

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.8 + phase: 0
(908 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
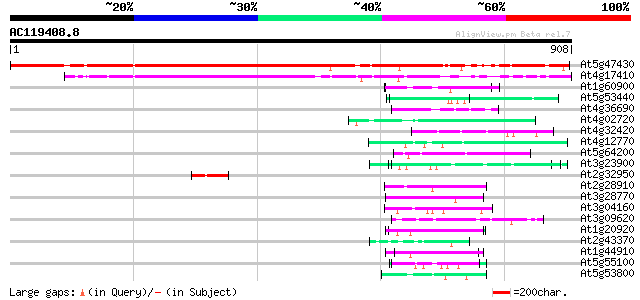
Score E
Sequences producing significant alignments: (bits) Value
At5g47430 DNA-binding protein-like 852 0.0
At4g17410 hypothetical protein 645 0.0
At1g60900 putative U2 snRNP auxiliary factor 65 1e-10
At5g53440 putative protein 55 2e-07
At4g36690 splicing factor like protein 54 5e-07
At4g02720 unknown protein 53 9e-07
At4g32420 cyclophilin (AtCYP95) 52 2e-06
At4g12770 auxilin-like protein 51 3e-06
At5g64200 unknown protein 51 3e-06
At3g23900 unknown protein 51 3e-06
At2g32950 photomorphogenesis repressor (COP1) 50 4e-06
At2g28910 unknown protein 49 1e-05
At3g28770 hypothetical protein 48 2e-05
At3g04160 hypothetical protein 48 2e-05
At3g09620 putative RNA helicase 48 3e-05
At1g20920 putative RNA helicase 47 4e-05
At2g43370 U1 small nuclear ribonucleoprotein 70 kDa like protein 47 6e-05
At1g44910 splicing factor like protein 47 6e-05
At5g55100 unknown protein 46 8e-05
At5g53800 unknown protein 46 8e-05
>At5g47430 DNA-binding protein-like
Length = 889
Score = 852 bits (2200), Expect = 0.0
Identities = 493/946 (52%), Positives = 589/946 (62%), Gaps = 99/946 (10%)
Query: 1 MAVYYKFKSARDYDSISMDGPFISVGTLKEKIFESKHLGRGTDFDLVVTNAQTNEEYLDE 60
MA+YYKFKSARDYD+I+MDGPFISVG LK+KIFE+KHLG G D D+VV+NAQTNEEYLDE
Sbjct: 1 MAIYYKFKSARDYDTIAMDGPFISVGILKDKIFETKHLGTGKDLDIVVSNAQTNEEYLDE 60
Query: 61 EMLIPKNTSVLIRRVPGRPRLPIVTEQEYYLKPLISQKVENKVVDTEPANSSLPADDMSA 120
MLIPKNTSVLIRRVPGRPR+ ++T QE +++NKV D + ++ P D SA
Sbjct: 61 AMLIPKNTSVLIRRVPGRPRITVITTQE--------PRIQNKVEDVQAETTNFPVADPSA 112
Query: 121 MKYVEDSDWDEFGNDLYSIPDQLPVQSINMIQE-APPTSTVDEESKIKALIDTPALDWQH 179
+ ++DEFG DLYSIPD Q I A VDEESKI+ALIDTPALDWQ
Sbjct: 113 ----PEDEYDEFGTDLYSIPDTQDAQHIIPRPHLATADDKVDEESKIQALIDTPALDWQQ 168
Query: 180 QGSD-FGAGRGFGRGMGGRMGGGRGFGLERKTPPQGYVCHRCKVPGHFIQHCPTNGDPNY 238
QG D FGAGRG+GRGM GRM G RGFG+ERKTPP GYVCHRC +PGHFIQHCPTNGDPNY
Sbjct: 169 QGQDTFGAGRGYGRGMPGRMNG-RGFGMERKTPPPGYVCHRCNIPGHFIQHCPTNGDPNY 227
Query: 239 DVKRVKQPTGIPRSMLMVNPQGSYALPNGSVAVLKPNEAAFEKEMEGMPSTTRSVGDLPP 298
DVKRVK PTGIP+SMLM P GSY+LP+G+VAVLKPNE AFEKEMEG+PSTTRSVG+LPP
Sbjct: 228 DVKRVKPPTGIPKSMLMATPDGSYSLPSGAVAVLKPNEDAFEKEMEGLPSTTRSVGELPP 287
Query: 299 ELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCVCGAMNVLADDLLPNKTLRD 358
EL CPLC VMKDA LTSKCC+KSFCDKCIRD+IISKSMCVCG +VLADDLLPNKTLRD
Sbjct: 288 ELKCPLCKEVMKDAALTSKCCYKSFCDKCIRDHIISKSMCVCGRSDVLADDLLPNKTLRD 347
Query: 359 TINRILESGNSSTENAGSTYQVQDMESARCPQPKIPSPTSSAASKGGLKISPVYDGTTNI 418
TINRILE+GN STEN GS + D+ESARCP PK SPT+S ASKG K + +
Sbjct: 348 TINRILEAGNDSTENVGSVGHIPDLESARCPPPKALSPTTSVASKGEKKPVLSNNNDAST 407
Query: 419 QDTAVETKVVSAPPQTSEHVKIPRAGDVSEATHESKSVKE-PVSQGSAQVVEEEVQQKLV 477
+E +++ P+ S V + + D E+T S VKE VS+ + Q +EE+QQ++
Sbjct: 408 LKAPMEVAEITSAPRASAEVNVEKPVDACESTQGSVIVKEATVSKLNTQAPKEEMQQQVA 467
Query: 478 PTEAGKKKKKKKVRMPTNDFQWKPPHDLGAENYMMQMGPPP------------------G 519
E KKKKKK R+P ND QW P DL +YMMQMGP P G
Sbjct: 468 AGEPAGKKKKKKPRVPGNDMQWNPVPDLAGPDYMMQMGPGPQYFNGMQPGFNGVQPGFNG 527
Query: 520 YNPYWNGMQPCMDGFMAPYAGPM-HMMDYGHGPYDMPFPNGMPHDPFANGMPHDPFGMQG 578
P +NG P +GF P+ G M M YG P DM F GM N M DPF QG
Sbjct: 528 VQPGFNGFHPGFNGFGGPFPGAMPPFMGYGLNPMDMGFGGGM------NMMHPDPFMAQG 581
Query: 579 YMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVE-----RDFS 633
+ P I PPPHRDLAE MN+ M R+E EAR A+ RKRENERR E RD
Sbjct: 582 FGFPNI-PPPHRDLAEMGNRMNLQRAMMGRDEAEARNAEMLRKRENERRPEGGKMFRDGE 640
Query: 634 KDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPD--SSHREVEPPRPT 691
R +S S +K++ PP DY + R +R SP+ + + + P R +
Sbjct: 641 NSRMMMNNGTSASASSINPNKSRQAPPPPIHDYDRRRRPEKRLSPEHPPTRKNISPSRDS 700
Query: 692 KRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRR--------HHHRTEASSKKSTD 743
KRKS+ ER DR RD + RHQD D HDR + RR HHR E
Sbjct: 701 KRKSERYPDER-DRQRDRE-RSRHQDVDREHDRTRDRRDEDRSRDHRHHRGETER----- 753
Query: 744 PVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSR 803
++ RK SEP SSEP P T AE + K+SVF+R
Sbjct: 754 --SQHHHRKRSEP-------PSSEP---------PVPATKAE------IENNLKSSVFAR 789
Query: 804 ISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYES 863
ISFP EE ++ K+RK+ +SS+T T +AS SA A T + + + + DYES
Sbjct: 790 ISFPEEE--TSSGKRRKVPSSSSTSV-TDPSASASAAAAVGTSVHRHSSRKEIEVADYES 846
Query: 864 SDDDEDDERHFKRRPSRYEPSPPPPVDDWVEE----GRHSRGTRDR 905
SD+D RHFKR+PSRY SPP V D E+ + +G R R
Sbjct: 847 SDED----RHFKRKPSRYARSPPVVVSDVSEDKLRYSKRGKGERSR 888
>At4g17410 hypothetical protein
Length = 744
Score = 645 bits (1663), Expect = 0.0
Identities = 398/838 (47%), Positives = 494/838 (58%), Gaps = 117/838 (13%)
Query: 90 YLKPLISQKVENKVVDTEPANSSLPADDMSAMKYVEDSDWDEFGNDLYSIPDQLPVQSIN 149
+ +P + KVEN D N+ + AD VED ++DEFGNDLYSIPD V S N
Sbjct: 4 FCRPRVEDKVENVQADM---NNVITADASP----VED-EFDEFGNDLYSIPDAPAVHSNN 55
