

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.7 + phase: 0 /pseudo
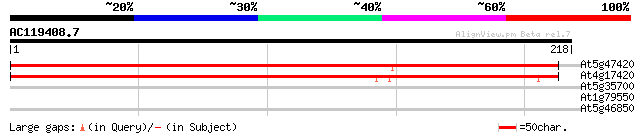
(218 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g47420 unknown protein 336 5e-93
At4g17420 unknown protein 317 2e-87
At5g35700 fimbrin 29 1.7
At1g79550 phosphoglycerate kinase (EC 2.7.2.3) like protein 29 2.2
At5g46850 unknown protein 27 6.4
>At5g47420 unknown protein
Length = 282
Score = 336 bits (862), Expect = 5e-93
Identities = 163/214 (76%), Positives = 188/214 (87%), Gaps = 1/214 (0%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQQD I PFQILGGESQVVQIMLK EK+IAKP SMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQQDTITPFQILGGESQVVQIMLKSEEKVIAKPASMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
MEN Y PE EVG+ QW+ GK+V+++V+RN+G +DGFVGIAAPY ARILPIDLA F GEIL
Sbjct: 61 MENTYTPEQEVGVLQWILGKSVSSVVLRNTGQNDGFVGIAAPYLARILPIDLAMFGGEIL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQRGRNVV-AGAEVFLRQKLSGQGLAFILGGGSVVQKILEV 179
CQPDAFLCSV+DVKV N+VDQR RN+V AGAE FLRQ+LSGQGLAFIL GGSVVQK+LEV
Sbjct: 121 CQPDAFLCSVHDVKVVNSVDQRARNIVAAGAEGFLRQRLSGQGLAFILAGGSVVQKVLEV 180
Query: 180 GEVLAVDVSCIVAVTSTVDIQIKYNGPARRTMFG 213
GEV ++DVSCI A+T ++D++IK N P RR +FG
Sbjct: 181 GEVFSIDVSCIAALTPSIDVRIKNNAPFRRALFG 214
>At4g17420 unknown protein
Length = 285
Score = 317 bits (813), Expect = 2e-87
Identities = 159/217 (73%), Positives = 185/217 (84%), Gaps = 4/217 (1%)
Query: 1 MAAPFFSTPFQPYVYQSQQDAIIPFQILGGESQVVQIMLKPHEKIIAKPGSMCFMSGSVE 60
MAAPFFSTPFQPYVYQSQ+D I PFQILGGE+QVVQIMLKP EK+IAKPGSMC+MSGS+E
Sbjct: 1 MAAPFFSTPFQPYVYQSQEDTITPFQILGGEAQVVQIMLKPQEKVIAKPGSMCYMSGSIE 60
Query: 61 MENAYLPENEVGIWQWLFGKTVTNIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEIL 120
M+N Y PE EVG+ QW+ GK+V++IV+RN+G +DGFVGIAAP ARILP+DLA F G+IL
Sbjct: 61 MDNTYTPEQEVGVVQWILGKSVSSIVLRNTGQNDGFVGIAAPSLARILPLDLAMFGGDIL 120
Query: 121 CQPDAFLCSVNDVKVSNTVDQ--RGRNV-VAGAEVFLRQKLSGQGLAFILGGGSVVQKIL 177
CQPDAFLCSV+DVKV NTV Q R RN+ AGAE LRQ+LSGQGLAFI+ GGSVVQK L
Sbjct: 121 CQPDAFLCSVHDVKVVNTVYQRHRARNIAAAGAEGVLRQRLSGQGLAFIIAGGSVVQKNL 180
Query: 178 EVGEVLAVDVSCIVAVTSTVDIQIKYN-GPARRTMFG 213
EVGEVL +DVSCI A+T +++ QIKYN P RR +FG
Sbjct: 181 EVGEVLTIDVSCIAALTPSINFQIKYNAAPVRRAVFG 217
>At5g35700 fimbrin
Length = 687
Score = 29.3 bits (64), Expect = 1.7
Identities = 16/44 (36%), Positives = 22/44 (49%)
Query: 102 PYFARILPIDLATFNGEILCQPDAFLCSVNDVKVSNTVDQRGRN 145
P+ LPID AT L + LC + +V V T+D+R N
Sbjct: 140 PFLKSYLPIDPATNAFFDLVKDGVLLCKLINVAVPGTIDERAIN 183
>At1g79550 phosphoglycerate kinase (EC 2.7.2.3) like protein
Length = 401
Score = 28.9 bits (63), Expect = 2.2
Identities = 15/35 (42%), Positives = 22/35 (62%), Gaps = 2/35 (5%)
Query: 148 AGAEVFLRQ--KLSGQGLAFILGGGSVVQKILEVG 180
AG E +Q +LSG+G+ I+GGG V + +VG
Sbjct: 334 AGTEAVAKQLAELSGKGVTTIIGGGDSVAAVEKVG 368
>At5g46850 unknown protein
Length = 296
Score = 27.3 bits (59), Expect = 6.4
Identities = 17/64 (26%), Positives = 31/64 (47%), Gaps = 3/64 (4%)
Query: 84 NIVVRNSGPSDGFVGIAAPYFARILPIDLATFNGEILCQPDAFLCSVNDVKVSNTVDQRG 143
++ + + +G + P AR P+D + ++ +PD FLCS + V N +R
Sbjct: 174 HVEISSRNYENGEISFKLPLHARYQPLDDSGYSRVEFGEPDLFLCSSH---VQNQRRERR 230
Query: 144 RNVV 147
R +V
Sbjct: 231 RCLV 234
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,689,483
Number of Sequences: 26719
Number of extensions: 187706
Number of successful extensions: 345
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 339
Number of HSP's gapped (non-prelim): 5
length of query: 218
length of database: 11,318,596
effective HSP length: 95
effective length of query: 123
effective length of database: 8,780,291
effective search space: 1079975793
effective search space used: 1079975793
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC119408.7


