

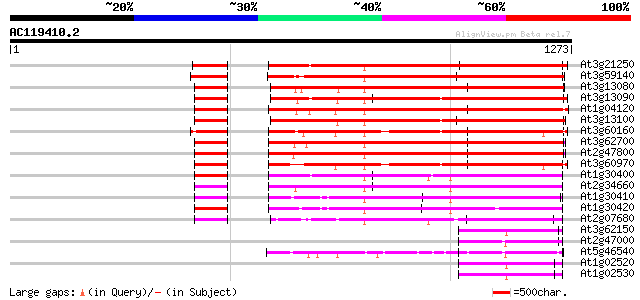
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119410.2 - phase: 1 /pseudo
(1273 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g21250 unknown protein 877 0.0
At3g59140 ABC transporter-like protein 615 e-176
At3g13080 ABC transporter, putative 593 e-169
At3g13090 ABC transporter like protein 588 e-168
At1g04120 multi-drug resistance protein 586 e-167
At3g13100 ABC transporter, putative 583 e-166
At3g60160 multi resistance protein homolog 565 e-161
At3g62700 ABC transporter-like protein 558 e-159
At2g47800 MRP-like ABC transporter 545 e-155
At3g60970 ABC transporter-like protein 545 e-155
At1g30400 AtMRP1 405 e-113
At2g34660 MRP-like ABC transporter 386 e-107
At1g30410 hypothetical protein 383 e-106
At1g30420 hypothetical protein 377 e-104
At2g07680 putative ABC transporter 335 1e-91
At3g62150 P-glycoprotein-like proetin 154 3e-37
At2g47000 putative ABC transporter 152 9e-37
At5g46540 multidrug resistance p-glycoprotein 148 2e-35
At1g02520 P-glycoprotein, putative 146 6e-35
At1g02530 hypothetical protein 144 4e-34
>At3g21250 unknown protein
Length = 1305
Score = 877 bits (2267), Expect = 0.0
Identities = 462/697 (66%), Positives = 546/697 (78%), Gaps = 21/697 (3%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHK- 642
CV +L++KTVILVTHQVEFLSEVD IL +G + QSG YE LL GTAF+ LV+AH
Sbjct: 606 CVEDSLKEKTVILVTHQVEFLSEVDQILVMEEGTITQSGKYEELLMMGTAFQQLVNAHND 665
Query: 643 -VTINDLNQNS---EVLSNPQDSHGFYLT---KNQSEGEISSIQGSIGAQLTQEEEKVIG 695
VT+ L N ++ +D +T K + E E + I G QLTQEEEK G
Sbjct: 666 AVTVLPLASNESLGDLRKEGKDREIRNMTVVEKIEEEIEKTDIPG---VQLTQEEEKESG 722
Query: 696 NVGWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLATAIEIPKVTDTTLIGVYA 755
VG KP DYI S G + +LGQ F+ Q +S +WLA AI IPK+T+T LIGVY+
Sbjct: 723 YVGMKPFLDYIGVSRGWCLLWSSVLGQVGFVVFQAASTYWLAFAIGIPKITNTMLIGVYS 782
Query: 756 LLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP-------ASSDL 808
++S S FVY R+ A LGLKAS AFFS FT ++F APMLFFDSTP ASSDL
Sbjct: 783 IISTLSAGFVYARAITTAHLGLKASKAFFSGFTNAVFKAPMLFFDSTPVGRILTRASSDL 842
Query: 809 SILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATAREL 868
++LD+D+P++ V A+E+ + ++ VTWQV+I+A+ A+ A +Q YY A+AREL
Sbjct: 843 NVLDYDVPFAFIFVVAPAVELTAALLIMTYVTWQVIIIALLALAATKVVQDYYLASAREL 902
Query: 869 IRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLV 928
IRINGTTKAPVMN+AAET LGVVT+RAF +RFFKNYL LVD DA LFF SN AMEW++
Sbjct: 903 IRINGTTKAPVMNYAAETSLGVVTIRAFGTAERFFKNYLNLVDADAVLFFLSNAAMEWVI 962
Query: 929 LRIEALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYII 988
LRIE L N+T+ T ALLLIL+P+ Y++PG VGLSLSYALTL Q+F TRW+ LSN II
Sbjct: 963 LRIETLQNVTLFTCALLLILIPKGYIAPGLVGLSLSYALTLTQTQVFLTRWYCTLSNSII 1022
Query: 989 SVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKG 1048
SVERIKQ+++IP EPPAI+D+ RPPSSWPS G I LQ L++RYRPNAPLVLKGI+CTF+
Sbjct: 1023 SVERIKQYMNIPEEPPAIIDDKRPPSSWPSNGTIHLQELKIRYRPNAPLVLKGISCTFRE 1082
Query: 1049 GSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTL 1108
G+RVGVVGRTGSGKSTLISALFRLVEP+ G ILIDGI+I +GLKDLRMKLSIIPQEPTL
Sbjct: 1083 GTRVGVVGRTGSGKSTLISALFRLVEPASGCILIDGIDISKIGLKDLRMKLSIIPQEPTL 1142
Query: 1109 FKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFC 1168
F+G IRTNLDPLG+YSDDEIWKA+EKCQLK TIS LP+ LDSSVSDEG NWS+GQRQLFC
Sbjct: 1143 FRGCIRTNLDPLGVYSDDEIWKALEKCQLKTTISNLPNKLDSSVSDEGENWSVGQRQLFC 1202
Query: 1169 LGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMV 1228
LGRVLLKRN+ILVLDEATASIDSATDAI+QRIIR+EF +CTVITVAHRVPTVIDSDMVMV
Sbjct: 1203 LGRVLLKRNKILVLDEATASIDSATDAIIQRIIREEFADCTVITVAHRVPTVIDSDMVMV 1262
Query: 1229 LSYGKLVEYDEPSKLMDTNSSFSKLVAEYWSSCRKNS 1265
LS+G LVEY+EPSKLM+T+S FSKLVAEYW+SCR NS
Sbjct: 1263 LSFGDLVEYNEPSKLMETDSYFSKLVAEYWASCRGNS 1299
Score = 96.7 bits (239), Expect = 8e-20
Identities = 46/78 (58%), Positives = 61/78 (77%)
Query: 416 SVNALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKI 475
S A+ IQ G F W+ E+ P L++++LEIK QK+AVC PVG+ KSS L+A+LGEIPK+
Sbjct: 435 SGTAVDIQVGNFGWEPETKIPTLRNIHLEIKHGQKVAVCGPVGAGKSSLLHAVLGEIPKV 494
Query: 476 SGTVYVGGTLAYVSQSSW 493
SGTV V G++AYVSQ+SW
Sbjct: 495 SGTVKVFGSIAYVSQTSW 512
Score = 68.9 bits (167), Expect = 2e-11
Identities = 53/235 (22%), Positives = 106/235 (44%), Gaps = 15/235 (6%)
Query: 1022 IDLQGLEVRYRPNAPL-VLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDI 1080
+D+Q + P + L+ I K G +V V G G+GKS+L+ A+ + G +
Sbjct: 439 VDIQVGNFGWEPETKIPTLRNIHLEIKHGQKVAVCGPVGAGKSSLLHAVLGEIPKVSGTV 498
Query: 1081 LIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKET 1140
+ G ++ + Q + G+IR N+ A++ C L +
Sbjct: 499 KVFG-------------SIAYVSQTSWIQSGTIRDNILYGKPMESRRYNAAIKACALDKD 545
Query: 1141 ISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAIL-QR 1199
++ + + G N S GQ+Q L R + + +LD+ +++D+ T +L +
Sbjct: 546 MNGFGHGDLTEIGQRGINLSGGQKQRIQLARAVYADADVYLLDDPFSAVDAHTAGVLFHK 605
Query: 1200 IIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLV 1254
+ +E TVI V H+V + + D ++V+ G + + + +L+ ++F +LV
Sbjct: 606 CVEDSLKEKTVILVTHQVEFLSEVDQILVMEEGTITQSGKYEELLMMGTAFQQLV 660
>At3g59140 ABC transporter-like protein
Length = 1389
Score = 615 bits (1587), Expect = e-176
Identities = 324/689 (47%), Positives = 458/689 (66%), Gaps = 24/689 (3%)
Query: 585 QYCVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAH 641
Q VM AL K V+LVTHQV+FL D++L DG++ ++ +Y+ LL F+ LV+AH
Sbjct: 703 QEYVMDALAGKAVLLVTHQVDFLPAFDSVLLMSDGEITEADTYQELLARSRDFQDLVNAH 762
Query: 642 KVTINDLNQNSEVLSNPQDSHGFYLTKNQSE-GEISSIQGSI--GAQLTQEEEKVIGNVG 698
+ T ++ + NP TK E + S Q + ++L ++EE+ G+ G
Sbjct: 763 RETAG--SERVVAVENP--------TKPVKEINRVISSQSKVLKPSRLIKQEEREKGDTG 812
Query: 699 WKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLATAIEIPKVTDTTLIGVYALLS 758
+P Y+N + G + + L Q F Q N W+A ++ P+V+ LI VY L+
Sbjct: 813 LRPYIQYMNQNKGYIFFFIASLAQVTFAVGQILQNSWMAANVDNPQVSTLKLILVYLLIG 872
Query: 759 ISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP-------ASSDLSIL 811
+ S + VRS ++ +K+S + FS S+F APM F+DSTP SSDLSI+
Sbjct: 873 LCSVLCLMVRSVCVVIMCMKSSASLFSQLLNSLFRAPMSFYDSTPLGRILSRVSSDLSIV 932
Query: 812 DFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRI 871
D D+P+ L V ++ + V+A VTWQVL V+VP + +QKYY TA+EL+RI
Sbjct: 933 DLDVPFGLIFVVASSVNTGCSLGVLAIVTWQVLFVSVPMVYLAFRLQKYYFQTAKELMRI 992
Query: 872 NGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRI 931
NGTT++ V N AE+ G +T+RAF+ +RFFK L L+DT+AS FFHS A EWL+ R+
Sbjct: 993 NGTTRSYVANHLAESVAGAITIRAFDEEERFFKKSLTLIDTNASPFFHSFAANEWLIQRL 1052
Query: 932 EALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVE 991
E + + + + A +ILLP S G +G++LSY L+LN ++ + L+N+IISVE
Sbjct: 1053 ETVSAIVLASTAFCMILLPTGTFSSGFIGMALSYGLSLNMGLVYSVQNQCYLANWIISVE 1112
Query: 992 RIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSR 1051
R+ Q+ H+ E P +++ RPP +WP G++++ L++RYR +PLVLKGI+CTF+GG +
Sbjct: 1113 RLNQYTHLTPEAPEVIEETRPPVNWPVTGRVEISDLQIRYRRESPLVLKGISCTFEGGHK 1172
Query: 1052 VGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKG 1111
+G+VGRTGSGK+TLISALFRLVEP G I++DG++I +G+ DLR + IIPQ+PTLF G
Sbjct: 1173 IGIVGRTGSGKTTLISALFRLVEPVGGKIVVDGVDISKIGVHDLRSRFGIIPQDPTLFNG 1232
Query: 1112 SIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGR 1171
++R NLDPL +SD EIW+ + KCQLKE + + + LDS V ++G NWS+GQRQLFCLGR
Sbjct: 1233 TVRFNLDPLCQHSDAEIWEVLGKCQLKEVVQEKENGLDSLVVEDGSNWSMGQRQLFCLGR 1292
Query: 1172 VLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSY 1231
+L+R+R+LVLDEATASID+ATD ILQ+ IR+EF +CTVITVAHR+PTV+D MV+ +S
Sbjct: 1293 AVLRRSRVLVLDEATASIDNATDLILQKTIRREFADCTVITVAHRIPTVMDCTMVLSISD 1352
Query: 1232 GKLVEYDEPSKLM-DTNSSFSKLVAEYWS 1259
G++VEYDEP KLM D NS F KLV EYWS
Sbjct: 1353 GRIVEYDEPMKLMKDENSLFGKLVKEYWS 1381
Score = 83.2 bits (204), Expect = 9e-16
Identities = 63/241 (26%), Positives = 109/241 (45%), Gaps = 27/241 (11%)
Query: 1015 SWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVE 1074
SW KG +PN L+ ++ K G +V V G GSGKSTL++A+
Sbjct: 545 SWEEKGST---------KPN----LRNVSLEVKFGEKVAVCGEVGSGKSTLLAAIL---- 587
Query: 1075 PSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEK 1134
G C G D ++ + Q + G+IR N+ G+ + + ++K
Sbjct: 588 ---------GETPCVSGTIDFYGTIAYVSQTAWIQTGTIRDNILFGGVMDEHRYRETIQK 638
Query: 1135 CQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSAT- 1193
L + + LP + + + G N S GQ+Q L R L + I +LD+ +++D+ T
Sbjct: 639 SSLDKDLELLPDGDQTEIGERGVNLSGGQKQRIQLARALYQDADIYLLDDPFSAVDAHTA 698
Query: 1194 DAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKL 1253
++ Q + V+ V H+V + D V+++S G++ E D +L+ + F L
Sbjct: 699 SSLFQEYVMDALAGKAVLLVTHQVDFLPAFDSVLLMSDGEITEADTYQELLARSRDFQDL 758
Query: 1254 V 1254
V
Sbjct: 759 V 759
Score = 81.3 bits (199), Expect = 3e-15
Identities = 40/85 (47%), Positives = 61/85 (71%), Gaps = 1/85 (1%)
Query: 410 KNLTQCSVNALQIQDGTFIWDHE-SMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAI 468
K ++ + NA+ I+ +F W+ + S P L++V+LE+K+ +K+AVC VGS KS+ L AI
Sbjct: 527 KQRSEGNQNAIIIKSASFSWEEKGSTKPNLRNVSLEVKFGEKVAVCGEVGSGKSTLLAAI 586
Query: 469 LGEIPKISGTVYVGGTLAYVSQSSW 493
LGE P +SGT+ GT+AYVSQ++W
Sbjct: 587 LGETPCVSGTIDFYGTIAYVSQTAW 611
>At3g13080 ABC transporter, putative
Length = 1514
Score = 593 bits (1530), Expect = e-169
Identities = 320/699 (45%), Positives = 459/699 (64%), Gaps = 34/699 (4%)
Query: 592 LRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKVTIN-- 646
L K+VI VTHQVEFL D IL DG++ Q+G Y ++L SGT F L+ AH+ +
Sbjct: 812 LCSKSVIYVTHQVEFLPAADLILVMKDGRISQAGKYNDILNSGTDFMELIGAHQEALAVV 871