Query: 150 MIQEAPPTSTVDEESKIKALIDTPALDWQHQGSD-FGAGRGFGRGMGGRMGGGRGFGLER 208
+ ++ P DEE+K+KALIDTPALDW QG+D FG GRG+GRGM GRMGG RGFG+ER
Sbjct: 56 LCHDSAPAD--DEETKLKALIDTPALDWHQQGADSFGPGRGYGRGMAGRMGG-RGFGMER 112
Query: 209 KTPPQGYVCHRCKVPGHFIQHCPTNGDPNYDVKRVKQPTGIPRSMLMVNPQGSYALPNGS 268
TPP GYVCHRC V GHFIQHC TNG+PN+DVKRVK PTGIP+SMLM P GSY+LP+G+
Sbjct: 113 TTPPPGYVCHRCNVSGHFIQHCSTNGNPNFDVKRVKPPTGIPKSMLMATPNGSYSLPSGA 172
Query: 269 VAVLKPNEAAFEKEMEGMPSTTRSVGDLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCI 328
VAVLKPNE AFEKEMEG+ STTRSVG+ PPEL CPLC VM+DA L SKCC KS+CDKCI
Sbjct: 173 VAVLKPNEDAFEKEMEGLTSTTRSVGEFPPELKCPLCKEVMRDAALASKCCLKSYCDKCI 232
Query: 329 RDYIISKSMCVCGAMNVLADDLLPNKTLRDTINRILESGNSSTENAGSTYQVQDMESARC 388
RD+II+KSMCVCGA +VLADDLLPNKTLRDTINRILESGNSS ENAGS QVQDMES RC
Sbjct: 233 RDHIIAKSMCVCGATHVLADDLLPNKTLRDTINRILESGNSSAENAGSMCQVQDMESVRC 292
Query: 389 PQPKIPSPTSSAASKGGLKISPVYDGTTNIQDTAVETKVVSAPPQTSEHVKIPRAGDVSE 448
P PK SPT+SAAS G K +P + T+ ++E +++ ++E VK+ + D S
Sbjct: 293 PPPKALSPTTSAASGGEKKPAPSNNNETSTLKPSIEIAEITSAWASAEIVKVEKPVDASA 352
Query: 449 ATHESKSVKEP-VSQGSAQVVEEEVQQKLVPTEAGKKKKKKKVRMPTNDFQWKPPHDLGA 507
S + KE VSQ + Q +EE+ Q++ E GK+KKKK T DL
Sbjct: 353 NIQGSSNGKEAAVSQLNTQPPKEEMPQQVASGEQGKRKKKKPRMSGT---------DLAG 403
Query: 508 ENYMMQMGPPPGYNPYWNGMQPCMDGFMAPYAGPM---HMMDYGHGPYDMPFPNGMPHDP 564
+YMM MGP PG N Y+NG QP +G + G + +G + PFP MP P
Sbjct: 404 PDYMMPMGPGPG-NQYFNGFQPGFNGVQHGFNGVQPGFNGFHHGFNGFPGPFPGAMP--P 460
Query: 565 FAN----GMPH-DPFGMQGYMMPPIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADAR 619
F G+ H DPF QG+ P IPPP +RDLAE MN+ P M REEFEA+K + +
Sbjct: 461 FVGYGFGGVIHPDPFAAQGFGFPNIPPP-YRDLAEMGNRMNLQHPIMGREEFEAKKTEMK 519
Query: 620 RKRENERR------VERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRS 673
RKRENE R V RD K R +S S +K K++ PP S + R RS
Sbjct: 520 RKRENEIRRSEGGNVVRDSEKSRIMN---NSAVTSSPVKPKSRQGPPPPISSDYDRRRRS 576
Query: 674 ERPSPD--SSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHH 731
+R SP+ SS R PPR + RKS+ DRH D D HDR + R
Sbjct: 577 DRSSPERQSSRRFTSPPRSSSRKSER---------------DRHHDLDSEHDRRRDR--- 618
Query: 732 HRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAA 791
+ T RK + +S KSS T +
Sbjct: 619 --------------PRETDRKH-----RKRSEKSSSDPTVEI------------------ 641
Query: 792 ADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGG 851
D K++VF+RISFP E ++ K+RK S SS + SV+ + H S
Sbjct: 642 -DDNNKSNVFTRISFPEE----SSGKQRKTSKSSPAPPES----SVAPVSSGRRHHSRRE 692
Query: 852 RKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPP-PPVDDWVEEGRHSRGTRDRKHR 908
R+ M +Y+SSDD++ RHFKR+PSRY+ SP P D E RHS+ ++ + R
Sbjct: 693 RE----MVEYDSSDDED---RHFKRKPSRYKRSPSVAPSDAGDEHFRHSKRSKGERAR 743
>At1g60900 putative U2 snRNP auxiliary factor
Length = 589
Score = 65.5 bits (158), Expect = 1e-10
Identities = 52/188 (27%), Positives = 84/188 (44%), Gaps = 18/188 (9%)
Query: 607 SREEFEARKADAR-RKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASD 665
SRE + +K ++ +++NE ++D +KDRD ++ + K++ + D
Sbjct: 36 SRESHDLKKDSSKISEKDNENGRDKDGNKDRDREKDRD--------REKSRDRDREKSRD 87
Query: 666 YHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDR-DYDYHDRHQDRDYXHDR 724
++R RS+ D HR+ R D S E RD D D+H R +DRD R
Sbjct: 88 RDRDRERSKDRQRDRHHRD--------RHRDRSRERSEKRDDLDDDHHRRSRDRDRRRSR 139
Query: 725 HQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSA 784
+ R HR + S+ + +S S + + +S+SR S +K S AP
Sbjct: 140 DRDREVRHRRRSRSRSRSRSERRSRSEHRHKSEHRSRSRSRSRSKSKRRSGFDMAPPDML 199
Query: 785 EAAAAAAA 792
A A AAA
Sbjct: 200 AATAVAAA 207
Score = 50.1 bits (118), Expect = 6e-06
Identities = 45/180 (25%), Positives = 78/180 (43%), Gaps = 18/180 (10%)
Query: 608 REEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYH 667
++ R D + R+ E+ +R+ S+DRD RE S D +SK + +H
Sbjct: 52 KDNENGRDKDGNKDRDREKDRDREKSRDRD--REKSRDRDRDRERSKDR-----QRDRHH 104
Query: 668 QNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYH----DRHQDRDYXHD 723
++RHR +R S R+ +R D R DRDR+ + R + R
Sbjct: 105 RDRHR-DRSRERSEKRDDLDDDHHRRSRDRDRRRSRDRDREVRHRRRSRSRSRSRSERRS 163
Query: 724 RHQHRRHHHRTEASSKKSTDPVTKSTSRKS---SEPDIKSKSRKSSEPVTKSLSLSKTAP 780
R +HR H++E S+ + +KS R + PD+ + + ++ S+ + T P
Sbjct: 164 RSEHR---HKSEHRSRSRSRSRSKSKRRSGFDMAPPDMLAATAVAAAGQVPSVPTTATIP 220
>At5g53440 putative protein
Length = 1181
Score = 54.7 bits (130), Expect = 2e-07
Identities = 71/312 (22%), Positives = 111/312 (34%), Gaps = 58/312 (18%)
Query: 610 EFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQN 669
E + + D R R+ +R ERD +DRD RE DY +
Sbjct: 312 ESDRNERDRERTRDRDRDYERDRDRDRDRDRE-----------------RDRDRRDYEHD 354
Query: 670 RHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHD---------RHQDRDY 720
R+ D S R + D + +DR RDY YHD R DRD
Sbjct: 355 RYHDRDWDRDRSR-----DRDRDHERDRTHDREKDRSRDY-YHDGKRSKSDRERDNDRDV 408
Query: 721 XH-----DRHQHRRHHHRT------------EASSKKSTDPVTKSTSRKSSEPDIKSKSR 763
R++ RR R+ SS+ D R+ S ++ ++
