 26 3.0
At3g45380 putative protein 26 3.9
At2g43900 putative inositol polyphosphate 5'-phosphatase 25 5.1
At2g29680 cell division control protein 6 (cdc6) 25 5.1
At3g12700 unknown protein 25 6.7
At4g20750 unknown protein 25 8.7
At3g05790 putative mitochondrial LON ATP-dependent protease 25 8.7
At2g14400 putative retroelement pol polyprotein 25 8.7
>At3g27890 NADPH:quinone oxidoreductase (NQR)
Length = 196
Score = 128 bits (322), Expect = 4e-31
Identities = 60/87 (68%), Positives = 78/87 (88%)
Query: 5 VIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPP 64
+I+VAALSGS+RK S+H+GL+RAAI+L+K + G++IE+IDIS LP+ NTDLE GTYPP
Sbjct: 10 LIRVAALSGSLRKTSFHTGLLRAAIDLTKESVPGLQIEYIDISPLPLINTDLEVNGTYPP 69
Query: 65 LVEAFRHKILQADSVLFASPEYNYSVT 91
+VEAFR KIL+ADS+LFASPEYN+SV+
Sbjct: 70 VVEAFRQKILEADSILFASPEYNFSVS 96
>At2g17050 putative disease resistance protein
Length = 957
Score = 26.9 bits (58), Expect = 1.8
Identities = 11/21 (52%), Positives = 15/21 (71%)
Query: 32 SKGATEGIEIEFIDISTLPMY 52
SKG TE IE+ F+D+S L +
Sbjct: 355 SKGTTEDIEVIFLDMSNLKFF 375
>At4g33010 P-Protein - like protein
Length = 1037
Score = 26.2 bits (56), Expect = 3.0
Identities = 17/55 (30%), Positives = 26/55 (46%), Gaps = 3/55 (5%)
Query: 30 ELSKGATEGI---EIEFIDISTLPMYNTDLENEGTYPPLVEAFRHKILQADSVLF 81
E+S+G E + + D++ LPM N L +EGT A + IL+ F
Sbjct: 185 EISQGRLESLLNFQTVITDLTGLPMSNASLLDEGTAAAEAMAMCNNILKGKKKTF 239
>At2g26080 putative glycine dehydrogenase
Length = 1044
Score = 26.2 bits (56), Expect = 3.0
Identities = 17/55 (30%), Positives = 26/55 (46%), Gaps = 3/55 (5%)
Query: 30 ELSKGATEGI---EIEFIDISTLPMYNTDLENEGTYPPLVEAFRHKILQADSVLF 81
E+S+G E + + D++ LPM N L +EGT A + IL+ F
Sbjct: 191 EISQGRLESLLNYQTVITDLTGLPMSNASLLDEGTAAAEAMAMCNNILKGKKKTF 245
>At3g45380 putative protein
Length = 690
Score = 25.8 bits (55), Expect = 3.9
Identities = 18/78 (23%), Positives = 37/78 (47%), Gaps = 7/78 (8%)
Query: 15 IRKVSYHSGLIRAAIELSKGATEGIE-----IEFIDI--STLPMYNTDLENEGTYPPLVE 67
+ ++ + + L++ T+G + I+ ID+ ++ ++ DL+ E
Sbjct: 524 VHEIENEIQFVASITSLTQMMTKGFDETKDKIDAIDVRVQSIKLFVADLKEREHGKQTEE 583
Query: 68 AFRHKILQADSVLFASPE 85
A +H + D VLFASP+
Sbjct: 584 ASQHGHARDDDVLFASPD 601
>At2g43900 putative inositol polyphosphate 5'-phosphatase
Length = 1305
Score = 25.4 bits (54), Expect = 5.1
Identities = 16/36 (44%), Positives = 21/36 (57%), Gaps = 1/36 (2%)
Query: 4 AVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGI 39
AVIK+AA G I ++ H G IR +S G +GI
Sbjct: 524 AVIKMAAADGYIFSLATHGG-IRGWPVISPGPLDGI 558
>At2g29680 cell division control protein 6 (cdc6)
Length = 539
Score = 25.4 bits (54), Expect = 5.1
Identities = 24/75 (32%), Positives = 33/75 (44%), Gaps = 7/75 (9%)
Query: 10 ALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDISTLPMYNTDLENEGTYPPLVEAF 69
AL RKVS SG +R A+ + + A E +EIE + D E +G P
Sbjct: 366 ALEICARKVSAASGDMRKALCVCRSALEILEIEV-------RGSIDQEPKGPVPECQVVK 418
Query: 70 RHKILQADSVLFASP 84
++ A S F SP
Sbjct: 419 MDHMIAALSKTFKSP 433
>At3g12700 unknown protein
Length = 439
Score = 25.0 bits (53), Expect = 6.7
Identities = 14/50 (28%), Positives = 23/50 (46%), Gaps = 7/50 (14%)
Query: 22 SGLIRAAIELSKGATEGIEIEF-------IDISTLPMYNTDLENEGTYPP 64
+GL R +EL + EG+ IE+ ++S LP L+ + P
Sbjct: 334 TGLARYLVELKRVKPEGVPIEYCFSFTSGFNVSKLPQLTFHLKGGARFEP 383
>At4g20750 unknown protein
Length = 105
Score = 24.6 bits (52), Expect = 8.7
Identities = 13/44 (29%), Positives = 22/44 (49%)
Query: 1 MATAVIKVAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFI 44
MA A +K +SG S + + + +GA+ GI +EF+
Sbjct: 1 MAVAALKRMRVSGLATSSSSSVPNWKGGVSMVQGASRGIGLEFV 44
>At3g05790 putative mitochondrial LON ATP-dependent protease
Length = 942
Score = 24.6 bits (52), Expect = 8.7
Identities = 10/38 (26%), Positives = 20/38 (52%)
Query: 8 VAALSGSIRKVSYHSGLIRAAIELSKGATEGIEIEFID 45
+AA ++ + + R EL++ EG+E+ F+D
Sbjct: 891 IAARRSQVKVIIFPEANRRDFDELARNVKEGLEVHFVD 928
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 24.6 bits (52), Expect = 8.7
Identities = 11/35 (31%), Positives = 18/35 (51%)
Query: 13 GSIRKVSYHSGLIRAAIELSKGATEGIEIEFIDIS 47
GSIR+V Y L + K + + I+F D++
Sbjct: 491 GSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLN 525
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,796,681
Number of Sequences: 26719
Number of extensions: 61297
Number of successful extensions: 129
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 123
Number of HSP's gapped (non-prelim): 11
length of query: 92
length of database: 11,318,596
effective HSP length: 68
effective length of query: 24
effective length of database: 9,501,704
effective search space: 228040896
effective search space used: 228040896
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC121234.11