Query: 647 ---DLNQNSEVLSNPQDS---------HGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVI 694
D N SE + Q++ +++ ++ S++ Q+ QEEE+
Sbjct: 872 DSVDANSVSEKSALGQENVIVKDAIAVDEKLESQDLKNDKLESVEPQ--RQIIQEEEREK 929
Query: 695 GNVGWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLATAIEIPK-----VTDTT 749
G+V W YI + G + ++LGQ F LQ SN+W+A A + + V +T
Sbjct: 930 GSVALDVYWKYITLAYGGALVPFILLGQVLFQLLQIGSNYWMAWATPVSEDVQAPVKLST 989
Query: 750 LIGVYALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP------ 803
L+ VY L+ S+ + +R+ G K +T F IF +PM FFDSTP
Sbjct: 990 LMIVYVALAFGSSLCILLRATLLVTAGYKTATELFHKMHHCIFRSPMSFFDSTPSGRIMS 1049
Query: 804 -ASSDLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQ 862
AS+D S +D ++PY VAI I+++ +I V++ V+W V +V +P + A I+ Q+YY
Sbjct: 1050 RASTDQSAVDLELPYQFGSVAITVIQLIGIIGVMSQVSWLVFLVFIPVVAASIWYQRYYI 1109
Query: 863 ATARELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNV 922
A AREL R+ G KAP++ +ET G T+R+F+ RF + ++L D + F++
Sbjct: 1110 AAARELSRLVGVCKAPLIQHFSETISGATTIRSFSQEFRFRSDNMRLSDGYSRPKFYTAG 1169
Query: 923 AMEWLVLRIEALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRW-FS 981
AMEWL R++ L +LT + + + L+ +P + P GL+++Y L+LN Q W W
Sbjct: 1170 AMEWLCFRLDMLSSLTFVFSLVFLVSIPTGVIDPSLAGLAVTYGLSLNTLQA-WLIWTLC 1228
Query: 982 NLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKG 1041
NL N IISVERI Q+ +P+EPP ++++NRP SWPS+G+++++ L+VRY P+ PLVL+G
Sbjct: 1229 NLENKIISVERILQYASVPSEPPLVIESNRPEQSWPSRGEVEIRDLQVRYAPHMPLVLRG 1288
Query: 1042 ITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSI 1101
ITCTFKGG R G+VGRTGSGKSTLI LFR+VEPS G+I IDG+NI ++GL DLR++LSI
Sbjct: 1289 ITCTFKGGLRTGIVGRTGSGKSTLIQTLFRIVEPSAGEIRIDGVNILTIGLHDLRLRLSI 1348
Query: 1102 IPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSL 1161
IPQ+PT+F+G++R+NLDPL Y+DD+IW+A++KCQL + + K LDSSVS+ G NWS+
Sbjct: 1349 IPQDPTMFEGTMRSNLDPLEEYTDDQIWEALDKCQLGDEVRKKEQKLDSSVSENGDNWSM 1408
Query: 1162 GQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVI 1221
GQRQL CLGRVLLKR++ILVLDEATAS+D+ATD ++Q+ +R+ F +CTVIT+AHR+ +VI
Sbjct: 1409 GQRQLVCLGRVLLKRSKILVLDEATASVDTATDNLIQKTLREHFSDCTVITIAHRISSVI 1468
Query: 1222 DSDMVMVLSYGKLVEYDEPSKLM-DTNSSFSKLVAEYWS 1259
DSDMV++LS G + EYD P +L+ D +SSFSKLVAEY S
Sbjct: 1469 DSDMVLLLSNGIIEEYDTPVRLLEDKSSSFSKLVAEYTS 1507
Score = 80.5 bits (197), Expect = 6e-15
Identities = 37/75 (49%), Positives = 53/75 (70%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
A+++ + T WD S +P LKD+N ++ K+AVC VGS KSS L ++LGE+PK+SG+
Sbjct: 639 AVEVINSTLSWDVSSSNPTLKDINFKVFPGMKVAVCGTVGSGKSSLLSSLLGEVPKVSGS 698
Query: 479 VYVGGTLAYVSQSSW 493
+ V GT AYV+QS W
Sbjct: 699 LKVCGTKAYVAQSPW 713
Score = 65.5 bits (158), Expect = 2e-10
Identities = 50/220 (22%), Positives = 102/220 (45%), Gaps = 14/220 (6%)
Query: 1039 LKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMK 1098
LK I G +V V G GSGKS+L+S+L V G + + G
Sbjct: 658 LKDINFKVFPGMKVAVCGTVGSGKSSLLSSLLGEVPKVSGSLKVCGTK------------ 705
Query: 1099 LSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGN 1158
+ + Q P + G I N+ + K +E C L + + L + + + G N
Sbjct: 706 -AYVAQSPWIQSGKIEDNILFGKPMERERYDKVLEACSLSKDLEILSFGDQTVIGERGIN 764
Query: 1159 WSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDA-ILQRIIRQEFEECTVITVAHRV 1217
S GQ+Q + R L + I + D+ +++D+ T + + + ++ +VI V H+V
Sbjct: 765 LSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHTGSHLFKEVLLGLLCSKSVIYVTHQV 824
Query: 1218 PTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEY 1257
+ +D+++V+ G++ + + + ++++ + F +L+ +
Sbjct: 825 EFLPAADLILVMKDGRISQAGKYNDILNSGTDFMELIGAH 864
>At3g13090 ABC transporter like protein
Length = 1466
Score = 588 bits (1517), Expect = e-168
Identities = 318/697 (45%), Positives = 450/697 (63%), Gaps = 29/697 (4%)
Query: 592 LRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKVTINDL 648
LR KTVI VTHQVEFL E D IL DGK+ Q+G Y +L SGT F LV AH + +
Sbjct: 773 LRHKTVIYVTHQVEFLPEADLILVMKDGKITQAGKYHEILDSGTDFMELVGAHTEALATI 832
Query: 649 NQ-----NSEVLSNPQDSHGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVIGNVGWKPLW 703
+ SE + +++ + + Q G + G QL QEEE+ G VG+
Sbjct: 833 DSCETGYASEKSTTDKENEVLHHKEKQENGSDNKPSG----QLVQEEEREKGKVGFTVYK 888
Query: 704 DYINYSNGTLMSCLVILGQCCFLALQTSSNFWLATAIEI-----PKVTDTTLIGVYALLS 758
Y+ + G + L+++ Q F L SN+W+ + P V+ TLI VY LL+
Sbjct: 889 KYMALAYGGAVIPLILVVQVLFQLLSIGSNYWMTWVTPVSKDVEPPVSGFTLILVYVLLA 948
Query: 759 ISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP-------ASSDLSIL 811
++S+ + +R+ A+ G K +T F+ IF A M FFD+TP AS+D S+
Sbjct: 949 VASSFCILIRALLVAMTGFKMATELFTQMHLRIFRASMSFFDATPMGRILNRASTDQSVA 1008
Query: 812 DFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRI 871
D +P VAI AI +L +I VI V WQVLIV +P + A + ++YY + AREL R+
Sbjct: 1009 DLRLPGQFAYVAIAAINILGIIGVIVQVAWQVLIVFIPVVAACAWYRQYYISAARELARL 1068
Query: 872 NGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRI 931
G +++PV++ +ET G+ T+R+F+ RF + ++L D + L FHS AMEWL R+
Sbjct: 1069 AGISRSPVVHHFSETLSGITTIRSFDQEPRFRGDIMRLSDCYSRLKFHSTGAMEWLCFRL 1128
Query: 932 EALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQ--IFWTRWFSNLSNYIIS 989
E L ++ ++L+ P+ ++P GL+++YAL LN Q + WT +L N +IS
Sbjct: 1129 ELLSTFAFASSLVILVSAPEGVINPSLAGLAITYALNLNTLQATLIWT--LCDLENKMIS 1186
Query: 990 VERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGG 1049
VER+ Q+ +IP+EPP +++ RP SWPS+G+I + L+VRY P+ P+VL G+TCTF GG
Sbjct: 1187 VERMLQYTNIPSEPPLVIETTRPEKSWPSRGEITICNLQVRYGPHLPMVLHGLTCTFPGG 1246
Query: 1050 SRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLF 1109
+ G+VGRTG GKSTLI LFR+VEP+ G+I IDGINI S+GL DLR +LSIIPQ+PT+F
Sbjct: 1247 LKTGIVGRTGCGKSTLIQTLFRIVEPAAGEIRIDGINILSIGLHDLRSRLSIIPQDPTMF 1306
Query: 1110 KGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCL 1169
+G+IR+NLDPL Y+DD+IW+A++ CQL + + K LDS VS+ G NWS+GQRQL CL
Sbjct: 1307 EGTIRSNLDPLEEYTDDQIWEALDNCQLGDEVRKKELKLDSPVSENGQNWSVGQRQLVCL 1366
Query: 1170 GRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVL 1229
GRVLLKR+++LVLDEATASID+ATD ++Q +R F +CTVIT+AHR+ +VIDSDMV++L
Sbjct: 1367 GRVLLKRSKLLVLDEATASIDTATDNLIQETLRHHFADCTVITIAHRISSVIDSDMVLLL 1426
Query: 1230 SYGKLVEYDEPSKLMDTNSS-FSKLVAEYWSSCRKNS 1265
G + E+D P++L++ SS FSKLVAEY +S S
Sbjct: 1427 DQGLIKEHDSPARLLEDRSSLFSKLVAEYTTSSESKS 1463
Score = 85.1 bits (209), Expect = 2e-16
Identities = 93/441 (21%), Positives = 198/441 (44%), Gaps = 29/441 (6%)
Query: 824 IVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRIN-GTTKAPVMNF 882
I+ ++V + ++++ I A PA + L+ + Y A E + + +K M
Sbjct: 407 ILVLQVSLALWILYKSLGLGSIAAFPATI-LVMLANYPFAKLEEKFQSSLMKSKDNRMKK 465
Query: 883 AAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASL---FFHSNVAMEWLVLRIEALLNLTV 939
+E L + ++ +F L+L +A F +++ A+ ++ + ++ T
Sbjct: 466 TSEVLLNMKILKLQGWEMKFLSKILELRHIEAGWLKKFVYNSSAINSVLWAAPSFISATA 525
Query: 940 ITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVERIKQFIHI 999
A LLL + L G++ +L+ L G S + +S+ RI F+ +
Sbjct: 526 FGACLLLKIP----LESGKILAALATFRILQGPIYKLPETISMIVQTKVSLNRIASFLCL 581
Query: 1000 PAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPL-VLKGITCTFKGGSRVGVVGRT 1058
+V R PS S+ +++ + ++P+ L+ + G V + G
Sbjct: 582 DDLQQDVV--GRLPSG-SSEMAVEISNGTFSWDDSSPIPTLRDMNFKVSQGMNVAICGTV 638
Query: 1059 GSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLD 1118
GSGKS+L+S++ V G++ + G + + I Q P + G + N+
Sbjct: 639 GSGKSSLLSSILGEVPKISGNLKVCG-------------RKAYIAQSPWIQSGKVEENI- 684
Query: 1119 PLGLYSDDEIW-KAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRN 1177
G + E + + +E C L + + LP + + + G N S GQ+Q + R L +
Sbjct: 685 LFGKPMEREWYDRVLEACSLNKDLEILPFHDQTVIGERGINLSGGQKQRIQIARALYQDA 744
Query: 1178 RILVLDEATASIDSATDA-ILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1236
I + D+ +++D+ T + + + ++ TVI V H+V + ++D+++V+ GK+ +
Sbjct: 745 DIYLFDDPFSAVDAHTGSHLFKEVLLGLLRHKTVIYVTHQVEFLPEADLILVMKDGKITQ 804
Query: 1237 YDEPSKLMDTNSSFSKLVAEY 1257
+ +++D+ + F +LV +
Sbjct: 805 AGKYHEILDSGTDFMELVGAH 825
Score = 81.6 bits (200), Expect = 3e-15
Identities = 37/75 (49%), Positives = 51/75 (67%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
A++I +GTF WD S P L+D+N ++ +A+C VGS KSS L +ILGE+PKISG
Sbjct: 600 AVEISNGTFSWDDSSPIPTLRDMNFKVSQGMNVAICGTVGSGKSSLLSSILGEVPKISGN 659
Query: 479 VYVGGTLAYVSQSSW 493
+ V G AY++QS W
Sbjct: 660 LKVCGRKAYIAQSPW 674
>At1g04120 multi-drug resistance protein
Length = 1514
Score = 586 bits (1510), Expect = e-167
Identities = 330/725 (45%), Positives = 452/725 (61%), Gaps = 47/725 (6%)
Query: 588 VMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKVT 644
+++AL +KTV+ VTHQVEFL D IL +G++IQSG Y++LL++GT F+ LVSAH
Sbjct: 790 ILSALAEKTVVFVTHQVEFLPAADLILVLKEGRIIQSGKYDDLLQAGTDFKALVSAHHEA 849
Query: 645 INDLN--------------QNSEVLSNPQ----DSHGFYLTKNQSEGEISS--------- 677
I ++ ++S VL NP+ ++ L K EG +S
Sbjct: 850 IEAMDIPSPSSEDSDENPIRDSLVLHNPKSDVFENDIETLAKEVQEGGSASDLKAIKEKK 909
Query: 678 --IQGSIGAQLTQEEEKVIGNVGWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFW 735
+ S QL QEEE+V G V K Y+ + + L+IL Q F LQ +SN+W
Sbjct: 910 KKAKRSRKKQLVQEEERVKGKVSMKVYLSYMGAAYKGALIPLIILAQAAFQFLQIASNWW 969
Query: 736 LA-----TAIEIPKVTDTTLIGVYALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTS 790
+A T + KV T L+ VY L+ S+ F++VR+ A GL A+ F + S
Sbjct: 970 MAWANPQTEGDESKVDPTLLLIVYTALAFGSSVFIFVRAALVATFGLAAAQKLFLNMLRS 1029
Query: 791 IFNAPMLFFDSTPA-------SSDLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQV 843
+F APM FFDSTPA S D S++D DIP+ L A I++ ++ V+ +VTWQV
Sbjct: 1030 VFRAPMSFFDSTPAGRILNRVSIDQSVVDLDIPFRLGGFASTTIQLCGIVAVMTNVTWQV 1089