Sbjct: 409 SRLDDQSGRYKDRRDGRRSPDYQDYQDVITGSRSSRVEPDGDMTRPERQLSSSVVQEENG 468
Query: 764 KSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRI--SFPSEEEAAAAAKKRKL 821
+S+ +TK S + A + ++ A++ + FP+E AA A R +
Sbjct: 469 NASDQITKGASSREVAELSGGSERGTRQKVSEKTANMEDGVLGEFPAERSFAAKASPRPM 528
Query: 822 ---SASSTT---EASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDERHFK 875
S SST+ + A S + T H N R A ++ D+ E F
Sbjct: 529 VERSPSSTSLERRYNNRGGARRSIEVEETGH-RNNARDYSATEEERHLVDETSQAELSFN 587
Query: 876 RRPSRYEPSPPP 887
+ ++ S PP
Sbjct: 588 NKANQNNSSFPP 599
Score = 53.9 bits (128), Expect = 4e-07
Identities = 42/152 (27%), Positives = 59/152 (38%), Gaps = 23/152 (15%)
Query: 615 KADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSE 674
K + R + E + E D + D R+ + K S + N H E
Sbjct: 233 KGEKSRDKYREDKEEEDIKQKGDKQRDDRPTKEHLRSDEKLTRDESKKKSKFQDNDHGHE 292
Query: 675 RPSPDSSHREVEPPRPTKRKSDHSERERE---DRDRDY-------------------DY- 711
S + E E R R+SD +ER+RE DRDRDY DY
Sbjct: 293 PDSELDGYHERERNRDYDRESDRNERDRERTRDRDRDYERDRDRDRDRDRERDRDRRDYE 352
Query: 712 HDRHQDRDYXHDRHQHRRHHHRTEASSKKSTD 743
HDR+ DRD+ DR + R H + + + D
Sbjct: 353 HDRYHDRDWDRDRSRDRDRDHERDRTHDREKD 384
Score = 38.9 bits (89), Expect = 0.013
Identities = 38/159 (23%), Positives = 64/159 (39%), Gaps = 24/159 (15%)
Query: 609 EEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ 668
EE+ + + R+ + E +R KD D G E S VSS KS+ + D +
Sbjct: 66 EEYTSSSSKRRKGKSGESGSDRWNGKDDDKG-ESSKKTKVSSEKSRKR-----DEGDGEE 119
Query: 669 NRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHR 728
+ S + D HRE S E + D+++D Y + D+ Y D
Sbjct: 120 TKKSSGKS--DGKHRE----------SSRRESKDVDKEKDRKYKEGKSDKFYDGD----- 162
Query: 729 RHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSE 767
HH+++A S K+ +E + +SR+ +
Sbjct: 163 -DHHKSKAGSDKTESKAQDHARSPGTENYTEKRSRRKRD 200
>At4g36690 splicing factor like protein
Length = 573
Score = 53.5 bits (127), Expect = 5e-07
Identities = 54/177 (30%), Positives = 73/177 (40%), Gaps = 30/177 (16%)
Query: 619 RRKRENERRVERDFSKDRDFGREVSSVGDVS-SIKSKTKPIPPSSASDYHQNRHRSERPS 677
R R++ER R ++R+ GR+ D S +S+ + S ++R ER
Sbjct: 39 RESRDHERETSRSKDREREKGRDKDRERDSEVSRRSRDRDGEKSKERSRDKDRDHRERHH 98
Query: 678 PDSSHREVEPPRPTKRKSDHSEREREDRDR---DYDYHDRHQDRDYXHDRHQHRRHHHRT 734
S HR DHS E R+R D D + R +DRD HDR + R R+
Sbjct: 99 RSSRHR------------DHSRERGERRERGGRDDDDYRRSRDRD--HDRRRDDRGGRRS 144
Query: 735 EASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAA 791
S +S D +S R S KSK R S + AP SA AA AA
Sbjct: 145 RRSRSRSKD---RSERRTRSRSPSKSKQRVSGFDM---------APPASAMLAAGAA 189
>At4g02720 unknown protein
Length = 422
Score = 52.8 bits (125), Expect = 9e-07
Identities = 70/315 (22%), Positives = 108/315 (34%), Gaps = 37/315 (11%)
Query: 549 HGPYDMPFPNGM----PHDPFANGMPHDPFGMQGYMMPPIPPPPHRDLAEFSMGMNVPPP 604
HG D + NG D G+ H+ + Q + R A+F N P P
Sbjct: 14 HGYLDRDYRNGRRSGSDSDEELKGLSHEEYRRQKRLKM-------RKSAKFCFWENTPSP 66
Query: 605 AMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSAS 664
+ E AD + + R + K+R K K+ S S
Sbjct: 67 PRDQNEDSDENADEIQDKNGGERDDNSKGKER---------------KGKSDS---ESES 108
Query: 665 DYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDR 724
D ++R R + S R+ ++ + S+ E EDR R + +
Sbjct: 109 DGLRSRKRKSKSSRSKRRRKRSYDSDSESEGSESDSEEEDRRRRRKSSSKRKKSRSSRSF 168
Query: 725 HQHRRHHHRTEAS-SKKSTDPVTK---STSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAP 780
+ R H +T+ S S +S+D +K S S E D KSKS++ + S SK
Sbjct: 169 RKKRSHRRKTKYSDSDESSDEDSKAEISASSSGEEEDTKSKSKRRKKSSDSSSKRSKGEK 228
Query: 781 TTSAEAAAAAAADRK-QKASVFSRISFPSEEEAAAAAKKR---KLSASSTTEASTAATAS 836
T S + D K Q +EE K+ K +S+ E
Sbjct: 229 TKSGSDSDGTEEDSKMQVDETVKNTELELDEEELKKFKEMIELKKKSSAVDEEEEEGDVG 288
Query: 837 VSAKAPSTTHLSNGG 851
+ H+S GG
Sbjct: 289 PMPLPKAEGHISYGG 303
Score = 33.1 bits (74), Expect = 0.73
Identities = 44/198 (22%), Positives = 76/198 (38%), Gaps = 32/198 (16%)
Query: 713 DRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPD-IKSKSRKSSEPVTK 771
D+++D D D Q + R + S K K S SE D ++S+ RKS +K
Sbjct: 69 DQNEDSDENADEIQDKNGGERDDNSKGKER----KGKSDSESESDGLRSRKRKSKSSRSK 124
Query: 772 SLSLSKTAPTTSAEAAAAAAAD----RKQKASVFSRISFPSEEEAAAAAKKRKLSASSTT 827
+ +E + + + + R++K+S + S S + +RK S +
Sbjct: 125 RRRKRSYDSDSESEGSESDSEEEDRRRRRKSSSKRKKSRSSRSFRKKRSHRRKTKYSDSD 184
Query: 828 EASTA-ATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDERHFKRRPSRYEPSPP 886
E+S + A +SA SS ++ED + KRR + S
Sbjct: 185 ESSDEDSKAEISAS----------------------SSGEEEDTKSKSKRRKKSSDSSSK 222
Query: 887 PPVDDWVEEGRHSRGTRD 904
+ + G S GT +
Sbjct: 223 RSKGEKTKSGSDSDGTEE 240
>At4g32420 cyclophilin (AtCYP95)
Length = 837
Score = 51.6 bits (122), Expect = 2e-06
Identities = 57/258 (22%), Positives = 106/258 (40%), Gaps = 32/258 (12%)
Query: 651 IKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYD 710
++ K + SS S+ + S S ++ P S H +++ R D
Sbjct: 196 VRRKKRRRHSSSESESSSDSETDSSESDSESDSDLSSPSFLS-SSSHERQKKRKRSSKKD 254
Query: 711 YHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKS---SEPDIKSKSRKSSE 767
H R + RD H++ + R R + S++S D + S S S+ +++ ++K
Sbjct: 255 KHRRSKQRDKRHEKKRSMR-DKRPKRKSRRSPDSLEDSNSGSEASLSDVNVEIGAKKRKH 313
Query: 768 PVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSR------ISFPSEEEAA-------- 813
V++ +AP EA + RK + +R IS + E+ +
Sbjct: 314 RVSR--RTGNSAPAVEKEAESLHQGKRKGPDLLENRGLRSNGISDAASEQISDRQPDIVD 371
Query: 814 ---AAAKKRKLSASSTTEASTAATASVS-AKAPSTTHLSNGG-------RKSKAVMDDYE 862
+ ++ R LS T ST+ + S +K+PS++ NGG R+ K + +
Sbjct: 372 DHPSKSRSRSLSPKRTVSKSTSVSPRRSQSKSPSSSPRWNGGRSPAKGSRQVKNLTNSRR 431
Query: 863 SSDDDEDDERHFKRRPSR 880
S E+ RH +R P++
Sbjct: 432 ESPGSEEKGRHVRRSPTK 449
>At4g12770 auxilin-like protein
Length = 909
Score = 51.2 bits (121), Expect = 3e-06
Identities = 73/342 (21%), Positives = 127/342 (36%), Gaps = 29/342 (8%)
Query: 582 PPIPPPPHRDLAEFSMGMN---VPPPAMSREEFEARKADARRKRENERRVERDFSKDRDF 638
PP PPP R +N +P A + +A ++ DFS R+
Sbjct: 371 PPTRPPPPRPTRPIKKKVNEPSIPTSAYHSHVPSSGRASVNSPTASQMDELDDFSIGRNQ 430
Query: 639 ----GREVSSVGDVSSIKSKTKPIPPSSASDYHQN-----RHRSERPSPDSSHREVEPPR 689
G S G+ S + S T ++ D RH ER RE E +
Sbjct: 431 TAANGYPDPSSGEDSDVFS-TAAASAAAMKDAMDKAEAKFRHAKER-------REKESLK 482
Query: 690 PTK-RKSDHSE----REREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDP 744
++ R+ DH+E RERE R++ +R+ ++ Q R R E K+
Sbjct: 483 ASRSREGDHTENYDSRERELREKQVRLDRERAEREAEMEKTQAREREER-EREQKRIERE 541
Query: 745 VTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRI 804
+ +R++ E + +++ + + T A A AA ++ A R
Sbjct: 542 RERLLARQAVERATREARERAATEAHAKVQRAAVGKVTDARERAERAAVQRAHAEARERA 601
Query: 805 SFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESS 864
+ + E+A AA + + A++ A + A + G ++A + +
Sbjct: 602 AAGAREKAEKAAAEARERANAEVREKEAKVRAERAAVERAAAEARGRAAAQAKAKQQQEN 661
Query: 865 DDDEDDERHFKRRPS---RYEPSPPPPVDDWVEEGRHSRGTR 903
++D D + RPS R +PP P D +G +R
Sbjct: 662 NNDLDSFFNSVSRPSSVPRQRTNPPDPFQDSWNKGGSFESSR 703
>At5g64200 unknown protein
Length = 303
Score = 50.8 bits (120), Expect = 3e-06
Identities = 60/234 (25%), Positives = 98/234 (41%), Gaps = 15/234 (6%)
Query: 621 KRENERRVERDFSKDRDFGREVS----SVGDVSSIKSKTKPIPPSSASDYHQNRHRSERP 676
K E + VER + D GRE++ G + SK + + P S ++R RS R
Sbjct: 67 KDEAHKAVERLDGRVVD-GREITVQFAKYGPNAEKISKGRVVEPPPKS--RRSRSRSPRR 123
Query: 677 SPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDY-XHDRHQHRRHHHRTE 735
S P R + R+S R D R+ DY R + R Y +RH+ + HR
Sbjct: 124 SRSPRRSRSPPRRRSPRRSRSPRRRSRDDYREKDYRKRSRSRSYDRRERHEEKDRDHRRR 183
Query: 736 ASSKKSTDPVTKSTSRKSSEPDIKSKSRK-SSEPVTKSLSLSKTAPTTSAEAAAAAAADR 794
S +S P K R + + +S SR S+ P +S S T+ A +
Sbjct: 184 TRS-RSASPDEKRRVRGRYDNESRSHSRSLSASPARRSPRSSSPQKTSPAREVSPDKRSN 242
Query: 795 KQKASVFSRIS--FPSEEEAAAA---AKKRKLSASSTTEASTAATASVSAKAPS 843
++ S +S P+ ++A+ + + +R+ + S + S A V A + S
Sbjct: 243 ERSPSPRRSLSPRSPALQKASPSKEMSPERRSNERSPSPGSPAPLRKVDAASRS 296
Score = 35.8 bits (81), Expect = 0.11
Identities = 43/183 (23%), Positives = 65/183 (35%), Gaps = 24/183 (13%)
Query: 603 PPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSS 662
P SR+++ + R + + R ER KDRD R S S
Sbjct: 145 PRRRSRDDYREKDYRKRSRSRSYDRRERHEEKDRDHRRRTRS----------------RS 188
Query: 663 ASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXH 722
AS + R R + SH P +R S ++ R+ R +R
Sbjct: 189 ASPDEKRRVRGRYDNESRSHSRSLSASPARRSPRSSSPQKTSPAREVSPDKRSNERSPSP 248
Query: 723 DRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTT 782
R R +AS K P R+S+E +S S S P+ K + S++
Sbjct: 249 RRSLSPRSPALQKASPSKEMSP-----ERRSNE---RSPSPGSPAPLRKVDAASRSQSPY 300
Query: 783 SAE 785
+AE
Sbjct: 301 AAE 303
>At3g23900 unknown protein
Length = 989
Score = 50.8 bits (120), Expect = 3e-06
Identities = 66/284 (23%), Positives = 113/284 (39%), Gaps = 24/284 (8%)
Query: 619 RRKRENERRVE-----RDFSKDRDFGREVSSVGDVS-SIKSKTKPIPPSS--ASDYHQNR 670
RR R R VE +D S+D + S +SKT+ +S A H+ R
Sbjct: 653 RRTRSRSRSVEDSADIKDKSRDEELKHHKKRSRSRSREDRSKTRDTSRNSDEAKQKHRQR 712
Query: 671 HRSERPSPDS-SHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRR 729
RS D+ SH V+ + S HS+R + D DYD +R R +
Sbjct: 713 SRSRSLENDNGSHENVDVAQDNDLNSRHSKRRSKSLDEDYDMKERR-----GRSRSRSLE 767
Query: 730 HHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAA 789
+R+ +K D T S R+S ++ K + E + S K + S + +
Sbjct: 768 TKNRSSRKNKLDEDRNTGSRRRRSRSKSVEGKRSYNKE----TRSRDKKSKRRSGRRSRS 823
Query: 790 AAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSN 849
+++ KQ + S + E++ ++ ++ S S + E ++ S + S
Sbjct: 824 PSSEGKQGRDIRSSPGYSDEKK----SRHKRHSRSRSIEKKNSSRDKRSKRHERLRSSSP 879
Query: 850 GGRKSKAVMD-DYESSDDDEDDERHFKRRPSRYEP-SPPPPVDD 891
G K + SS+D + +RH + + +P S VDD
Sbjct: 880 GRDKRRGDRSLSPVSSEDHKIKKRHSGSKSVKEKPHSDYEKVDD 923
Score = 50.1 bits (118), Expect = 6e-06
Identities = 68/331 (20%), Positives = 105/331 (31%), Gaps = 29/331 (8%)
Query: 583 PIPPPPHRDLAEFSMGMNVPPPAMSREEFEARKADARRKRENERRVERDFSKDRDFGR-- 640