Query: 844 LIVAVPAMVALIFIQKYYQATARELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFF 903
++ VP VA ++QKYY A++REL+RI K+P+++ E+ G T+R F RF
Sbjct: 1090 FLLVVPVAVACFWMQKYYMASSRELVRIVSIQKSPIIHLFGESIAGAATIRGFGQEKRFI 1149
Query: 904 KNYLKLVDTDASLFFHSNVAMEWLVLRIEALLNLTVITAALLLILLPQRYLSPGRVGLSL 963
K L L+D FF S A+EWL LR+E L L +LL+ P + P GL++
Sbjct: 1150 KRNLYLLDCFVRPFFCSIAAIEWLCLRMELLSTLVFAFCMVLLVSFPHGTIDPSMAGLAV 1209
Query: 964 SYALTLNGAQIFWTRWFSNLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKID 1023
+Y L LNG W F L N IIS+ERI Q+ I E PAI+++ RPPSSWP+ G I+
Sbjct: 1210 TYGLNLNGRLSRWILSFCKLENKIISIERIYQYSQIVGEAPAIIEDFRPPSSWPATGTIE 1269
Query: 1024 LQGLEVRYRPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILID 1083
L ++VRY N P VL G++C F GG ++G+VGRTGSGKSTLI ALFRL+EP+ G I ID
Sbjct: 1270 LVDVKVRYAENLPTVLHGVSCVFPGGKKIGIVGRTGSGKSTLIQALFRLIEPTAGKITID 1329
Query: 1084 GINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISK 1143
I+I +GL DLR +L IIPQ+PTLF+G+IR NLDPL +SDD+IW+A++K QL + +
Sbjct: 1330 NIDISQIGLHDLRSRLGIIPQDPTLFEGTIRANLDPLEEHSDDKIWEALDKSQLGDVVRG 1389
Query: 1144 LPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQ 1203
LDS V + G NWS+GQRQL LGR LLK+ +ILVLDEATAS+D+ATD ++Q+IIR
Sbjct: 1390 KDLKLDSPVLENGDNWSVGQRQLVSLGRALLKQAKILVLDEATASVDTATDNLIQKIIRT 1449
Query: 1204 EFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLM-DTNSSFSKLVAEYWSSCR 1262
EFE+CTV T+AHR+PTVIDSD+V+VLS G++ E+D P++L+ D +S F KLV EY S R
Sbjct: 1450 EFEDCTVCTIAHRIPTVIDSDLVLVLSDGRVAEFDTPARLLEDKSSMFLKLVTEY--SSR 1507
Query: 1263 KNSLP 1267
+P
Sbjct: 1508 STGIP 1512
Score = 84.3 bits (207), Expect = 4e-16
Identities = 39/75 (52%), Positives = 52/75 (69%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
A++I+DG F WD S P L + ++++ ++AVC VGS KSSF+ ILGEIPKISG
Sbjct: 621 AIEIKDGVFCWDPFSSRPTLSGIQMKVEKGMRVAVCGTVGSGKSSFISCILGEIPKISGE 680
Query: 479 VYVGGTLAYVSQSSW 493
V + GT YVSQS+W
Sbjct: 681 VRICGTTGYVSQSAW 695
Score = 76.6 bits (187), Expect = 8e-14
Identities = 58/230 (25%), Positives = 102/230 (44%), Gaps = 14/230 (6%)
Query: 1039 LKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMK 1098
L GI + G RV V G GSGKS+ IS + + G++ I G
Sbjct: 640 LSGIQMKVEKGMRVAVCGTVGSGKSSFISCILGEIPKISGEVRICGTT------------ 687
Query: 1099 LSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGN 1158
+ Q + G+I N+ + ++ C LK+ I + + + G N
Sbjct: 688 -GYVSQSAWIQSGNIEENILFGSPMEKTKYKNVIQACSLKKDIELFSHGDQTIIGERGIN 746
Query: 1159 WSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQR-IIRQEFEECTVITVAHRV 1217
S GQ+Q L R L + I +LD+ +++D+ T + L R I E TV+ V H+V
Sbjct: 747 LSGGQKQRVQLARALYQDADIYLLDDPFSALDAHTGSDLFRDYILSALAEKTVVFVTHQV 806
Query: 1218 PTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEYWSSCRKNSLP 1267
+ +D+++VL G++++ + L+ + F LV+ + + +P
Sbjct: 807 EFLPAADLILVLKEGRIIQSGKYDDLLQAGTDFKALVSAHHEAIEAMDIP 856
>At3g13100 ABC transporter, putative
Length = 1248
Score = 583 bits (1504), Expect = e-166
Identities = 307/687 (44%), Positives = 454/687 (65%), Gaps = 20/687 (2%)
Query: 592 LRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKVTINDL 648
LR+KTVI VTHQ+EFL E D IL DG++ Q+G Y +L+SGT F LV AH + +
Sbjct: 551 LRNKTVIYVTHQLEFLPEADLILVMKDGRITQAGKYNEILESGTDFMELVGAHTDALAAV 610
Query: 649 NQNSEVLSNPQDSHGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVIGNVGWKPLWDYINY 708
+ + ++ Q + + E + S QL QEEE+ G VG+ Y+
Sbjct: 611 DSYEKGSASAQSTTSKESKVSNDEEKQEEDLPSPKGQLVQEEEREKGKVGFTVYQKYMKL 670
Query: 709 SNGTLMSCLVILGQCCFLALQTSSNFWLATAIEI-----PKVTDTTLIGVYALLSISSTS 763
+ G + ++++ Q F L SN+W+A + P V+ +TLI VY L+ +S+
Sbjct: 671 AYGGALVPIILVVQILFQVLNIGSNYWMAWVTPVSKDVKPLVSGSTLILVYVFLATASSF 730
Query: 764 FVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP-------ASSDLSILDFDIP 816
+ VR+ +A+ G K +T F+ IF A M FFD+TP AS+D S +D +P
Sbjct: 731 CILVRAMLSAMTGFKIATELFNQMHFRIFRASMSFFDATPIGRILNRASTDQSAVDLRLP 790
Query: 817 YSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRINGTTK 876
+ +AI A+ +L +I V+ V WQVLIV +P + A + ++YY + AREL R++G ++
Sbjct: 791 SQFSNLAIAAVNILGIIGVMGQVAWQVLIVFIPVIAACTWYRQYYISAARELARLSGISR 850
Query: 877 APVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRIEALLN 936
+P++ +ET G+ T+R+F+ RF + ++L D + L FH+ AMEWL R++ L
Sbjct: 851 SPLVQHFSETLSGITTIRSFDQEPRFRTDIMRLNDCYSRLRFHAISAMEWLCFRLDLLST 910
Query: 937 LTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQ--IFWTRWFSNLSNYIISVERIK 994
+ + ++L+ +P+ ++P GL+++YAL LN Q + WT +L N +ISVER+
Sbjct: 911 VAFALSLVILVSVPEGVINPSFAGLAVTYALNLNSLQATLIWT--LCDLENKMISVERML 968
Query: 995 QFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRVGV 1054
Q+I IP+EP ++++ RP SWP +G+I + L+VRY P+ P+VL+G+TCTF+GG + G+
Sbjct: 969 QYIDIPSEPSLVIESTRPEKSWPCRGEITICNLQVRYGPHLPMVLRGLTCTFRGGLKTGI 1028
Query: 1055 VGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIR 1114
VGRTG GKSTLI LFR+VEP+ G+I IDGINI ++GL DLR +LSIIPQEPT+F+G++R
Sbjct: 1029 VGRTGCGKSTLIQTLFRIVEPAAGEIRIDGINILTIGLHDLRSRLSIIPQEPTMFEGTVR 1088
Query: 1115 TNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLL 1174
+NLDPL Y+DD+IW+A++KCQL + I K LDS VS+ G NWS+GQRQL CLGRVLL
Sbjct: 1089 SNLDPLEEYADDQIWEALDKCQLGDEIRKKELKLDSPVSENGQNWSVGQRQLVCLGRVLL 1148
Query: 1175 KRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKL 1234
KR+++L+LDEATAS+D+ATD ++Q +RQ F CTVIT+AHR+ +VIDSDMV++L G +
Sbjct: 1149 KRSKVLILDEATASVDTATDTLIQETLRQHFSGCTVITIAHRISSVIDSDMVLLLDQGLI 1208
Query: 1235 VEYDEPSKLM-DTNSSFSKLVAEYWSS 1260
E+D P++L+ D +SSFSKLVAEY +S
Sbjct: 1209 EEHDSPARLLEDKSSSFSKLVAEYTAS 1235
Score = 77.8 bits (190), Expect = 4e-14
Identities = 36/74 (48%), Positives = 48/74 (64%)
Query: 420 LQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGTV 479
+++ +G F WD S P LKD+ +I IA+C VGS KSS L +ILGE+PKISG +
Sbjct: 379 VEVSNGAFSWDDSSPIPTLKDIRFKIPHGMNIAICGTVGSGKSSLLSSILGEVPKISGNL 438
Query: 480 YVGGTLAYVSQSSW 493
V G AY++QS W
Sbjct: 439 KVCGRKAYIAQSPW 452
Score = 72.8 bits (177), Expect = 1e-12
Identities = 51/245 (20%), Positives = 116/245 (46%), Gaps = 15/245 (6%)
Query: 1015 SWPSKGKIDLQGLEVRYRPNAPL-VLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLV 1073
S SK +++ + ++P+ LK I G + + G GSGKS+L+S++ V
Sbjct: 372 SGSSKMDVEVSNGAFSWDDSSPIPTLKDIRFKIPHGMNIAICGTVGSGKSSLLSSILGEV 431
Query: 1074 EPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVE 1133
G++ + G + + I Q P + G + N+ + + +E
Sbjct: 432 PKISGNLKVCG-------------RKAYIAQSPWIQSGKVEENILFGKPMQREWYQRVLE 478
Query: 1134 KCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSAT 1193
C L + + P + + + G N S GQ+Q + R L + I + D+ +++D+ T
Sbjct: 479 ACSLNKDLEVFPFRDQTVIGERGINLSGGQKQRIQIARALYQDADIYLFDDPFSAVDAHT 538
Query: 1194 DA-ILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSK 1252
+ + + ++ TVI V H++ + ++D+++V+ G++ + + ++++++ + F +
Sbjct: 539 GSHLFKEVLLGLLRNKTVIYVTHQLEFLPEADLILVMKDGRITQAGKYNEILESGTDFME 598
Query: 1253 LVAEY 1257
LV +
Sbjct: 599 LVGAH 603
>At3g60160 multi resistance protein homolog
Length = 1490
Score = 565 bits (1455), Expect = e-161
Identities = 319/710 (44%), Positives = 438/710 (60%), Gaps = 58/710 (8%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKV 643
C+M L+DKTV+ VTHQVEFL D IL +G+V+Q+G +E LLK FE+LV AH
Sbjct: 797 CLMGILKDKTVLYVTHQVEFLPAADLILVMQNGRVMQAGKFEELLKQNIGFEVLVGAHNE 856
Query: 644 TINDLNQNSEVLSNPQDSHGF--------------YLTKNQSEGEISSIQGSIGAQLTQE 689
++ +LS + S F T SE IS+ A+L Q+
Sbjct: 857 ALDS------ILSIEKSSRNFKEGSKDDTASIAESLQTHCDSEHNISTENKKKEAKLVQD 910
Query: 690 EEKVIGNVGWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLA-----TAIEIPK 744
EE G +G + Y+ G L+ +IL Q CF LQ +SN+W+A TA IPK
Sbjct: 911 EETEKGVIGKEVYLAYLTTVKGGLLVPFIILAQSCFQMLQIASNYWMAWTAPPTAESIPK 970
Query: 745 VTDTTLIGVYALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP- 803
+ ++ VYALL+ S+ V R+ A+ GL + FFS SIF APM FFDSTP
Sbjct: 971 LGMGRILLVYALLAAGSSLCVLARTILVAIGGLSTAETFFSRMLCSIFRAPMSFFDSTPT 1030
Query: 804 ------ASSDLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFI 857
AS+D S+LD ++ L A I+++ IFV++ V WQ
Sbjct: 1031 GRILNRASTDQSVLDLEMAVKLGWCAFSIIQIVGTIFVMSQVAWQ--------------- 1075
Query: 858 QKYYQATARELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLF 917
+YY TAREL R++G +AP+++ AE+ G T+RAF+ DRF + L L+D+ + +
Sbjct: 1076 -RYYTPTARELSRMSGVERAPILHHFAESLAGATTIRAFDQRDRFISSNLVLIDSHSRPW 1134
Query: 918 FHSNVAMEWLVLRIEALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQ--IF 975
FH AMEWL R+ L + + +LL+ LP+ ++P GL ++Y L+LN Q +
Sbjct: 1135 FHVASAMEWLSFRLNLLSHFVFAFSLVLLVTLPEGVINPSIAGLGVTYGLSLNVLQATVI 1194
Query: 976 WTRWFSNLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNA 1035
W N N +ISVERI Q+ IP+E P ++D +RP +WP+ G I + L+VRY +
Sbjct: 1195 WN--ICNAENKMISVERILQYSKIPSEAPLVIDGHRPLDNWPNVGSIVFRDLQVRYAEHF 1252
Query: 1036 PLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDL 1095
P VLK ITC F GG ++GVVGRTGSGKSTLI ALFR+VEPS+G I+ID ++I +GL DL
Sbjct: 1253 PAVLKNITCEFPGGKKIGVVGRTGSGKSTLIQALFRIVEPSQGTIVIDNVDITKIGLHDL 1312
Query: 1096 RMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDE 1155
R +L IIPQ+P LF G+IR NLDPL Y+D EIW+A++KCQL + I LD++V +
Sbjct: 1313 RSRLGIIPQDPALFDGTIRLNLDPLAQYTDHEIWEAIDKCQLGDVIRAKDERLDATVVEN 1372
Query: 1156 GGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAH 1215
G NWS+GQRQL CLGRVLLK++ ILVLDEATAS+DSATD ++Q+II QEF++ TV+T+AH
Sbjct: 1373 GENWSVGQRQLVCLGRVLLKKSNILVLDEATASVDSATDGVIQKIINQEFKDRTVVTIAH 1432
Query: 1216 RVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSS-FSKLVAEYWSSCRKN 1264
R+ TVI+SD+V+VLS G++ E+D P+KL+ S FSKL+ EY S R N
Sbjct: 1433 RIHTVIESDLVLVLSDGRIAEFDSPAKLLQREDSFFSKLIKEY--SLRSN 1480
Score = 85.9 bits (211), Expect = 1e-16
Identities = 57/227 (25%), Positives = 110/227 (48%), Gaps = 28/227 (12%)
Query: 1039 LKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMK 1098
L I K G +V V G GSGKS+L+S++ ++ +G + + G K
Sbjct: 648 LDDIELKVKSGMKVAVCGAVGSGKSSLLSSILGEIQKLKGTVRVSG-------------K 694
Query: 1099 LSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGN 1158
+ +PQ P + G+IR N+ +Y ++ + V+ C L + + + + + G N
Sbjct: 695 QAYVPQSPWILSGTIRDNILFGSMYESEKYERTVKACALIKDFELFSNGDLTEIGERGIN 754
Query: 1159 WSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEEC--------TV 1210
S GQ+Q + R + + I +LD+ +++D+ T R+ FE+C TV
Sbjct: 755 MSGGQKQRIQIARAVYQNADIYLLDDPFSAVDAHTG-------RELFEDCLMGILKDKTV 807
Query: 1211 ITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEY 1257
+ V H+V + +D+++V+ G++++ + +L+ N F LV +
Sbjct: 808 LYVTHQVEFLPAADLILVMQNGRVMQAGKFEELLKQNIGFEVLVGAH 854
Score = 82.0 bits (201), Expect = 2e-15
Identities = 43/84 (51%), Positives = 55/84 (65%), Gaps = 3/84 (3%)
Query: 410 KNLTQCSVNALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAIL 469
K+ T+ SV +I++G F W+ ES P L D+ L++K K+AVC VGS KSS L +IL
Sbjct: 623 KDHTELSV---EIENGAFSWEPESSRPTLDDIELKVKSGMKVAVCGAVGSGKSSLLSSIL 679
Query: 470 GEIPKISGTVYVGGTLAYVSQSSW 493
GEI K+ GTV V G AYV QS W
Sbjct: 680 GEIQKLKGTVRVSGKQAYVPQSPW 703
Score = 33.1 bits (74), Expect = 1.0
Identities = 19/77 (24%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 584 IQYCVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLL-KSGTAFELLVS 639
IQ + +D+TV+ + H++ + E D +L DG++ + S LL + + F L+
Sbjct: 1414 IQKIINQEFKDRTVVTIAHRIHTVIESDLVLVLSDGRIAEFDSPAKLLQREDSFFSKLIK 1473
Query: 640 AHKVTINDLNQNSEVLS 656
+ + N ++++LS
Sbjct: 1474 EYSLRSNHFAGSNDLLS 1490
>At3g62700 ABC transporter-like protein
Length = 1539
Score = 558 bits (1437), Expect = e-159
Identities = 308/728 (42%), Positives = 445/728 (60%), Gaps = 58/728 (7%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKV 643
CV AL+ KT++LVTHQV+FL VD IL DG ++QSG Y+ L+ SG F LV+AH+
Sbjct: 806 CVRGALKGKTILLVTHQVDFLHNVDRILVMRDGMIVQSGKYDELVSSGLDFGELVAAHET 865
Query: 644 TI------------------NDLNQNSEVLSNPQDSHGFYLTKNQS-------------- 671
++ + + Q S + +P+ + + S
Sbjct: 866 SMELVEAGSASATAANVPMASPITQRSISIESPRQPKSPKVHRTTSMESPRVLRTTSMES 925
Query: 672 -------EGEISSIQGSI----GAQLTQEEEKVIGNVGWKPLWDYINYSNGTLMSCLVIL 720
+ I S GS G++L +EEE+ +G V ++ Y + G LV+
Sbjct: 926 PRLSELNDESIKSFLGSNIPEDGSRLIKEEEREVGQVSFQVYKLYSTEAYGWWGMILVVF 985
Query: 721 GQCCFLALQTSSNFWLA---TAIEIPKVTDTTLIGVYALLSISSTSFVYVRSYFAALLGL 777
+ A +S++WLA +A T I VY +++ S V +R+++ LGL
Sbjct: 986 FSVAWQASLMASDYWLAYETSAKNEVSFDATVFIRVYVIIAAVSIVLVCLRAFYVTHLGL 1045
Query: 778 KASTAFFSSFTTSIFNAPMLFFDSTP-------ASSDLSILDFDIPYSLTCVAIVAIEVL 830
K + FF S+ +APM FFD+TP AS+D + +D IP+ + VA + +L
Sbjct: 1046 KTAQIFFKQILNSLVHAPMSFFDTTPSGRILSRASTDQTNVDIFIPFMIGLVATMYTTLL 1105
Query: 831 VMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRINGTTKAPVMNFAAETHLGV 890
+ V W + +P I+ + YY A++REL R++ TKAPV++ +E+ GV
Sbjct: 1106 SIFIVTCQYAWPTVFFIIPLGWLNIWYRGYYLASSRELTRLDSITKAPVIHHFSESIAGV 1165
Query: 891 VTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRIEALLNLTVITAALLLILLP 950
+T+RAF F + +K V+ + + FH+N + EWL R+E + + + +AL +++LP
Sbjct: 1166 MTIRAFKKQPMFRQENVKRVNANLRMDFHNNGSNEWLGFRLELIGSWVLCISALFMVMLP 1225
Query: 951 QRYLSPGRVGLSLSYALTLNGAQIFWTRWFSN-LSNYIISVERIKQFIHIPAEPPAIVDN 1009
+ P VGLSLSY L+LNG +FW + S + N ++SVERIKQF IPAE +
Sbjct: 1226 SNIIKPENVGLSLSYGLSLNGV-LFWAIYLSCFIENKMVSVERIKQFTDIPAEAKWEIKE 1284
Query: 1010 NRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISAL 1069
+RPP +WP KG I L+ ++VRYRPN PLVLKG+T KGG ++GVVGRTGSGKSTLI L
Sbjct: 1285 SRPPPNWPYKGNIRLEDVKVRYRPNTPLVLKGLTIDIKGGEKIGVVGRTGSGKSTLIQVL 1344
Query: 1070 FRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIW 1129
FRLVEPS G I+IDGI+IC++GL DLR + IIPQEP LF+G++R+N+DP YSD+EIW
Sbjct: 1345 FRLVEPSGGKIIIDGIDICTLGLHDLRSRFGIIPQEPVLFEGTVRSNIDPTEKYSDEEIW 1404
Query: 1130 KAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASI 1189
K++E+CQLK+ ++ P LDS V+D G NWS+GQRQL CLGRV+LKR+RIL LDEATAS+
Sbjct: 1405 KSLERCQLKDVVASKPEKLDSLVADNGENWSVGQRQLLCLGRVMLKRSRILFLDEATASV 1464
Query: 1190 DSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSS 1249
DS TDA++Q+IIR++F +CT+I++AHR+PTV+D D V+V+ GK EYD P +L++ S
Sbjct: 1465 DSQTDAMIQKIIREDFSDCTIISIAHRIPTVMDCDRVLVIDAGKAKEYDSPVRLLERQSL 1524
Query: 1250 FSKLVAEY 1257
F+ LV EY
Sbjct: 1525 FAALVQEY 1532
Score = 79.3 bits (194), Expect = 1e-14
Identities = 37/75 (49%), Positives = 54/75 (71%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
A++I+DG+F WD E PA++++N E+K + A+ VGS KSS L ++LGE+ K+SG
Sbjct: 638 AVEIKDGSFSWDDEDDEPAIENINFEVKKGELAAIVGTVGSGKSSLLASVLGEMHKLSGK 697
Query: 479 VYVGGTLAYVSQSSW 493
V V GT AYV+Q+SW
Sbjct: 698 VRVCGTTAYVAQTSW 712
Score = 67.0 bits (162), Expect = 7e-11
Identities = 49/226 (21%), Positives = 110/226 (47%), Gaps = 20/226 (8%)
Query: 1039 LKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMK 1098
++ I K G +VG GSGKS+L++++ + G + + G
Sbjct: 657 IENINFEVKKGELAAIVGTVGSGKSSLLASVLGEMHKLSGKVRVCGTT------------ 704
Query: 1099 LSIIPQEPTLFKGSIRTNLD---PLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDE 1155
+ + Q + G+++ N+ P+ +E+ K C L++ + + + + +
Sbjct: 705 -AYVAQTSWIQNGTVQDNILFGLPMNRSKYNEVLKV---CCLEKDMQIMEFGDQTEIGER 760
Query: 1156 GGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDA-ILQRIIRQEFEECTVITVA 1214
G N S GQ+Q L R + + + + +LD+ +++D+ T + I ++ +R + T++ V
Sbjct: 761 GINLSGGQKQRIQLARAVYQESDVYLLDDVFSAVDAHTGSDIFKKCVRGALKGKTILLVT 820
Query: 1215 HRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEYWSS 1260
H+V + + D ++V+ G +V+ + +L+ + F +LVA + +S
Sbjct: 821 HQVDFLHNVDRILVMRDGMIVQSGKYDELVSSGLDFGELVAAHETS 866
>At2g47800 MRP-like ABC transporter
Length = 1516
Score = 545 bits (1405), Expect = e-155
Identities = 303/702 (43%), Positives = 428/702 (60%), Gaps = 31/702 (4%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHK- 642
CV AL+ KTV+LVTHQV+FL VD IL DGK+++SG Y+ L+ SG F LV+AH+
Sbjct: 808 CVRGALKGKTVLLVTHQVDFLHNVDCILVMRDGKIVESGKYDELVSSGLDFGELVAAHET 867
Query: 643 -------------VTINDLNQNSEVLSNPQDSHGFYLTKNQSEGEISSIQGSI----GAQ 685
V + S S+P+ S + ++ I S GS G++
Sbjct: 868 SMELVEAGADSAAVATSPRTPTSPHASSPRTSMESPHLSDLNDEHIKSFLGSHIVEDGSK 927
Query: 686 LTQEEEKVIGNVGWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLA---TAIEI 742
L +EEE+ G V Y + G LV+ + +S++WLA +A
Sbjct: 928 LIKEEERETGQVSLGVYKQYCTEAYGWWGIVLVLFFSLTWQGSLMASDYWLAYETSAKNA 987
Query: 743 PKVTDTTLIGVYALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDST 802
+ I Y ++++ S V +RSY+ LGLK + FF SI +APM FFD+T
Sbjct: 988 ISFDASVFILGYVIIALVSIVLVSIRSYYVTHLGLKTAQIFFRQILNSILHAPMSFFDTT 1047
Query: 803 P-------ASSDLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALI 855
P AS+D + +D IP+ L V + +L + V W +P I
Sbjct: 1048 PSGRILSRASTDQTNVDILIPFMLGLVVSMYTTLLSIFIVTCQYAWPTAFFVIPLGWLNI 1107
Query: 856 FIQKYYQATARELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDAS 915
+ + YY A++REL R++ TKAP+++ +E+ GV+T+R+F + F + +K V+ +
Sbjct: 1108 WYRNYYLASSRELTRMDSITKAPIIHHFSESIAGVMTIRSFRKQELFRQENVKRVNDNLR 1167
Query: 916 LFFHSNVAMEWLVLRIEALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIF 975
+ FH+N + EWL R+E + + + +AL ++LLP + P VGLSLSY L+LN F
Sbjct: 1168 MDFHNNGSNEWLGFRLELVGSWVLCISALFMVLLPSNVIRPENVGLSLSYGLSLNSVLFF 1227
Query: 976 WTRWFSNLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNA 1035
+ N ++SVERIKQF IP+E PPS+WP G + L+ L+VRYRPN
Sbjct: 1228 AIYMSCFVENKMVSVERIKQFTDIPSESEWERKETLPPSNWPFHGNVHLEDLKVRYRPNT 1287
Query: 1036 PLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDL 1095
PLVLKGIT KGG +VGVVGRTGSGKSTLI LFRLVEPS G I+IDGI+I ++GL DL
Sbjct: 1288 PLVLKGITLDIKGGEKVGVVGRTGSGKSTLIQVLFRLVEPSGGKIIIDGIDISTLGLHDL 1347
Query: 1096 RMKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDE 1155
R + IIPQEP LF+G++R+N+DP YSD+EIWK++E+CQLK+ ++ P LDS V D
Sbjct: 1348 RSRFGIIPQEPVLFEGTVRSNIDPTEQYSDEEIWKSLERCQLKDVVATKPEKLDSLVVDN 1407
Query: 1156 GGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAH 1215
G NWS+GQRQL CLGRV+LKR+R+L LDEATAS+DS TDA++Q+IIR++F CT+I++AH
Sbjct: 1408 GENWSVGQRQLLCLGRVMLKRSRLLFLDEATASVDSQTDAVIQKIIREDFASCTIISIAH 1467
Query: 1216 RVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEY 1257
R+PTV+D D V+V+ GK E+D P++L++ S F+ LV EY
Sbjct: 1468 RIPTVMDGDRVLVIDAGKAKEFDSPARLLERPSLFAALVQEY 1509
Score = 76.6 bits (187), Expect = 8e-14
Identities = 35/75 (46%), Positives = 52/75 (68%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
A++++DG+F WD E PAL D+N ++K + A+ VGS KSS L ++LGE+ +ISG
Sbjct: 640 AVEVRDGSFSWDDEDNEPALSDINFKVKKGELTAIVGTVGSGKSSLLASVLGEMHRISGQ 699
Query: 479 VYVGGTLAYVSQSSW 493
V V G+ YV+Q+SW
Sbjct: 700 VRVCGSTGYVAQTSW 714
Score = 70.9 bits (172), Expect = 5e-12
Identities = 54/224 (24%), Positives = 108/224 (48%), Gaps = 16/224 (7%)
Query: 1039 LKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMK 1098
L I K G +VG GSGKS+L++++ + G + + G
Sbjct: 659 LSDINFKVKKGELTAIVGTVGSGKSSLLASVLGEMHRISGQVRVCG-------------S 705
Query: 1099 LSIIPQEPTLFKGSIRTNLDPLGLYSDDEIW-KAVEKCQLKETISKLPSLLDSSVSDEGG 1157
+ Q + G+++ N+ GL E + K + C L++ + + + + + G
Sbjct: 706 TGYVAQTSWIENGTVQDNI-LFGLPMVREKYNKVLNVCSLEKDLQMMEFGDKTEIGERGI 764
Query: 1158 NWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDA-ILQRIIRQEFEECTVITVAHR 1216
N S GQ+Q L R + + + +LD+ +++D+ T + I ++ +R + TV+ V H+