PI R +S P SR ++ R R RD S+ R +GR
Sbjct: 524 PISYRRRRRSPTYSPPFRRPRSHRSRSPLRYQRRSTYEGRRRSYRDSRDISESRRYGRSD 583
Query: 641 --EVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPR------PTK 692
SS S+ K + + R S R SS PR T
Sbjct: 584 EHHSSSSRRSRSVSPKKRKSGQEDSELSRLRRDSSSRGEKKSSRAGSRSPRRRKEVKSTP 643
Query: 693 RKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRK 752
R + ++ +R R R D +D D + +HH + S + T+ TSR
Sbjct: 644 RDDEENKVKRRTRSRSRSVEDSADIKDKSRD--EELKHHKKRSRSRSREDRSKTRDTSRN 701
Query: 753 SSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEA 812
S E K + R S + + + + R+ K S +E
Sbjct: 702 SDEAKQKHRQRSRSRSLENDNGSHENVDVAQDNDLNSRHSKRRSK----------SLDED 751
Query: 813 AAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDER 872
++R S S + E ++ T +SK+V + + ++
Sbjct: 752 YDMKERRGRSRSRSLETKNRSSRKNKLDEDRNTGSRRRRSRSKSVEGKRSYNKETRSRDK 811
Query: 873 HFKRRPSRYEPSPPPPVDDWVEEGRHSRGTR 903
KRR R SP EG+ R R
Sbjct: 812 KSKRRSGRRSRSPS-------SEGKQGRDIR 835
Score = 48.9 bits (115), Expect = 1e-05
Identities = 66/274 (24%), Positives = 95/274 (34%), Gaps = 14/274 (5%)
Query: 613 ARKADARRKRENERRVERD-FSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRH 671
A + A R E R++ D D + S S +S++K P S ++ R
Sbjct: 476 ATELAAARAAEISRKLRPDGVGNDEKEADQKSRSPSKSPARSRSKSKSPIS----YRRRR 531
Query: 672 RSERPSPD----SSHREVEPPRPTKRKSDHSEREREDRD-RDYDYHDRHQDRDYXHDRHQ 726
RS SP SHR P R +R+S + R R RD RD R+ D H
Sbjct: 532 RSPTYSPPFRRPRSHRSRSPLR-YQRRSTYEGRRRSYRDSRDISESRRYGRSDEHHSSSS 590
Query: 727 HRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEA 786
R + D R SS K SR S + + T
Sbjct: 591 RRSRSVSPKKRKSGQEDSELSRLRRDSSSRGEKKSSRAGSRSPRRRKEVKSTPRDDEENK 650
Query: 787 AAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTH 846
R + + I S +E KKR S S + T T+ S +A
Sbjct: 651 VKRRTRSRSRSVEDSADIKDKSRDEELKHHKKRSRSRSREDRSKTRDTSRNSDEAKQKHR 710
Query: 847 LSNGGRKSKAVMDDYESSD---DDEDDERHFKRR 877
+ R + +E+ D D++ + RH KRR
Sbjct: 711 QRSRSRSLENDNGSHENVDVAQDNDLNSRHSKRR 744
Score = 35.8 bits (81), Expect = 0.11
Identities = 29/158 (18%), Positives = 56/158 (35%), Gaps = 4/158 (2%)
Query: 617 DARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERP 676
++R + + ++ D+ GR S + + S+ + + + R RS+
Sbjct: 737 NSRHSKRRSKSLDEDYDMKERRGRSRSRSLETKNRSSRKNKLDEDRNTGSRRRRSRSKSV 796
Query: 677 SPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEA 736
S+ + R K K R R D Y ++ + H R+ +
Sbjct: 797 EGKRSYNKETRSRDKKSKRRSGRRSRSPSSEGKQGRDIRSSPGYSDEKKSRHKRHSRSRS 856
Query: 737 SSKKSTDPVTKSTS----RKSSEPDIKSKSRKSSEPVT 770
KK++ +S R SS K + +S PV+
Sbjct: 857 IEKKNSSRDKRSKRHERLRSSSPGRDKRRGDRSLSPVS 894
>At2g32950 photomorphogenesis repressor (COP1)
Length = 675
Score = 50.4 bits (119), Expect = 4e-06
Identities = 24/59 (40%), Positives = 36/59 (60%), Gaps = 2/59 (3%)
Query: 295 DLPPELHCPLCSNVMKDAVLTSKCCFKSFCDKCIRDYIISKSMCVCGAMNVLADDLLPN 353
DL +L CP+C ++KDA LT+ C SFC CI ++ +KS C C + ++ + L PN
Sbjct: 45 DLDKDLLCPICMQIIKDAFLTA--CGHSFCYMCIITHLRNKSDCPCCSQHLTNNQLYPN 101
>At2g28910 unknown protein
Length = 332
Score = 48.9 bits (115), Expect = 1e-05
Identities = 44/176 (25%), Positives = 74/176 (42%), Gaps = 15/176 (8%)
Query: 607 SREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDY 666
S EE E+ +D+ E ER + F K + SSV SS++ K K + S SD
Sbjct: 138 SEEEEESESSDSDVDSEMERIIAERFGKKKGG----SSVKKTSSVRKKKKRVSDESDSDS 193
Query: 667 HQNRHRSERPSPD--SSHR-------EVEPPRPTKRKSDHSERERE--DRDRDYDYHDRH 715
+ R S SSH+ E E +KR+ + R+R+ D D D DR
Sbjct: 194 DSGDRKRRRRSMKKRSSHKRRSLSESEDEEEGRSKRRKERRGRKRDEDDSDESEDEDDRR 253
Query: 716 QDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTK 771
R ++ + R + ++ S +S++ + R S++ S + V++
Sbjct: 254 VKRKSRKEKRRRRSRRNHSDDSDSESSEDDRRQKRRNKVAASSDSEANVSGDDVSR 309
Score = 33.1 bits (74), Expect = 0.73
Identities = 49/215 (22%), Positives = 66/215 (29%), Gaps = 33/215 (15%)
Query: 698 SEREREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPD 757
SE E E D D D +R R + SS K T V K R S E D
Sbjct: 138 SEEEEESESSDSDV-------DSEMERIIAERFGKKKGGSSVKKTSSVRKKKKRVSDESD 190
Query: 758 IKSKS--RKSSEPVTKSLSLSKTAPTTSAEAAAAAAADRKQKASVFSRISFPSEEEAAAA 815
S S RK K S K + +E EEE +
Sbjct: 191 SDSDSGDRKRRRRSMKKRSSHKRRSLSESE----------------------DEEEGRSK 228
Query: 816 AKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDERHFK 875
+K + + S + + + R+S+ D S+ EDD R +
Sbjct: 229 RRKERRGRKRDEDDSDESEDEDDRRVKRKSRKEKRRRRSRRNHSDDSDSESSEDDRRQKR 288
Query: 876 RRPSRYEPSPPPPV--DDWVEEGRHSRGTRDRKHR 908
R V DD GR S ++K R
Sbjct: 289 RNKVAASSDSEANVSGDDVSRVGRGSSKRSEKKSR 323
>At3g28770 hypothetical protein
Length = 2081
Score = 48.1 bits (113), Expect = 2e-05
Identities = 29/170 (17%), Positives = 77/170 (45%), Gaps = 10/170 (5%)
Query: 608 REEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYH 667
+++ E++ ++ ++K E+++ + K ++ ++ ++ + S +K + K S+
Sbjct: 946 KKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDS 1005