Sbjct: 765 NLSGGQKQRIQLARAVYQECDVYLLDDVFSAVDAHTGSDIFKKCVRGALKGKTVLLVTHQ 824
Query: 1217 VPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLVAEYWSS 1260
V + + D ++V+ GK+VE + +L+ + F +LVA + +S
Sbjct: 825 VDFLHNVDCILVMRDGKIVESGKYDELVSSGLDFGELVAAHETS 868
>At3g60970 ABC transporter-like protein
Length = 1037
Score = 545 bits (1404), Expect = e-155
Identities = 309/696 (44%), Positives = 424/696 (60%), Gaps = 67/696 (9%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKV 643
C+M L+DKTV+ VTHQVEFL D IL +G+V+Q+G +E LLK FE+L
Sbjct: 381 CLMGILKDKTVLYVTHQVEFLPAADLILVMQNGRVMQAGKFEELLKQNIGFEVL------ 434
Query: 644 TINDLNQNSEVLSNPQDSHGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVIGNVGWKPLW 703
T+ SE IS+ A+L Q+EE G +G +
Sbjct: 435 -----------------------TQCDSEHNISTENKKKEAKLVQDEETEKGVIGKEVYL 471
Query: 704 DYINYSNGTLMSCLVILGQCCFLALQTSSNFWLA-----TAIEIPKVTDTTLIGVYALLS 758
Y+ G L+ +IL Q CF LQ +SN+W+A TA IPK+ ++ VYALL+
Sbjct: 472 TYLTTVKGGLLVPFIILAQSCFQMLQIASNYWMAWTAPPTAESIPKLGMGRILLVYALLA 531
Query: 759 ISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTP-------ASSDLSIL 811
S+ V R+ A+ GL + FFS SIF APM +FDSTP AS+D S+L
Sbjct: 532 AGSSLCVLARTILVAIGGLSTAETFFSRMLCSIFRAPMSYFDSTPTGRILNRASTDQSVL 591
Query: 812 DFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRI 871
D ++ L A I+++ IFV++ V WQ +YY T REL R+
Sbjct: 592 DLEMAVKLGWCAFSIIQIVGTIFVMSQVAWQ----------------RYYTPTERELSRM 635
Query: 872 NGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRI 931
+G +AP+++ AE+ G T+RAF+ DRF + L L+D+ + +FH AMEWL R+
Sbjct: 636 SGVERAPILHHFAESLAGATTIRAFDQRDRFISSNLVLIDSHSRPWFHVASAMEWLSFRL 695
Query: 932 EALLNLTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQ--IFWTRWFSNLSNYIIS 989
L + + +LL+ LP+ ++P GL ++Y L+LN Q + W N N +IS
Sbjct: 696 NLLSHFVFAFSLVLLVTLPEGVINPSIAGLGVTYGLSLNVLQATVIWN--ICNAENKMIS 753
Query: 990 VERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGG 1049
VERI Q IP+E P ++D+ RP +WP+ G I + L+VRY + P VLK ITC F GG
Sbjct: 754 VERILQHSKIPSEAPLVIDDQRPLDNWPNVGSIVFRDLQVRYAEHFPAVLKNITCAFPGG 813
Query: 1050 SRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLF 1109
++GVVGRTGSGKSTLI ALFR+VEPS G I+ID ++I +GL DLR +L IIPQ+ LF
Sbjct: 814 KKIGVVGRTGSGKSTLIQALFRIVEPSHGTIVIDNVDITKIGLHDLRSRLGIIPQDNALF 873
Query: 1110 KGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCL 1169
G+IR NLDPL Y+D EIW+A++KCQL + I LD++V + G NWS+GQRQL CL
Sbjct: 874 DGTIRLNLDPLAQYTDREIWEALDKCQLGDVIRAKDEKLDATVVENGENWSVGQRQLVCL 933
Query: 1170 GRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVL 1229
GRVLLK++ ILVLDEATAS+DSATD ++Q+II QEF++ TV+T+AHR+ TVI+SD+V+VL
Sbjct: 934 GRVLLKKSNILVLDEATASVDSATDGVIQKIINQEFKDRTVVTIAHRIHTVIESDLVLVL 993
Query: 1230 SYGKLVEYDEPSKLMDTNSS-FSKLVAEYWSSCRKN 1264
S G++ E+D P+KL+ S FSKL+ EY S R N
Sbjct: 994 SDGRIAEFDSPAKLLQREDSFFSKLIKEY--SLRSN 1027
Score = 81.6 bits (200), Expect = 3e-15
Identities = 55/223 (24%), Positives = 107/223 (47%), Gaps = 28/223 (12%)
Query: 1039 LKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMK 1098
L I K G +V + G GSGKS+L S++ ++ +G + + G K
Sbjct: 232 LDDIELKVKSGMKVAICGAVGSGKSSLPSSILGEIQKLKGTVRVSG-------------K 278
Query: 1099 LSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGN 1158
+ +PQ P + G+IR N+ +Y ++ + V+ C L + + + + + G N
Sbjct: 279 QAYVPQSPWILSGTIRDNILFGSIYESEKYERTVKACALIKDFELFSNGDLTEIGERGIN 338
Query: 1159 WSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEEC--------TV 1210
S GQ+Q + R + + I +LD+ +++D+ T R+ FE+C TV
Sbjct: 339 MSGGQKQRIQIARAVYQNADIYLLDDPFSAVDAHTG-------RELFEDCLMGILKDKTV 391
Query: 1211 ITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKL 1253
+ V H+V + +D+++V+ G++++ + +L+ N F L
Sbjct: 392 LYVTHQVEFLPAADLILVMQNGRVMQAGKFEELLKQNIGFEVL 434
Score = 78.6 bits (192), Expect = 2e-14
Identities = 37/75 (49%), Positives = 50/75 (66%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
+++I++G F W+ ES P L D+ L++K K+A+C VGS KSS +ILGEI K+ GT
Sbjct: 213 SVEIENGAFSWEPESSRPTLDDIELKVKSGMKVAICGAVGSGKSSLPSSILGEIQKLKGT 272
Query: 479 VYVGGTLAYVSQSSW 493
V V G AYV QS W
Sbjct: 273 VRVSGKQAYVPQSPW 287
Score = 33.1 bits (74), Expect = 1.0
Identities = 19/77 (24%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 584 IQYCVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLL-KSGTAFELLVS 639
IQ + +D+TV+ + H++ + E D +L DG++ + S LL + + F L+
Sbjct: 961 IQKIINQEFKDRTVVTIAHRIHTVIESDLVLVLSDGRIAEFDSPAKLLQREDSFFSKLIK 1020
Query: 640 AHKVTINDLNQNSEVLS 656
+ + N ++++LS
Sbjct: 1021 EYSLRSNHFAGSNDLLS 1037
>At1g30400 AtMRP1
Length = 1622
Score = 405 bits (1041), Expect = e-113
Identities = 254/694 (36%), Positives = 385/694 (54%), Gaps = 31/694 (4%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFE-LLVSAHK 642
C+ L T +LVT+Q+ FLS+VD IL +G V + G+YE L SG F+ L+ +A K
Sbjct: 782 CIKRELGQTTRVLVTNQLHFLSQVDKILLVHEGTVKEEGTYEELCHSGPLFQRLMENAGK 841
Query: 643 VT-INDLNQNSEV----LSNPQDSHGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVIGNV 697
V ++ N +EV + ++ + L K+ E + S S+ L + EE+ G V
Sbjct: 842 VEDYSEENGEAEVDQTSVKPVENGNANNLQKDGIETKNSKEGNSV---LVKREERETGVV 898
Query: 698 GWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLA--TAIEIPKVTDTTLIG-VY 754
WK L Y N G + ++++ + SS+ WL+ T PK VY
Sbjct: 899 SWKVLERYQNALGGAWVVMMLVICYVLTQVFRVSSSTWLSEWTDSGTPKTHGPLFYNIVY 958
Query: 755 ALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTPA-------SSD 807
ALLS S + SY+ + L A+ + SI APM+FF + P + D
Sbjct: 959 ALLSFGQVSVTLINSYWLIMSSLYAAKKMHDAMLGSILRAPMVFFQTNPLGRIINRFAKD 1018
Query: 808 LSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARE 867
+ +D + + ++L + +I V+ L +P +V YYQ T+RE
Sbjct: 1019 MGDIDRTVAVFVNMFMGSIAQLLSTVILIGIVSTLSLWAIMPLLVVFYGAYLYYQNTSRE 1078
Query: 868 LIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWL 927
+ R++ TT++PV E G+ ++RA+ DR + + +D + + A WL
Sbjct: 1079 IKRMDSTTRSPVYAQFGEALNGLSSIRAYKAYDRMAEINGRSMDNNIRFTLVNMAANRWL 1138
Query: 928 VLRIEALLNLTVITAALLLILL------PQRYLSPGRVGLSLSYALTLNGAQIFWTRWFS 981
+R+E L L V A L ++ Q Y S +GL LSYAL++ + R S
Sbjct: 1139 GIRLEVLGGLMVWLTASLAVMQNGKAANQQAYAST--MGLLLSYALSITSSLTAVLRLAS 1196
Query: 982 NLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKG 1041
N + SVER+ +I IP+E P +++NNRPP WPS G I + + +RYRP P VL G
Sbjct: 1197 LAENSLNSVERVGNYIEIPSEAPLVIENNRPPPGWPSSGSIKFEDVVLRYRPELPPVLHG 1256
Query: 1042 ITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSI 1101
++ +VG+VGRTG+GKS+L++ALFR+VE +G ILID +I GL DLR L I
Sbjct: 1257 VSFLISPMDKVGIVGRTGAGKSSLLNALFRIVELEKGRILIDECDIGRFGLMDLRKVLGI 1316
Query: 1102 IPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSL 1161
IPQ P LF G++R NLDP ++D ++W+++E+ LK+TI + P LD+ V++ G N+S+
Sbjct: 1317 IPQAPVLFSGTVRFNLDPFSEHNDADLWESLERAHLKDTIRRNPLGLDAEVTEAGENFSV 1376
Query: 1162 GQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVI 1221
GQRQL L R LL+R++ILVLDEATA++D TD ++Q+ IR+EF+ CT++ +AHR+ T+I
Sbjct: 1377 GQRQLLSLARALLRRSKILVLDEATAAVDVRTDVLIQKTIREEFKSCTMLIIAHRLNTII 1436
Query: 1222 DSDMVMVLSYGKLVEYDEPSKLMDT-NSSFSKLV 1254
D D V+VL GK+ E+ P L+ SSFSK+V
Sbjct: 1437 DCDKVLVLDSGKVQEFSSPENLLSNGESSFSKMV 1470
Score = 79.7 bits (195), Expect = 1e-14
Identities = 96/443 (21%), Positives = 189/443 (41%), Gaps = 46/443 (10%)
Query: 824 IVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRINGTTKAPVMNFA 883
IVA+ +L +AS+ + +V + + + I K + T L R + + +MN
Sbjct: 428 IVALVLLYQQLGVASIIGALFLVLM-FPIQTVIISKTQKLTKEGLQRTD--KRIGLMN-- 482
Query: 884 AETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRIEALLNLTVITAA 943
E + TV+ + + F + D + S F + + + + + ++ L + +
Sbjct: 483 -EVLAAMDTVKCYAWENSFQSKVQTVRDDELSWFRKAQLLSAFNMFILNSIPVLVTVVSF 541
Query: 944 LLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVERIKQFIHI---- 999
+ LL L+P R SLS L + + N +S+ R+++ +
Sbjct: 542 GVFSLLGGD-LTPARAFTSLSLFSVLRFPLFMLPNIITQMVNANVSLNRLEEVLSTEERV 600
Query: 1000 -----PAEP--PAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGSRV 1052
P EP PAI N SW SK RP L I GS V
Sbjct: 601 LLPNPPIEPGQPAISIRNGY-FSWDSKAD----------RPT----LSNINLDIPLGSLV 645
Query: 1053 GVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGS 1112
VVG TG GK++LISA+ + P+R D + LR ++ +PQ +F +
Sbjct: 646 AVVGSTGEGKTSLISAMLGEL-PARSDATV-----------TLRGSVAYVPQVSWIFNAT 693
Query: 1113 IRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRV 1172
+R N+ + ++ + ++ L+ + LP + + + G N S GQ+Q + R
Sbjct: 694 VRDNILFGAPFDQEKYERVIDVTALQHDLELLPGGDLTEIGERGVNISGGQKQRVSMARA 753
Query: 1173 LLKRNRILVLDEATASIDS-ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSY 1231
+ + + +LD+ +++D+ + ++ I++E + T + V +++ + D ++++
Sbjct: 754 VYSNSDVCILDDPLSALDAHVGQQVFEKCIKRELGQTTRVLVTNQLHFLSQVDKILLVHE 813
Query: 1232 GKLVEYDEPSKLMDTNSSFSKLV 1254
G + E +L + F +L+
Sbjct: 814 GTVKEEGTYEELCHSGPLFQRLM 836
Score = 60.1 bits (144), Expect = 8e-09
Identities = 30/76 (39%), Positives = 47/76 (61%), Gaps = 1/76 (1%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIP-KISG 477
A+ I++G F WD ++ P L ++NL+I +AV G K+S + A+LGE+P +
Sbjct: 613 AISIRNGYFSWDSKADRPTLSNINLDIPLGSLVAVVGSTGEGKTSLISAMLGELPARSDA 672
Query: 478 TVYVGGTLAYVSQSSW 493
TV + G++AYV Q SW
Sbjct: 673 TVTLRGSVAYVPQVSW 688
>At2g34660 MRP-like ABC transporter
Length = 1623
Score = 386 bits (992), Expect = e-107
Identities = 240/694 (34%), Positives = 376/694 (53%), Gaps = 26/694 (3%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFE-LLVSAHK 642
C+ L KT +LVT+Q+ FLS+VD I+ +G V + G+YE L +G F+ L+ +A K
Sbjct: 782 CIKRELGQKTRVLVTNQLHFLSQVDRIVLVHEGTVKEEGTYEELSSNGPLFQRLMENAGK 841
Query: 643 VTIN-------DLNQNSEVLSNPQDSHGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVIG 695
V + +Q +E +++G + + + + + L ++EE+ G
Sbjct: 842 VEEYSEENGEAEADQTAEQPVANGNTNGLQMDGSDDKKSKEGNKKGGKSVLIKQEERETG 901
Query: 696 NVGWKPLWDYINYSNGTLMSCLVILGQCCFLALQTSSNFWLA--TAIEIPKVTDTTLIG- 752
V W+ L Y + G + +++L + +S+ WL+ T PK
Sbjct: 902 VVSWRVLKRYQDALGGAWVVMMLLLCYVLTEVFRVTSSTWLSEWTDAGTPKSHGPLFYNL 961
Query: 753 VYALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTPA-------S 805
+YALLS SY+ + L A+ + SI APM FF + P +
Sbjct: 962 IYALLSFGQVLVTLTNSYWLIMSSLYAAKKLHDNMLHSILRAPMSFFHTNPLGRIINRFA 1021
Query: 806 SDLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATA 865
DL +D + + ++L + +I V+ L +P +V YYQ TA
Sbjct: 1022 KDLGDIDRTVAVFVNMFMGQVSQLLSTVVLIGIVSTLSLWAIMPLLVLFYGAYLYYQNTA 1081
Query: 866 RELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAME 925
RE+ R++ +++PV E G+ T+RA+ DR + +D + + A
Sbjct: 1082 REVKRMDSISRSPVYAQFGEALNGLSTIRAYKAYDRMADINGRSMDNNIRFTLVNMGANR 1141
Query: 926 WLVLRIEALLNLTVITAALLLILLPQRYLSP----GRVGLSLSYALTLNGAQIFWTRWFS 981
WL +R+E L L + A ++ R + +GL LSYAL + R S
Sbjct: 1142 WLGIRLETLGGLMIWLTASFAVMQNGRAENQQAFASTMGLLLSYALNITSLLTGVLRLAS 1201
Query: 982 NLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKG 1041
N + +VER+ +I IP E P +++NNRPP WPS G I + + +RYRP P VL G
Sbjct: 1202 LAENSLNAVERVGNYIEIPPEAPPVIENNRPPPGWPSSGSIKFEDVVLRYRPQLPPVLHG 1261
Query: 1042 ITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSI 1101
++ +VG+VGRTG+GKS+L++ALFR+VE +G ILID ++ GL DLR L I
Sbjct: 1262 VSFFIHPTDKVGIVGRTGAGKSSLLNALFRIVEVEKGRILIDDCDVGKFGLMDLRKVLGI 1321
Query: 1102 IPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSL 1161
IPQ P LF G++R NLDP G ++D ++W+++E+ LK+TI + P LD+ VS+ G N+S+
Sbjct: 1322 IPQSPVLFSGTVRFNLDPFGEHNDADLWESLERAHLKDTIRRNPLGLDAEVSEAGENFSV 1381
Query: 1162 GQRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVI 1221
GQRQL L R LL+R++ILVLDEATA++D TDA++Q+ IR+EF+ CT++ +AHR+ T+I
Sbjct: 1382 GQRQLLSLSRALLRRSKILVLDEATAAVDVRTDALIQKTIREEFKSCTMLIIAHRLNTII 1441
Query: 1222 DSDMVMVLSYGKLVEYDEPSKLM-DTNSSFSKLV 1254
D D ++VL G++ E+ P L+ + SSFSK+V
Sbjct: 1442 DCDKILVLDSGRVQEFSSPENLLSNEGSSFSKMV 1475
Score = 82.8 bits (203), Expect = 1e-15
Identities = 101/445 (22%), Positives = 196/445 (43%), Gaps = 50/445 (11%)
Query: 824 IVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRINGTTKAPVMNFA 883
I+A+ +L +AS+ +L+V + + +I I K + T L R + + +MN
Sbjct: 428 IIALILLYQQLGVASLIGALLLVLMFPLQTVI-ISKMQKLTKEGLQRTD--KRIGLMN-- 482
Query: 884 AETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNV--AMEWLVLRIEALLNLTVIT 941
E + TV+ + + F + D + S F S + A+ +L +L +T+++
Sbjct: 483 -EVLAAMDTVKCYAWENSFQSKVQTVRDDELSWFRKSQLLGALNMFILNSIPVL-VTIVS 540
Query: 942 AALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVERIKQFI---- 997
+ +L L+P R SLS L + + N +S++R+++ +
Sbjct: 541 FGVFTLLGGD--LTPARAFTSLSLFAVLRFPLFMLPNIITQVVNANVSLKRLEEVLATEE 598
Query: 998 -----HIPAEP--PAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKGGS 1050
+ P EP PAI N SW SKG RP L I GS
Sbjct: 599 RILLPNPPIEPGEPAISIRNGY-FSWDSKGD----------RPT----LSNINLDVPLGS 643
Query: 1051 RVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFK 1110
V VVG TG GK++LISA+ + P+ D ++ LR ++ +PQ +F
Sbjct: 644 LVAVVGSTGEGKTSLISAILGEL-PATSDAIVT-----------LRGSVAYVPQVSWIFN 691
Query: 1111 GSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLG 1170
++R N+ + ++ +A++ LK + LP + + + G N S GQ+Q +
Sbjct: 692 ATVRDNILFGSPFDREKYERAIDVTSLKHDLELLPGGDLTEIGERGVNISGGQKQRVSMA 751
Query: 1171 RVLLKRNRILVLDEATASIDS-ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVL 1229
R + + + + D+ +++D+ + ++ I++E + T + V +++ + D ++++
Sbjct: 752 RAVYSNSDVYIFDDPLSALDAHVGQQVFEKCIKRELGQKTRVLVTNQLHFLSQVDRIVLV 811
Query: 1230 SYGKLVEYDEPSKLMDTNSSFSKLV 1254
G + E +L F +L+
Sbjct: 812 HEGTVKEEGTYEELSSNGPLFQRLM 836
Score = 58.9 bits (141), Expect = 2e-08
Identities = 30/76 (39%), Positives = 45/76 (58%), Gaps = 1/76 (1%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKIS-G 477
A+ I++G F WD + P L ++NL++ +AV G K+S + AILGE+P S
Sbjct: 613 AISIRNGYFSWDSKGDRPTLSNINLDVPLGSLVAVVGSTGEGKTSLISAILGELPATSDA 672
Query: 478 TVYVGGTLAYVSQSSW 493
V + G++AYV Q SW
Sbjct: 673 IVTLRGSVAYVPQVSW 688
>At1g30410 hypothetical protein
Length = 1495
Score = 383 bits (983), Expect = e-106
Identities = 233/687 (33%), Positives = 378/687 (54%), Gaps = 31/687 (4%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFELLVSAHKV 643
C+ LR KT +LVT+Q+ FL +D I+ +G + + G++ L KSG F+ L+
Sbjct: 783 CMKDELRGKTRVLVTNQLHFLPLMDKIILVSEGMIKEEGTFVELSKSGILFKKLME---- 838
Query: 644 TINDLNQNSEVLSNPQD--SHGFYLTKNQSEGEISSIQGSIGAQ--LTQEEEKVIGNVGW 699
++ EV +N ++ G +T + SE + S + + L ++EE+ G + W
Sbjct: 839 NAGKMDATQEVNTNDENILKLGPTVTVDVSERNLGSTKQGKRRRSVLIKQEERETGIISW 898
Query: 700 KPLWDYINYSNGTLMSCLVILGQCCFLA---LQTSSNFWLATAIEIPKVTDTT---LIGV 753
L Y + G L +++L C+LA L+ SS+ WL+ + + + I V
Sbjct: 899 NVLMRY-KEAVGGLWVVMILLA--CYLATEVLRVSSSTWLSIWTDQSTSKNYSPGFYIVV 955
Query: 754 YALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTPA-------SS 806
YALL + + S++ L A+ + +SI APMLFF + P S
Sbjct: 956 YALLGFGQVAVTFTNSFWLITSSLHAARRLHDAMLSSILRAPMLFFHTNPTGRVINRFSK 1015
Query: 807 DLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATAR 866
D+ +D ++ + ++L +I +V+ L +P ++ YYQ+T+R
Sbjct: 1016 DIGDIDRNVANLMNMFMNQLWQLLSTFALIGTVSTISLWAIMPLLILFYAAYLYYQSTSR 1075
Query: 867 ELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEW 926
E+ R++ T++P+ E G+ ++RA+ DR K K +D + + + W
Sbjct: 1076 EVRRLDSVTRSPIYAQFGEALNGLSSIRAYKAYDRMAKINGKSMDNNIRFTLANTSSNRW 1135
Query: 927 LVLRIEALLNLTVITAALLLILLPQRYLSPG----RVGLSLSYALTLNGAQIFWTRWFSN 982
L +R+E L + + A +L + +GL LSY L + R S
Sbjct: 1136 LTIRLETLGGVMIWLTATFAVLQNGNTNNQAGFASTMGLLLSYTLNITSLLSGVLRQASR 1195
Query: 983 LSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGI 1042
N + SVER+ +I +P+E I++NNRP WPS G I + + +RYRP P VL G+
Sbjct: 1196 AENSLNSVERVGNYIDLPSEATDIIENNRPVCGWPSGGSIKFEDVHLRYRPGLPPVLHGL 1255
Query: 1043 TCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSII 1102
T +VGVVGRTG+GKS++++ALFR+VE +G I+ID ++ GL D+R LSII
Sbjct: 1256 TFFVSPSEKVGVVGRTGAGKSSMLNALFRIVEVEKGRIMIDDCDVAKFGLTDVRRVLSII 1315
Query: 1103 PQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLG 1162
PQ P LF G++R N+DP ++D +W+A+ + +K+ IS+ P LD+ V + G N+S+G
Sbjct: 1316 PQSPVLFSGTVRFNIDPFSEHNDAGLWEALHRAHIKDVISRNPFGLDAEVCEGGENFSVG 1375
Query: 1163 QRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVID 1222
QRQL L R LL+R++ILVLDEATAS+D TD+++QR IR+EF+ CT++ +AHR+ T+ID
Sbjct: 1376 QRQLLSLARALLRRSKILVLDEATASVDVRTDSLIQRTIREEFKSCTMLVIAHRLNTIID 1435
Query: 1223 SDMVMVLSYGKLVEYDEPSKLMDTNSS 1249
D ++VLS G+++EYD P +L+ ++S
Sbjct: 1436 CDKILVLSSGQVLEYDSPQELLSRDTS 1462
Score = 69.7 bits (169), Expect = 1e-11
Identities = 70/330 (21%), Positives = 140/330 (42%), Gaps = 41/330 (12%)
Query: 937 LTVITAALLLILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVERIKQF 996
+TV++ + ++L L+P R SLS L S + N +S++RI++
Sbjct: 537 VTVVSFGVFVLLGGD--LTPARAFTSLSLFAVLRFPLNMLPNLLSQVVNANVSLQRIEEL 594
Query: 997 I---------HIPAEP--PAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCT 1045
+ + P +P PAI N SW SK L I
Sbjct: 595 LLSEERILAQNPPLQPGTPAISIKNGY-FSWDSK--------------TTKPTLSDINLE 639
Query: 1046 FKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQE 1105
G+ V +VG TG GK++LISA+ + + ++ +R ++ +PQ
Sbjct: 640 IPVGTLVAIVGGTGEGKTSLISAMLGELSHAETTSVV------------IRGSVAYVPQV 687
Query: 1106 PTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQ 1165
+F ++R N+ + + W+A++ L+ + LP + + + G N S GQ+Q
Sbjct: 688 SWIFNATVRENILFGSDFESERYWRAIDATALQHDLDLLPGRDLTEIGERGVNISGGQKQ 747
Query: 1166 LFCLGRVLLKRNRILVLDEATASIDS-ATDAILQRIIRQEFEECTVITVAHRVPTVIDSD 1224
+ R + + + + D+ +++D+ + ++ E T + V +++ + D
Sbjct: 748 RVSMARAVYSNSDVYIFDDPLSALDAHVAHQVFDSCMKDELRGKTRVLVTNQLHFLPLMD 807
Query: 1225 MVMVLSYGKLVEYDEPSKLMDTNSSFSKLV 1254
++++S G + E +L + F KL+
Sbjct: 808 KIILVSEGMIKEEGTFVELSKSGILFKKLM 837
Score = 59.3 bits (142), Expect = 1e-08
Identities = 30/76 (39%), Positives = 45/76 (58%), Gaps = 1/76 (1%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGT 478
A+ I++G F WD ++ P L D+NLEI +A+ G K+S + A+LGE+ T
Sbjct: 614 AISIKNGYFSWDSKTTKPTLSDINLEIPVGTLVAIVGGTGEGKTSLISAMLGELSHAETT 673
Query: 479 -VYVGGTLAYVSQSSW 493
V + G++AYV Q SW
Sbjct: 674 SVVIRGSVAYVPQVSW 689
>At1g30420 hypothetical protein
Length = 1488
Score = 377 bits (968), Expect = e-104
Identities = 240/693 (34%), Positives = 386/693 (55%), Gaps = 39/693 (5%)
Query: 587 CVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKSGTAFE-LLVSAHK 642
CV L+ KT +LVT+Q+ FL +D I+ +G + + G++ L KSGT F+ L+ +A K
Sbjct: 783 CVKHELKGKTRVLVTNQLHFLPLMDRIILVSEGMIKEEGNFAELSKSGTLFKKLMENAGK 842
Query: 643 V-TINDLNQNSEVLSNPQDSHGFYLTKNQSEGEISSIQ-GSIG-AQLTQEEEKVIGNVGW 699
+ ++N N E +S G +T + SE + SIQ G G + L ++EE+ G + W
Sbjct: 843 MDATQEVNTNDENISKL----GPTVTIDVSERSLGSIQQGKWGRSMLVKQEERETGIISW 898
Query: 700 KPLWDYINYSNGTLMSCLVILGQCCFLA---LQTSSNFWLA--TAIEIPK-VTDTTLIGV 753
+ Y N + G L +++L C+L L+ S+ WL+ T PK + I V
Sbjct: 899 DVVMRY-NKAVGGLWVVMILL--VCYLTTEVLRVLSSTWLSIWTDQSTPKSYSPGFYIVV 955
Query: 754 YALLSISSTSFVYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTPA-------SS 806
YALL + + S++ L A+ + SI APMLFF++ P S
Sbjct: 956 YALLGFGQVAVTFTNSFWLISSSLHAAKRLHDAMLNSILRAPMLFFETNPTGRVINRFSK 1015
Query: 807 DLSILDFDIPYSLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATAR 866
D+ +D ++ + ++L +I V+ L +P ++ YYQ+T+R