Query: 668 QNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDY------- 720
+++R ++ + + E + K+KS +RE +D + ++ + RD
Sbjct: 1006 ASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEE 1065
Query: 721 -XHDRHQHRRHHHRTEASSKKSTD--PVTKSTSRKSSEPDIKSKSRKSSE 767
++ + H + + K+ D + K +K + +SKSRK E
Sbjct: 1066 ETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEE 1115
Score = 37.4 bits (85), Expect = 0.039
Identities = 55/334 (16%), Positives = 113/334 (33%), Gaps = 29/334 (8%)
Query: 600 NVPPPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDV----------- 648
+V A E + ++ + +R ++ + RDF+ + D + S V
Sbjct: 866 SVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSGESVKYKKDEKKEGN 925
Query: 649 ---------SSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSE 699
+S K K K + N + E + + E++ K+++ SE
Sbjct: 926 KEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSE 985
Query: 700 REREDRDRDYDYHDRHQDRDYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDI- 758
+ ++ + D ++ + D + + + + + +++ KS +K E D
Sbjct: 986 NSKL-KEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSE 1044
Query: 759 --KSKSRKSSEPVTKSLSLSKTAPTT--SAEAAAAAAADRKQKASVFSRISFPSEEEAAA 814
KSK K ++ L K T E+ + ++ K S EE+
Sbjct: 1045 ERKSKKEKEE---SRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKE 1101
Query: 815 AAKKRKLSASSTTEASTAATASVSAKAPSTTHLSNGGRKSKAVMDDYESSDDDEDDERHF 874
K + + E + N +KS+ V + SD E E
Sbjct: 1102 KKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEE 1161
Query: 875 KRRPSRYEPSPPPPVDDWVEEGRHSRGTRDRKHR 908
K E S + +E + S+ + +K +
Sbjct: 1162 KSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEK 1195
Score = 34.7 bits (78), Expect = 0.25
Identities = 43/263 (16%), Positives = 102/263 (38%), Gaps = 22/263 (8%)
Query: 609 EEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ 668
EE ++RK + K++ E+ +++ +K ++ E V +K + S + +
Sbjct: 1106 EESKSRKKE-EDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKE------SDKKEKKE 1158
Query: 669 NRHRSERPSPDSSH---REVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRH 725
N +SE +SS EV+ K ++E+E ++ + ++++
Sbjct: 1159 NEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSV 1218
Query: 726 QHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAE 785
+ + T+ K D +T + + + K +E KS + ++ A + ++
Sbjct: 1219 EENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQ-ADSDESK 1277
Query: 786 AAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPSTT 845
AD S+ S+ +A + K ++ + ++A+T K ++
Sbjct: 1278 NEILMQAD--------SQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSV 1329
Query: 846 HLSNGGRKSKAVMDDYESSDDDE 868
+K K ++ DD+
Sbjct: 1330 ---AENKKQKETKEEKNKPKDDK 1349
Score = 29.6 bits (65), Expect = 8.0
Identities = 42/264 (15%), Positives = 97/264 (35%), Gaps = 35/264 (13%)
Query: 610 EFEARKADARRKRENERRVERDFSKDR-----DFGREVSSVGDVSSIKSKTKPIPPSSAS 664
E E+++A+ ++K + + + D SK+ D + S S +SK + + + +
Sbjct: 1253 ESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQ 1312
Query: 665 DYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDR 724
Q + +R +S E + + TK + + + ++++ + + +
Sbjct: 1313 ATTQRNNEEDRKK-QTSVAENKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAE 1371
Query: 725 HQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSA 784
+Q + S + + + ++ S+ S D ++ S +S + T +
Sbjct: 1372 NQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNE 1431
Query: 785 EAAAAAAADRKQKASVFSRISFPSEEEAAAAAKKRKLSASSTTEASTAATASVSAKAPST 844
E DRK++ SV ++E K K +TTE
Sbjct: 1432 E-------DRKKQTSVAEN---KKQKETKEEKNKPKDDKKNTTE---------------- 1465
Query: 845 THLSNGGRKSKAVMDDYESSDDDE 868
+GG+K + E+ + +
Sbjct: 1466 ---QSGGKKESMESESKEAENQQK 1486
>At3g04160 hypothetical protein
Length = 712
Score = 48.1 bits (113), Expect = 2e-05
Identities = 53/209 (25%), Positives = 87/209 (41%), Gaps = 37/209 (17%)
Query: 607 SREEFEARKADARRKRENER-----RVERDFSKD--RDFGREVSSVGDVSSIKSKTKPIP 659
+REE A + D +R+R + R R R D ++ E+ G + + K +P
Sbjct: 504 TREELLAEERDYKRRRMSYRGKKVKRTPRQVLHDMIEEYTEEIKLAGGIGCFE---KGMP 560
Query: 660 PSSASDYHQNRHRSER----PSPDSSHR-----EVEPPRPTKRKSDHSER---------E 701
S S ++ S+ PS D + ++E P ++ SD +R +
Sbjct: 561 LQSRSPIGNDQKESDFGYSIPSTDKQWKGENRADIEYPIDNRQNSDKVKRHDEYDSGSSQ 620
Query: 702 REDRDRDYDYHDRHQD--RDYXHDRHQHRRHHH--RTEASSKKSTDPVTKSTSRKSSEPD 757
R+ R Y + DR D RD D+H RR RT+ S + +SR+ S D
Sbjct: 621 RQQSHRSYKHSDRRDDKLRDRRKDKHNDRRDDEFTRTKRHSIEGESYQNYRSSREKSSSD 680
Query: 758 IKSK-----SRKSSEPVTKSLSLSKTAPT 781
K+K R+S +P ++L + PT
Sbjct: 681 YKTKRDDPYDRRSQQPRNQNLFEDRYIPT 709
>At3g09620 putative RNA helicase
Length = 989
Score = 47.8 bits (112), Expect = 3e-05
Identities = 57/255 (22%), Positives = 105/255 (40%), Gaps = 33/255 (12%)
Query: 618 ARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPS 677
++ ++EN+R KDRD ++ + D + ++S+ K + + + R R +R S
Sbjct: 5 SKSRKENDR-------KDRDRSKKENGRRDTTEMRSRVKRC---DSEEEERIRIRRDRKS 54
Query: 678 PDSSHREVEPPRPTKRKSDHSEREREDRDRD---YDYHDRHQDRDYXHDRHQHRRHHHRT 734