Sbjct: 1016 DIGDIDRNVANLMNMFMNQLWQLLSTFALIGIVSTISLWAIMPLLILFYATYIYYQSTSR 1075
Query: 867 ELIRINGTTKAPVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEW 926
E+ R++ T++P+ E G+ ++RA+ DR K K +D + S + W
Sbjct: 1076 EVRRLDSVTRSPIYALFGEALNGLSSIRAYKAYDRMAKINGKSMDNNIRFTLASTSSNRW 1135
Query: 927 LVLRIEALLNLTVITAALLLIL----LPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSN 982
L +R E+L + + A +L + + +GL LSY L + R S
Sbjct: 1136 LTIRSESLGGVMIWLTATFAVLRYGNAENQAVFASTMGLLLSYTLNITTLLSGVLRQASK 1195
Query: 983 LSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGI 1042
N + SVER+ +I +P+E AI++NNRP S WPS+G I + + +RYRP P VL G+
Sbjct: 1196 AENSLNSVERVGNYIDLPSEATAIIENNRPVSGWPSRGSIQFEDVHLRYRPGLPPVLHGL 1255
Query: 1043 TCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSII 1102
+ +VGVVGRTG+GKS++++AL+R+VE +G ILID ++ GL DLR K +
Sbjct: 1256 SFFVYPSEKVGVVGRTGAGKSSMLNALYRIVELEKGRILIDDYDVAKFGLTDLRRKQFFL 1315
Query: 1103 PQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLG 1162
G++R N+DP ++D ++W+A+E+ +K+ I + P LD+ VS+ G N+S+G
Sbjct: 1316 -------LGTVRFNIDPFSEHNDADLWEALERAHIKDVIDRNPFGLDAEVSEGGENFSVG 1368
Query: 1163 QRQLFCLGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVID 1222
QRQL L R LL+R++IL LDEATAS+D TD+++QR IR+EF+ CT++ +AHR+ T+ID
Sbjct: 1369 QRQLLSLARALLRRSKILFLDEATASVDVRTDSLIQRTIREEFKSCTMLIIAHRLNTIID 1428
Query: 1223 SDMVMVLSYGKLVEYDEPSKLMDTN-SSFSKLV 1254
D ++VLS G+++EYD P +L+ + S+F K+V
Sbjct: 1429 CDKILVLSSGQVLEYDSPQELLSRDTSAFFKMV 1461
Score = 76.3 bits (186), Expect = 1e-13
Identities = 74/336 (22%), Positives = 143/336 (42%), Gaps = 42/336 (12%)
Query: 934 LLNLTVITAALL---LILLPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISV 990
+LN T + L+ + +L L+P R SLS L S N +S+
Sbjct: 529 ILNSTPVVVTLVSFGVYVLLGGDLTPARAFTSLSLFAVLRSPLSTLPNLISQAVNANVSL 588
Query: 991 ERIKQFI---------HIPAEP--PAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVL 1039
+RI++ + + P +P PAI N SW SK + L
Sbjct: 589 QRIEELLLSEERILAQNPPLQPGAPAISIKNGY-FSWDSK--------------TSKPTL 633
Query: 1040 KGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKL 1099
I GS V +VG TG GK++LISA+ + ++ D+R +
Sbjct: 634 SDINLEIPVGSLVAIVGGTGEGKTSLISAM------------LGELSHAETSSVDIRGSV 681
Query: 1100 SIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNW 1159
+ +PQ +F ++R N+ + + W+A++ L+ + P + + + G N
Sbjct: 682 AYVPQVSWIFNATLRENILFGSDFESERYWRAIDVTALQHDLDLFPGRDRTEIGERGVNI 741
Query: 1160 SLGQRQLFCLGRVLLKRNRILVLDEATASIDS-ATDAILQRIIRQEFEECTVITVAHRVP 1218
S GQ+Q + R + + I + D+ +++D+ + ++ E + T + V +++
Sbjct: 742 SGGQKQRVSMARAVYSNSDIYIFDDPFSALDAHVAHQVFDSCVKHELKGKTRVLVTNQLH 801
Query: 1219 TVIDSDMVMVLSYGKLVEYDEPSKLMDTNSSFSKLV 1254
+ D ++++S G + E ++L + + F KL+
Sbjct: 802 FLPLMDRIILVSEGMIKEEGNFAELSKSGTLFKKLM 837
Score = 58.5 bits (140), Expect = 2e-08
Identities = 29/76 (38%), Positives = 46/76 (60%), Gaps = 1/76 (1%)
Query: 419 ALQIQDGTFIWDHESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKI-SG 477
A+ I++G F WD ++ P L D+NLEI +A+ G K+S + A+LGE+ +
Sbjct: 614 AISIKNGYFSWDSKTSKPTLSDINLEIPVGSLVAIVGGTGEGKTSLISAMLGELSHAETS 673
Query: 478 TVYVGGTLAYVSQSSW 493
+V + G++AYV Q SW
Sbjct: 674 SVDIRGSVAYVPQVSW 689
>At2g07680 putative ABC transporter
Length = 1146
Score = 335 bits (858), Expect = 1e-91
Identities = 226/687 (32%), Positives = 367/687 (52%), Gaps = 53/687 (7%)
Query: 592 LRDKTVILVTHQVEFLSEVDT----ILD-GKVIQSGSYENLLKS-GTAFELLVSAHKVTI 645
L KT ++ TH ++ +S + T ++D GKV SGS ++ KS F L +
Sbjct: 485 LNKKTRVMCTHNIQ-VSNLSTHMIVVMDKGKVNWSGSVTDMPKSISPTFSLTNEFDMSSP 543
Query: 646 NDLNQNSEVLSNPQDSHGFYLTKNQSEGEISSIQGSIGAQLTQEEEKVIGNVGWKPLWDY 705
N L + E LS +D EIS A + + EE+ G V +Y
Sbjct: 544 NHLTKRKETLSIKEDG----------VDEISEA----AADIVKLEERKEGRVEMMVYRNY 589
Query: 706 INYSNGTLMSCLVILGQCCFLALQTSSNFWLATAIEIPKVTDTTLIG-VYALLSISSTSF 764
+S G ++ ++++ + ++ WL+ + D T G V + I ++
Sbjct: 590 AVFS-GWFITIVILVSAVLMQGSRNGNDLWLSYWV------DKTGKGMVLCIFCIINSIL 642
Query: 765 VYVRSYFAALLGLKASTAFFSSFTTSIFNAPMLFFDSTPA-------SSDLSILDFDIPY 817
VR++ A GLKA+ ++ + + NAP FFD TP+ SSDL +D +P+
Sbjct: 643 TLVRAFSFAFGGLKAAVHVHNALISKLINAPTQFFDQTPSGRILNRFSSDLYTIDDSLPF 702
Query: 818 SLTCVAIVAIEVLVMIFVIASVTWQVLIVAVPAMVALIFIQKYYQATARELIRINGTTKA 877
L + + +L +I V++ V L++ +P +Q +Y++T+REL R++ +++
Sbjct: 703 ILNILLANFVGLLGIIVVLSYVQVLFLLLLLPFWYIYSKLQVFYRSTSRELRRLDSVSRS 762
Query: 878 PVMNFAAETHLGVVTVRAFNMVDRFFKNYLKLVDTDASLFFHSNVAMEWLVLRIEALLNL 937
P+ ET G T+RAF + F +++ + + +A WL LR++ L ++
Sbjct: 763 PIYASFTETLDGSSTIRAFKSEEHFVGRFIEHLTLYQRTSYSEIIASLWLSLRLQLLGSM 822
Query: 938 TVITAALLLIL-----LPQRYLSPGRVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVER 992
V+ A++ +L P + +PG VGL+LSYA L F+ ++SVER
Sbjct: 823 IVLFVAVMAVLGSGGNFPISFGTPGLVGLALSYAAPLVSLLGSLLTSFTETEKEMVSVER 882
Query: 993 IKQFIHIPAE----PPAIVDNNRPPSSWPSKGKIDLQGLEVRYRPNAPLVLKGITCTFKG 1048
+ Q++ +P E P ++ D WP G ++ + +RY P L I+ T +G
Sbjct: 883 VLQYMDVPQEEVSGPQSLSDK------WPVHGLVEFHNVTMRYISTLPPALTQISFTIQG 936
Query: 1049 GSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLRMKLSIIPQEPTL 1108
G VGV+GRTG+GKS++++ALFRL G+IL+DG NI + +++LR L+++PQ P L
Sbjct: 937 GMHVGVIGRTGAGKSSILNALFRLTPVCSGEILVDGKNISHLPIRELRSCLAVVPQSPFL 996
Query: 1109 FKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFC 1168
F+GS+R NLDPLGL D IW+ ++KC++K + + LDS V + G ++S+GQRQL C
Sbjct: 997 FQGSLRDNLDPLGLSEDWRIWEILDKCKVKAAVESVGG-LDSYVKESGCSFSVGQRQLLC 1055
Query: 1169 LGRVLLKRNRILVLDEATASIDSATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMV 1228
L R LLK ++IL LDE TA+ID T ++L I E + TVIT+AHR+ TV+D D +++
Sbjct: 1056 LARALLKSSKILCLDECTANIDVHTASLLHNTISSECKGVTVITIAHRISTVVDLDSILI 1115
Query: 1229 LSYGKLVEYDEPSKLM-DTNSSFSKLV 1254
L G LVE +P L+ D +S+FS V
Sbjct: 1116 LDRGILVEQGKPQHLLQDDSSTFSSFV 1142
Score = 70.5 bits (171), Expect = 6e-12
Identities = 52/201 (25%), Positives = 98/201 (47%), Gaps = 16/201 (7%)
Query: 1037 LVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPSRGDILIDGINICSMGLKDLR 1096
L +K ++ GS V V+G GSGK++L+++L + G IL++G
Sbjct: 328 LTIKQVSLRVPKGSFVAVIGEVGSGKTSLLNSLLGEMRCVHGSILLNG------------ 375
Query: 1097 MKLSIIPQEPTLFKGSIRTNLDPLGLYSDDEIWKAVEKCQLKETISKLPSLLDSSVSDEG 1156
++ +PQ P L G++R N+ + ++ + C L IS + + + D+G
Sbjct: 376 -SVAYVPQVPWLLSGTVRENILFGKPFDSKRYFETLSACALDVDISLMVGGDMACIGDKG 434
Query: 1157 GNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATDA-ILQR-IIRQEFEECTVITVA 1214
N S GQR F L R + + + +LD+ +++DS ILQR ++ + T +
Sbjct: 435 LNLSGGQRARFALARAVYHGSDMYLLDDVLSAVDSQVGCWILQRALLGPLLNKKTRVMCT 494
Query: 1215 HRVP-TVIDSDMVMVLSYGKL 1234
H + + + + M++V+ GK+
Sbjct: 495 HNIQVSNLSTHMIVVMDKGKV 515
Score = 48.5 bits (114), Expect = 2e-05
Identities = 25/78 (32%), Positives = 44/78 (56%), Gaps = 3/78 (3%)
Query: 419 ALQIQDGTFIWD---HESMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKI 475
A+ ++D + W E + +K V+L + +AV VGS K+S L ++LGE+ +
Sbjct: 308 AVCVEDASCTWSSNVEEDYNLTIKQVSLRVPKGSFVAVIGEVGSGKTSLLNSLLGEMRCV 367
Query: 476 SGTVYVGGTLAYVSQSSW 493
G++ + G++AYV Q W
Sbjct: 368 HGSILLNGSVAYVPQVPW 385
Score = 30.4 bits (67), Expect = 6.8
Identities = 16/51 (31%), Positives = 25/51 (48%)
Query: 433 SMSPALKDVNLEIKWRQKIAVCAPVGSEKSSFLYAILGEIPKISGTVYVGG 483
++ PAL ++ I+ + V G+ KSS L A+ P SG + V G
Sbjct: 922 TLPPALTQISFTIQGGMHVGVIGRTGAGKSSILNALFRLTPVCSGEILVDG 972
>At3g62150 P-glycoprotein-like proetin
Length = 1292
Score = 154 bits (389), Expect = 3e-37
Identities = 90/244 (36%), Positives = 143/244 (57%), Gaps = 13/244 (5%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
+G I+L + Y RP + +G + + GS V +VG++GSGKST++S + R +P
Sbjct: 400 RGDIELNNVNFSYPARPEEQ-IFRGFSLSISSGSTVALVGQSGSGKSTVVSLIERFYDPQ 458
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDD-----EIWKA 1131
G++ IDGIN+ LK +R K+ ++ QEP LF SI+ N+ Y + EI KA
Sbjct: 459 SGEVRIDGINLKEFQLKWIRSKIGLVSQEPVLFTSSIKENI----AYGKENATVEEIRKA 514
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
E + I KLP LD+ V + G S GQ+Q + R +LK RIL+LDEAT+++D+
Sbjct: 515 TELANASKFIDKLPQGLDTMVGEHGTQLSGGQKQRIAVARAILKDPRILLLDEATSALDA 574
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLM-DTNSSF 1250
++ I+Q + + T + VAHR+ TV ++DM+ V+ GK+VE S+L+ D ++
Sbjct: 575 ESERIVQEALDRIMVNRTTVVVAHRLSTVRNADMIAVIHQGKIVEKGSHSELLRDPEGAY 634
Query: 1251 SKLV 1254
S+L+
Sbjct: 635 SQLI 638
Score = 128 bits (321), Expect = 2e-29
Identities = 74/231 (32%), Positives = 132/231 (57%), Gaps = 5/231 (2%)
Query: 1019 KGKIDLQGLEVRYRPNAP--LVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
KG I+L+ + +Y P+ P + + + + + G + +VG +GSGKST+I+ L R +P
Sbjct: 1045 KGDIELRHISFKY-PSRPDVQIFQDLCLSIRAGKTIALVGESGSGKSTVIALLQRFYDPD 1103
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNL--DPLGLYSDDEIWKAVEK 1134
G I +DG+ I ++ LK LR + ++ QEP LF +IR N+ G ++ EI A E
Sbjct: 1104 SGQITLDGVEIKTLQLKWLRQQTGLVSQEPVLFNETIRANIAYGKGGDATETEIVSAAEL 1163
Query: 1135 CQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATD 1194
IS L D+ V + G S GQ+Q + R ++K ++L+LDEAT+++D+ ++
Sbjct: 1164 SNAHGFISGLQQGYDTMVGERGVQLSGGQKQRVAIARAIVKDPKVLLLDEATSALDAESE 1223
Query: 1195 AILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMD 1245
++Q + + T + VAHR+ T+ ++D++ V+ G +VE + L++
Sbjct: 1224 RVVQDALDRVMVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGKHETLIN 1274
>At2g47000 putative ABC transporter
Length = 1286
Score = 152 bits (385), Expect = 9e-37
Identities = 88/244 (36%), Positives = 145/244 (59%), Gaps = 13/244 (5%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
KG I+L+ + Y RP+ + +G + G+ V +VG++GSGKST++S + R +P
Sbjct: 381 KGDIELKDVYFTYPARPDEQ-IFRGFSLFISSGTTVALVGQSGSGKSTVVSLIERFYDPQ 439
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLY-----SDDEIWKA 1131
GD+LIDGIN+ LK +R K+ ++ QEP LF SI+ N+ Y + +EI A
Sbjct: 440 AGDVLIDGINLKEFQLKWIRSKIGLVSQEPVLFTASIKDNI----AYGKEDATTEEIKAA 495
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
E + + KLP LD+ V + G S GQ+Q + R +LK RIL+LDEAT+++D+
Sbjct: 496 AELANASKFVDKLPQGLDTMVGEHGTQLSGGQKQRIAVARAILKDPRILLLDEATSALDA 555
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLM-DTNSSF 1250
++ ++Q + + T + VAHR+ TV ++DM+ V+ GK+VE ++L+ D ++
Sbjct: 556 ESERVVQEALDRIMVNRTTVVVAHRLSTVRNADMIAVIHQGKIVEKGSHTELLKDPEGAY 615
Query: 1251 SKLV 1254
S+L+
Sbjct: 616 SQLI 619
Score = 130 bits (326), Expect = 6e-30
Identities = 77/231 (33%), Positives = 133/231 (57%), Gaps = 5/231 (2%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
KG I+L+ + +Y RP+ + + + + + G V +VG +GSGKST+I+ L R +P
Sbjct: 1039 KGDIELRHVSFKYPARPDVQ-IFQDLCLSIRAGKTVALVGESGSGKSTVIALLQRFYDPD 1097
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNL--DPLGLYSDDEIWKAVEK 1134
G+I +DG+ I S+ LK LR + ++ QEP LF +IR N+ G S+ EI + E
Sbjct: 1098 SGEITLDGVEIKSLRLKWLRQQTGLVSQEPILFNETIRANIAYGKGGDASESEIVSSAEL 1157
Query: 1135 CQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSATD 1194
IS L D+ V + G S GQ+Q + R ++K ++L+LDEAT+++D+ ++
Sbjct: 1158 SNAHGFISGLQQGYDTMVGERGIQLSGGQKQRVAIARAIVKDPKVLLLDEATSALDAESE 1217
Query: 1195 AILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMD 1245
++Q + + T I VAHR+ T+ ++D++ V+ G +VE + L++
Sbjct: 1218 RVVQDALDRVMVNRTTIVVAHRLSTIKNADVIAVVKNGVIVEKGKHDTLIN 1268
>At5g46540 multidrug resistance p-glycoprotein
Length = 1248
Score = 148 bits (374), Expect = 2e-35
Identities = 171/724 (23%), Positives = 314/724 (42%), Gaps = 67/724 (9%)
Query: 584 IQYCVMTALRDKTVILVTHQVEFLSEVDTIL---DGKVIQSGSYENLLKS--GTAFELLV 638
+Q ++ + +T ++V H++ + D I GKVI+ G+++ ++K GT +L
Sbjct: 534 VQDALVKLMLSRTTVVVAHRLTTIRTADMIAVVQQGKVIEKGTHDEMIKDPEGTYSQL-- 591
Query: 639 SAHKVTINDLNQNSEVLSNPQDSHGFYLTKNQSEGEI----------SSIQGSIGAQLTQ 688
V + + ++ E + + L S+ + S + G I T+
Sbjct: 592 ----VRLQEGSKKEEAIDKEPEKCEMSLEIESSDSQNGIHSGTLTSPSGLPGVISLDQTE 647
Query: 689 EEEKVIGNV-------GWKPLWDYINYSNGTLMSCLVI--LGQCCFLALQTSSNFWLATA 739
E + I + G + + + N +S L++ L + L+
Sbjct: 648 EFHENISSTKTQTVKKGKEVSLRRLAHLNKPEISVLLLGSLAAVIHGIVFPVQGLLLSRT 707
Query: 740 IEI-----PKVTDTTLIGVYALLSISSTSFVYV--RSYFAALLGLKASTAFFSSFTTSIF 792
I I K+ + +L +++ T + + ++Y A+ G K S +
Sbjct: 708 IRIFFEPSNKLKNDSLFWALIFVALGLTDLIVIPLQNYLFAIAGAKLIKRIRSLSFDRVL 767
Query: 793 NAPMLFFDSTPASSDLSILDFDIPYSLTCVAIVAIEVLVMI----------FVIA-SVTW 841
+ + +FD T SS ++ + + V + +VL +I F+IA + W
Sbjct: 768 HQDISWFDDTKNSS--GVIGARLSTDASTVKSIVGDVLGLIMQNMATIIGAFIIAFTANW 825
Query: 842 QVLIVAVPAMVALIFIQKYYQATARELIRINGTTK-APVMNFAAETHLGVVTVRAFNMVD 900
+ ++A+ + ++F Q YYQ K A++ + TV +F D
Sbjct: 826 LLALMAL-LVAPVMFFQGYYQIKFITGFGAKARGKYEEASQVASDAVSSIRTVASFCAED 884
Query: 901 RFFKNYLKLVDTDASLFFHSNVAMEWLVLRIEALLNLTVITAALLL---ILLPQRYLSPG 957
+ Y + D F + + + L L VI + L L+ R + G
Sbjct: 885 KVMDLYQEKCDEPKQQGFKLGLVSG--LCYGGSYLALYVIESVCFLGGSWLIQNRRATFG 942
Query: 958 RVGLSLSYALTLNGAQIFWTRWFSNLSNYIISVERIKQFIHIPAEPPAIVDNNRPPSSWP 1017
+ +ALTL + T + N + + I P I ++ + P
Sbjct: 943 EF-FQVFFALTLTAVGVTQTSTMAPDINK--AKDSAASIFDILDSKPKIDSSSEKGTILP 999
Query: 1018 S-KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVE 1074
G I+LQ + RY RP+ + + T G V +VG +GSGKST+IS L R +
Sbjct: 1000 IVHGDIELQHVSFRYPMRPDIQ-IFSDLCLTISSGQTVALVGESGSGKSTVISLLERFYD 1058
Query: 1075 PSRGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNL--DPLGLYSDDEIWKAV 1132
P G IL+D + I S+ L LR ++ ++ QEP LF +I +N+ +G +++EI A
Sbjct: 1059 PDSGKILLDQVEIQSLKLSWLREQMGLVSQEPVLFNETIGSNIAYGKIGGATEEEIITAA 1118
Query: 1133 EKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDSA 1192
+ + IS LP ++SV + G S GQ+Q + R +LK +IL+LDEAT+++D+
Sbjct: 1119 KAANVHNFISSLPQGYETSVGERGVQLSGGQKQRIAIARAILKDPKILLLDEATSALDAE 1178
Query: 1193 TDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLMD-TNSSFS 1251
++ ++Q + Q T + VAH + T+ D+DM+ V+ G + E LM+ + +++
Sbjct: 1179 SERVVQDALDQVMVNRTTVVVAHLLTTIKDADMIAVVKNGVIAESGRHETLMEISGGAYA 1238
Query: 1252 KLVA 1255
LVA
Sbjct: 1239 SLVA 1242
Score = 147 bits (370), Expect = 5e-35
Identities = 90/246 (36%), Positives = 146/246 (58%), Gaps = 17/246 (6%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
KG I+L+ + RY RP+ + + G + T G V +VG++GSGKST+IS + R +P
Sbjct: 354 KGDIELRDVYFRYPARPDVQIFV-GFSLTVPNGMTVALVGQSGSGKSTVISLIERFYDPE 412
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLY-----SDDEIWKA 1131
G++LIDGI++ +K +R K+ ++ QEP LF +IR N+ +Y SD EI A
Sbjct: 413 SGEVLIDGIDLKKFQVKWIRSKIGLVSQEPILFATTIRENI----VYGKKDASDQEIRTA 468
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
++ I KLP L++ V + G S GQ+Q + R +LK +IL+LDEAT+++D+
Sbjct: 469 LKLANASNFIDKLPQGLETMVGEHGTQLSGGQKQRIAIARAILKNPKILLLDEATSALDA 528
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE---YDEPSKLMDTNS 1248
++ I+Q + + T + VAHR+ T+ +DM+ V+ GK++E +DE K D
Sbjct: 529 ESERIVQDALVKLMLSRTTVVVAHRLTTIRTADMIAVVQQGKVIEKGTHDEMIK--DPEG 586
Query: 1249 SFSKLV 1254
++S+LV
Sbjct: 587 TYSQLV 592
>At1g02520 P-glycoprotein, putative
Length = 1278
Score = 146 bits (369), Expect = 6e-35
Identities = 87/244 (35%), Positives = 140/244 (56%), Gaps = 13/244 (5%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
+G I+L+ + Y RP+ + G + G+ +VG +GSGKST+IS + R +P
Sbjct: 378 RGDIELKDVHFSYPARPDEE-IFDGFSLFIPSGATAALVGESGSGKSTVISLIERFYDPK 436
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDD-----EIWKA 1131
G +LIDG+N+ LK +R K+ ++ QEP LF SI N+ Y + EI A
Sbjct: 437 SGAVLIDGVNLKEFQLKWIRSKIGLVSQEPVLFSSSIMENI----AYGKENATVEEIKAA 492
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
E + I KLP LD+ V + G S GQ+Q + R +LK RIL+LDEAT+++D+
Sbjct: 493 TELANAAKFIDKLPQGLDTMVGEHGTQLSGGQKQRIAIARAILKDPRILLLDEATSALDA 552
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLM-DTNSSF 1250
++ ++Q + + T + VAHR+ TV ++DM+ V+ GK+VE S+L+ D+ ++
Sbjct: 553 ESERVVQEALDRVMVNRTTVIVAHRLSTVRNADMIAVIHRGKMVEKGSHSELLKDSEGAY 612
Query: 1251 SKLV 1254
S+L+
Sbjct: 613 SQLI 616
Score = 129 bits (323), Expect = 1e-29
Identities = 77/225 (34%), Positives = 128/225 (56%), Gaps = 9/225 (4%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
KG I+L+ L Y RP+ + + + T + G V +VG +GSGKST+IS L R +P
Sbjct: 1029 KGDIELRHLSFTYPARPDIQ-IFRDLCLTIRAGKTVALVGESGSGKSTVISLLQRFYDPD 1087
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDD-----EIWKA 1131
G I +DG+ + + LK LR ++ ++ QEP LF +IR N+ G S++ EI A
Sbjct: 1088 SGHITLDGVELKKLQLKWLRQQMGLVGQEPVLFNDTIRANI-AYGKGSEEAATESEIIAA 1146
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
E + IS + D+ V + G S GQ+Q + R ++K +IL+LDEAT+++D+
Sbjct: 1147 AELANAHKFISSIQQGYDTVVGERGIQLSGGQKQRVAIARAIVKEPKILLLDEATSALDA 1206
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1236
++ ++Q + + T I VAHR+ T+ ++D++ V+ G + E
Sbjct: 1207 ESERVVQDALDRVMVNRTTIVVAHRLSTIKNADVIAVVKNGVIAE 1251
>At1g02530 hypothetical protein
Length = 1273
Score = 144 bits (362), Expect = 4e-34
Identities = 85/244 (34%), Positives = 141/244 (56%), Gaps = 13/244 (5%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
+G I+L+ + Y RP+ + G + G+ +VG +GSGKST+I+ + R +P
Sbjct: 365 RGDIELKDVHFSYPARPDEE-IFDGFSLFIPSGATAALVGESGSGKSTVINLIERFYDPK 423
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDD-----EIWKA 1131
G++LIDGIN+ LK +R K+ ++ QEP LF SI N+ Y + EI A
Sbjct: 424 AGEVLIDGINLKEFQLKWIRSKIGLVCQEPVLFSSSIMENI----AYGKENATLQEIKVA 479
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
E + I+ LP LD+ V + G S GQ+Q + R +LK R+L+LDEAT+++D+
Sbjct: 480 TELANAAKFINNLPQGLDTKVGEHGTQLSGGQKQRIAIARAILKDPRVLLLDEATSALDT 539
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVEYDEPSKLM-DTNSSF 1250
++ ++Q + + T + VAHR+ TV ++DM+ V+ GK+VE S+L+ D+ ++
Sbjct: 540 ESERVVQEALDRVMVNRTTVVVAHRLSTVRNADMIAVIHSGKMVEKGSHSELLKDSVGAY 599
Query: 1251 SKLV 1254
S+L+
Sbjct: 600 SQLI 603
Score = 128 bits (321), Expect = 2e-29
Identities = 75/225 (33%), Positives = 128/225 (56%), Gaps = 9/225 (4%)
Query: 1019 KGKIDLQGLEVRY--RPNAPLVLKGITCTFKGGSRVGVVGRTGSGKSTLISALFRLVEPS 1076
KG I+L+ L Y RP + + + T + G V +VG +GSGKST+IS L R +P
Sbjct: 1024 KGDIELRHLSFTYPARPGIQ-IFRDLCLTIRAGKTVALVGESGSGKSTVISLLQRFYDPD 1082
Query: 1077 RGDILIDGINICSMGLKDLRMKLSIIPQEPTLFKGSIRTNLDPLGLYSDD-----EIWKA 1131
G I +DG+ + + LK LR ++ ++ QEP LF +IR N+ G S++ EI A
Sbjct: 1083 SGQITLDGVELKKLQLKWLRQQMGLVGQEPVLFNDTIRANI-AYGKGSEEAATESEIIAA 1141
Query: 1132 VEKCQLKETISKLPSLLDSSVSDEGGNWSLGQRQLFCLGRVLLKRNRILVLDEATASIDS 1191
E + IS + D+ V ++G S GQ+Q + R ++K +IL+LDEAT+++D+
Sbjct: 1142 AELANAHKFISSIQQGYDTVVGEKGIQLSGGQKQRVAIARAIVKEPKILLLDEATSALDA 1201
Query: 1192 ATDAILQRIIRQEFEECTVITVAHRVPTVIDSDMVMVLSYGKLVE 1236
++ ++Q + + T + VAHR+ T+ ++D++ ++ G + E
Sbjct: 1202 ESERLVQDALDRVIVNRTTVVVAHRLSTIKNADVIAIVKNGVIAE 1246
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.346 0.150 0.508
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,062,653
Number of Sequences: 26719
Number of extensions: 995276
Number of successful extensions: 5704
Number of sequences better than 10.0: 113
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 5204
Number of HSP's gapped (non-prelim): 333
length of query: 1273
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1162
effective length of database: 8,352,787
effective search space: 9705938494
effective search space used: 9705938494
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC119410.2