D E E R +KR+ + R R +RDRD Y DR + R+ ++ + ++ R+
Sbjct: 55 SDFEEEEYE--RDSKRRGEDKGRGRRERDRDRGKYLKRDRER-REREKEKGRKKQKKERS 111
Query: 735 EASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAAAAAADR 794
+ +D V RK +E +SR + V K + + A R
Sbjct: 112 REDCNEESDDVKCGLKRKRTE-----RSRHGDDDVEKKTRDEQV--EDEQKQLAEEVEKR 164
Query: 795 KQKASVFSRISFPSEE-----EAAAAAKKRKLSASSTTEASTAATASVSAKAPSTTHLSN 849
+++ + + +EE + K L S E + + V T L N
Sbjct: 165 RRRVQEWQELKRQNEEAQIESKGPETGKAWTLDGESDDEVKSDSEMDVD----RDTKLEN 220
Query: 850 GGRKSKAVMDDYESS 864
GG +K V + E++
Sbjct: 221 GG-DAKMVASENETA 234
>At1g20920 putative RNA helicase
Length = 1166
Score = 47.4 bits (111), Expect = 4e-05
Identities = 44/169 (26%), Positives = 69/169 (40%), Gaps = 12/169 (7%)
Query: 609 EEFEARKADARRKRENE-----RRVERDFSKDRDFGREVSSVGDVSSIKSKTK---PIPP 660
EE E RK R +R + RR ER S D + E D + K +
Sbjct: 69 EEREKRKEKERERRRRDKDRVKRRSERRKSSDSEDDVEEEDERDKRRVNEKERGHREHER 128
Query: 661 SSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSER--EREDRDRDYDYHDRHQDR 718
D ++R R ER + + R +R+ ER ERE R+R+ DR ++R
Sbjct: 129 DRGKDRKRDREREERKDKEREREKDRERREREREEREKERVKERERREREDGERDR-RER 187
Query: 719 DYXHDRHQHRRHHHRTEASSKKSTDPVTKSTSRKSSE-PDIKSKSRKSS 766
+ ++R E +++S D V + R+ E + K K R+ S
Sbjct: 188 EKERGSRRNRERERSREVGNEESDDDVKRDLKRRRKEGGERKEKEREKS 236
Score = 46.2 bits (108), Expect = 8e-05
Identities = 40/166 (24%), Positives = 65/166 (39%), Gaps = 15/166 (9%)
Query: 614 RKADARRKRENERRVERDFSKDRDFGREVSSVG-------DVSSIKSKTKPIPPS-SASD 665
R+ D R+KR E D+ +D D RE D +K +++ S S D
Sbjct: 45 REKDRRKKRVKSSDSEDDYDRDDDEEREKRKEKERERRRRDKDRVKRRSERRKSSDSEDD 104
Query: 666 YHQNRHRSERP--SPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHD 723
+ R +R + HRE E R RK D ER+D++R +R +DR+
Sbjct: 105 VEEEDERDKRRVNEKERGHREHERDRGKDRKRDREREERKDKER-----EREKDRERRER 159
Query: 724 RHQHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPV 769
+ R E ++ D R+ +++ R+ S V
Sbjct: 160 EREEREKERVKERERREREDGERDRREREKERGSRRNRERERSREV 205
Score = 41.6 bits (96), Expect = 0.002
Identities = 57/282 (20%), Positives = 106/282 (37%), Gaps = 29/282 (10%)
Query: 609 EEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ 668
EE + +K+ R R NER+ ++ K R+ R K + K D
Sbjct: 18 EEADLKKSRRDRDRSNERKKDKGSEKRREKDRR----------KKRVKSSDSEDDYDRDD 67
Query: 669 NRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQHR 728
+ R +R + R + R KR+S+ R+ D + D + D +D+ +++ +
Sbjct: 68 DEEREKRKEKERERRRRDKDR-VKRRSER--RKSSDSEDDVEEEDE-RDKRRVNEKERGH 123
Query: 729 RHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKSLSLSKTAPTTSAEAAA 788
R H R +K D + K E + K + R+ E + K E
Sbjct: 124 REHERDRGKDRK-RDREREERKDKERERE-KDRERREREREEREKERVKERERREREDGE 181
Query: 789 AAAADR-KQKASVFSRI--------SFPSEEEAAAAAKKRKLSASSTTEASTAATASVSA 839
+R K++ S +R + S+++ K+R+ E + S+
Sbjct: 182 RDRREREKERGSRRNRERERSREVGNEESDDDVKRDLKRRRKEGGERKEKEREKSVGRSS 241
Query: 840 K---APSTTHLSNGGRKSKAVMDDYESSDDDED-DERHFKRR 877
+ +P + + G K + + E D+ + DE KRR
Sbjct: 242 RHEDSPKRKSVEDNGEKKEKKTREEELEDEQKKLDEEVEKRR 283
Score = 39.3 bits (90), Expect = 0.010
Identities = 60/298 (20%), Positives = 106/298 (35%), Gaps = 33/298 (11%)
Query: 608 REEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYH 667
R E E + D RR+RE ER R+ ++R+ REV + +K K
Sbjct: 174 RREREDGERD-RREREKERGSRRN--RERERSREVGNEESDDDVKRDLKRRRKEGGERKE 230
Query: 668 QNRHRSERPS---PDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDR 724
+ R +S S DS R+ K++ E E ED + D + + R +
Sbjct: 231 KEREKSVGRSSRHEDSPKRKSVEDNGEKKEKKTREEELEDEQKKLD-EEVEKRRRRVQEW 289
Query: 725 HQHRRHHHRTEASSK------------------KSTDPVTKSTSRKSSEPDIKSKSRKSS 766
+ +R E+ SK +S D + +E D+ +++ +
Sbjct: 290 QELKRKKEEAESESKGDADGNEPKAGKAWTLEGESDDEEGHPEEKSETEMDVDEETKPEN 349
Query: 767 EPVTKSLSLSKTAPTTSAEAAAAAAADRKQ---KASVFSRISFPSEEEAAAAAKKRKLSA 823
+ K + L T +E+ A D ++ + + + P E+ A ++
Sbjct: 350 DGDAKMVDLENETAATVSESGGDGAVDEEEIDPLDAFMNTMVLPEVEKFCNGAPPPAVND 409
Query: 824 SSTTEASTAATASVSAKAPSTTHLSN--GGRKSKAVMDDYESSDD---DEDDERHFKR 876
+ + K L G S + + ++ DD DEDDE KR
Sbjct: 410 GTLDSKMNGKESGDRPKKGFNKALGRIIQGEDSDSDYSEPKNDDDPSLDEDDEEFMKR 467
>At2g43370 U1 small nuclear ribonucleoprotein 70 kDa like protein
Length = 333
Score = 46.6 bits (109), Expect = 6e-05
Identities = 47/176 (26%), Positives = 69/176 (38%), Gaps = 35/176 (19%)
Query: 583 PIPPPPHRDLAEFSMGMNVPPPA--MSREEFEARKADARRKRENERRVERDFSKDRDFGR 640
P+ P PH DL + +G+ +PP MSR + + RRK R E ++ R
Sbjct: 178 PLRPIPHEDLKK--LGIQLPPEGRYMSRTQIPS---PPRRKGSVSDREE-------EYYR 225
Query: 641 EVSSVGDVSSIKSKTKPIPPSSASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSER 700
E SSV K + SS YH HRS + S R + R+ S+R
Sbjct: 226 EKSSVEREEEFKER------SSLRSYHS--HRSSAHTHSSHRRRSKDREECSREESRSDR 277
Query: 701 EREDRDRDYDYHD-----------RHQDRDYXHDRHQH--RRHHHRTEASSKKSTD 743
+ R + Y D + + D RH+H HH R+ + S+D
Sbjct: 278 KERARGMEDRYGDNKGEVSGSKRSKRSEEDRSRKRHKHLPSHHHRRSYSQDHHSSD 333
>At1g44910 splicing factor like protein
Length = 958
Score = 46.6 bits (109), Expect = 6e-05
Identities = 39/157 (24%), Positives = 68/157 (42%), Gaps = 13/157 (8%)
Query: 623 ENERRVERDFSKDRDFGREVSSVG----DVSSIKSKTKPIPPSSASD-YHQNRHRSE--- 674
E+ +++ + + R G E S G ++S++ K K + + + R E
Sbjct: 770 EDSKQLVEESQEYRSIGDESVSQGLFEEYITSLQEKAKEKERKRDEEKVRKEKERDEKEK 829
Query: 675 RPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQD---RDYXHDRHQHRRHH 731
R D RE E R ++ + S+RE D + D + H+D + DR RRHH
Sbjct: 830 RKDKDKERREKEREREKEKGKERSKREESDGETAMDVSEGHKDEKRKGKDRDRKHRRRHH 889
Query: 732 HRT--EASSKKSTDPVTKSTSRKSSEPDIKSKSRKSS 766
+ + + SS + +K +SRK KS+ +S
Sbjct: 890 NNSDEDVSSDRDDRDESKKSSRKHGNDRKKSRKHANS 926
Score = 45.1 bits (105), Expect = 2e-04
Identities = 34/152 (22%), Positives = 62/152 (40%), Gaps = 23/152 (15%)
Query: 608 REEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYH 667
R+E E RK + +RE ER E++ K+R SK + +A D
Sbjct: 824 RDEKEKRKDKDKERREKEREREKEKGKER----------------SKREESDGETAMDVS 867
Query: 668 QNRHRSERPSPDSSHREVEPPRPTKRKSDHSEREREDRDRDYDYHDRHQDRDYXHDRHQH 727
+ +R D + +R+ ++ E DRD + R + +DR +
Sbjct: 868 EGHKDEKRKGKDRDRKH-------RRRHHNNSDEDVSSDRDDRDESKKSSRKHGNDRKKS 920
Query: 728 RRHHHRTEASSKKSTDPVTKSTSRKSSEPDIK 759
R+H + E+ S+ K +SR+S +++
Sbjct: 921 RKHANSPESESENRHKRQKKESSRRSGNDELE 952
>At5g55100 unknown protein
Length = 844
Score = 46.2 bits (108), Expect = 8e-05
Identities = 48/190 (25%), Positives = 71/190 (37%), Gaps = 33/190 (17%)
Query: 616 ADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKT-----------KPIPPSSAS 664
++ R KR R +RD + G E S D + ++KT K S+
Sbjct: 611 SERRSKRNYRSRSQRDEDGKMEQGEEEESSMDEVTEETKTDKKHSCSRKRHKHKTRYSSK 670
Query: 665 DYH-QNRHRSERPSPDSSHREVEPPRPTKRKSDH------SEREREDRDR------DYDY 711
D H +++H+ E S D H + SD S+ E E R R D DY
Sbjct: 671 DRHSRDKHKHESSSDDEYHSRSRHRHRHSKSSDRHELYDSSDNEGEHRHRSSKHSKDVDY 730
Query: 712 HD---RHQDRDYXHDRH------QHRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKSKS 762
H R H++H +H H HR+ + + V KSS+ K +
Sbjct: 731 SKDKRSHHHRSRKHEKHRDSSDDEHHHHRHRSSRRKHEDSSDVEHGHRHKSSKRIKKDEK 790
Query: 763 RKSSEPVTKS 772
E V+KS
Sbjct: 791 TVEEETVSKS 800
Score = 44.7 bits (104), Expect = 2e-04
Identities = 39/158 (24%), Positives = 65/158 (40%), Gaps = 22/158 (13%)
Query: 618 ARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSSASDYHQ--------- 668
+R++ +++ R RD + SS D +S+ + S +SD H+
Sbjct: 657 SRKRHKHKTRYSSKDRHSRDKHKHESSSDDEYHSRSRHRH-RHSKSSDRHELYDSSDNEG 715
Query: 669 -NRHRSERPSPDSSHREVEPPRPTKRKSDHSERERE---DRDRDYDYHDRHQDRDYXHDR 724
+RHRS + S D + + KR H R+ E D D +H RH+ H+
Sbjct: 716 EHRHRSSKHSKDVDYSK------DKRSHHHRSRKHEKHRDSSDDEHHHHRHRSSRRKHED 769
Query: 725 HQ--HRRHHHRTEASSKKSTDPVTKSTSRKSSEPDIKS 760
H H++ KK V + T KS + D+K+
Sbjct: 770 SSDVEHGHRHKSSKRIKKDEKTVEEETVSKSDQSDLKA 807
Score = 30.0 bits (66), Expect = 6.1
Identities = 36/156 (23%), Positives = 56/156 (35%), Gaps = 30/156 (19%)
Query: 642 VSSVGDVSSIKSKTKPIP---PSSASDYHQNRHRSERPSPDSSHREV------------- 685
+S +G ++ + + IP S +D + RS+R S R+
Sbjct: 581 ISGLGAKAAKERDSSSIPYVAESKLADDGNSERRSKRNYRSRSQRDEDGKMEQGEEEESS 640
Query: 686 --EPPRPTKRKSDHS-EREREDRDRDYDYHDRHQDRDYXHDR------HQHRRHHHRTEA 736
E TK HS R+R Y DRH + H+ H RH HR
Sbjct: 641 MDEVTEETKTDKKHSCSRKRHKHKTRYSSKDRHSRDKHKHESSSDDEYHSRSRHRHRHSK 700
Query: 737 SSKKSTDPVTKSTSRKSSEPDIKSKSRKSSEPVTKS 772
SS + + +E + + +S K S+ V S
Sbjct: 701 SSDRH-----ELYDSSDNEGEHRHRSSKHSKDVDYS 731
>At5g53800 unknown protein
Length = 339
Score = 46.2 bits (108), Expect = 8e-05
Identities = 42/183 (22%), Positives = 69/183 (36%), Gaps = 22/183 (12%)
Query: 603 PPAMSREEFEARKADARRKRENERRVERDFSKDRDFGREVSSVGDVSSIKSKTKPIPPSS 662
PP ++ E E R + RE +D +++D R S K +TK S
Sbjct: 9 PPENTKREVEDRDIRRKSSREKPSGSGKDSGEEKDVSRRRES-------KRRTKDGNDSG 61
Query: 663 ASDYHQNRHRSERPSPDSSHREVEPPRPTKRKSDHSERERED---------RDRDYDYHD 713
+ ++ SE+ S ++ R + RKS S R R D + + +Y D
Sbjct: 62 SESGLESGSESEKEERRRSRKD-RGKRKSDRKSSRSRRRRRDYSSSSSDSESESESEYSD 120
Query: 714 RHQDRDYXHDRHQHRRHHHRTEASS-----KKSTDPVTKSTSRKSSEPDIKSKSRKSSEP 768
+ R + R+ R E ++ D ++ S K + K K +K SE
Sbjct: 121 SEESESEDERRRRKRKRKEREEEEKERKRRRREKDKKKRNKSDKDGDKKRKEKKKKKSEK 180
Query: 769 VTK 771
V K
Sbjct: 181 VKK 183
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.130 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,059,191
Number of Sequences: 26719
Number of extensions: 1161822
Number of successful extensions: 9349
Number of sequences better than 10.0: 427
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 311
Number of HSP's that attempted gapping in prelim test: 6608
Number of HSP's gapped (non-prelim): 1977
length of query: 908
length of database: 11,318,596
effective HSP length: 108
effective length of query: 800
effective length of database: 8,432,944
effective search space: 6746355200
effective search space used: 6746355200
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC119408.8


