

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119408.12 + phase: 0 /partial
(467 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
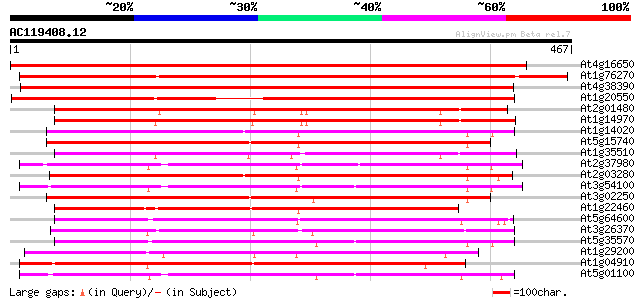
Score E
Sequences producing significant alignments: (bits) Value
At4g16650 growth regulator like protein 715 0.0
At1g76270 putative auxin-independent growth promoter (At1g76270) 538 e-153
At4g38390 putative growth regulator protein 528 e-150
At1g20550 hypothetical protein 463 e-131
At2g01480 similar to axi 1 protein from Nicotiana tabacum 326 2e-89
At1g14970 auxin-independent growth promoter, putative 324 6e-89
At1g14020 growth regulator like protein 308 5e-84
At5g15740 unknown protein 303 1e-82
At1g35510 putative growth regulator protein (At1g35510) 303 2e-82
At2g37980 similar to axi 1 protein from Nicotiana tabacum 301 4e-82
At2g03280 axi 1 -like protein 301 7e-82
At3g54100 unknown protein 298 4e-81
At3g02250 putative auxin-independent growth promoter 296 2e-80
At1g22460 Similar to auxin-independent growth promoter 296 2e-80
At5g64600 auxin-independent growth promoter-like protein 295 3e-80
At3g26370 unknown protein 288 3e-78
At5g35570 unknown protein 285 3e-77
At1g29200 hypothetical protein 276 1e-74
At1g04910 Similar to auxin-independent growth promoter (axi 1) 276 2e-74
At5g01100 unknown protein 265 4e-71
>At4g16650 growth regulator like protein
Length = 549
Score = 715 bits (1846), Expect = 0.0
Identities = 338/431 (78%), Positives = 389/431 (89%), Gaps = 1/431 (0%)
Query: 1 DGNDHAPIDIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDA 60
D + PID+W+S+YSK++YGC ERGR+F PA++E+ SNGYLLIAASGGLNQQRTGITDA
Sbjct: 85 DMSRREPIDVWKSKYSKFFYGCSERGRNFLPAVQEQSSNGYLLIAASGGLNQQRTGITDA 144
Query: 61 VVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDKVMRSMD 120
VVVARILNATLVVPELDH+S+WKDDSDF++IFDVNWFI+ LAKDVTIVKRVPD+VMR+M+
Sbjct: 145 VVVARILNATLVVPELDHHSYWKDDSDFSDIFDVNWFISSLAKDVTIVKRVPDRVMRAME 204
Query: 121 KPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLANDLDDELQKLRCRVNYHAL 180
KPPYT RVPRKS EYYLDQVLPIL RR VLQLTKFDYRLANDLD+++QKLRCRVNYHAL
Sbjct: 205 KPPYTTRVPRKSTLEYYLDQVLPILTRRHVLQLTKFDYRLANDLDEDMQKLRCRVNYHAL 264
Query: 181 RFTKPIRQLGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIRKRW 240
RFTK I+ +G ++V RMRKMA R+IAVHLRFEPDMLAFSGC FGGGEKER EL EIRKRW
Sbjct: 265 RFTKRIQSVGMKVVKRMRKMAKRFIAVHLRFEPDMLAFSGCDFGGGEKERAELAEIRKRW 324
Query: 241 TTLPDLSPDGERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLF 300
TLPDL P ERKRGKCPLTPHEVGLMLRALG+TNDTY+YVASGEIYGG++T++PLR+LF
Sbjct: 325 DTLPDLDPLEERKRGKCPLTPHEVGLMLRALGFTNDTYIYVASGEIYGGEKTLKPLRELF 384
Query: 301 PNIYTKEMLAEEELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHK 360
PN YTKEMLA +ELKP L +SSRLAA+DYIV DES+VF+ NNNGNMA+ILAG+RRYMGHK
Sbjct: 385 PNFYTKEMLANDELKPLLPYSSRLAAIDYIVSDESDVFITNNNGNMAKILAGRRRYMGHK 444
Query: 361 RTIRPNAKKLSALFMARHEMDWDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYPSTCVCE 420
RTIRPNAKKLSALFM R +M+W TF++KVK+CQRGFMG+P E K GRGEFHEYP +C+C+
Sbjct: 445 RTIRPNAKKLSALFMDREKMEWQTFAKKVKSCQRGFMGDPDEFKPGRGEFHEYPQSCICQ 504
Query: 421 KPF-IDELSED 430
+PF D+ S D
Sbjct: 505 RPFSYDKTSTD 515
>At1g76270 putative auxin-independent growth promoter (At1g76270)
Length = 572
Score = 538 bits (1387), Expect = e-153
Identities = 259/457 (56%), Positives = 348/457 (75%), Gaps = 5/457 (1%)
Query: 9 DIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILN 68
DIW S+ +++++GC F + ++ YL+IA SGGLNQQRTGI DAVV ARILN
Sbjct: 77 DIWRSRNAEFFFGCSNASSKFANSKAVTRNDRYLVIATSGGLNQQRTGIVDAVVAARILN 136
Query: 69 ATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDKVMRSMDKPPYTMRV 128
ATLVVP+LD S+WKD SDF++IFDV+WFI++L+ DV I+K++P K R+ MRV
Sbjct: 137 ATLVVPKLDQKSYWKDASDFSHIFDVDWFISFLSGDVRIIKQLPLKGGRTWSTS--RMRV 194
Query: 129 PRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLANDLDDELQKLRCRVNYHALRFTKPIRQ 188
PRK + Y+++VLP+LL+R +QL KFDYRL+N L D+LQKLRCRVNYHAL+FT PI
Sbjct: 195 PRKCNERCYINRVLPVLLKRHAVQLNKFDYRLSNKLSDDLQKLRCRVNYHALKFTDPILT 254
Query: 189 LGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIRKRWTTLPDLSP 248
+G +V RMR + +IA+HLR+EPDMLAFSGCY+GGG+KER+EL IR+RW TL +P
Sbjct: 255 MGNELVRRMRLRSKHFIALHLRYEPDMLAFSGCYYGGGDKERRELAAIRRRWKTLHINNP 314
Query: 249 DGERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEM 308
+ +R++G+CPLTP EVGLMLRALGY +D ++YVASGE+YGG+E++ PL+ LFP+ Y+K+
Sbjct: 315 EKQRRQGRCPLTPEEVGLMLRALGYGSDVHIYVASGEVYGGEESLAPLKALFPHFYSKDT 374
Query: 309 LA-EEELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKRTIRPNA 367
+A +EEL+PF S+SSR+AA+D++VCDES+VFV NNNGNMA+ILAG+RRY+GHK T+RPNA
Sbjct: 375 IATKEELEPFSSYSSRMAALDFLVCDESDVFVTNNNGNMAKILAGRRRYLGHKPTVRPNA 434
Query: 368 KKLSALFMARHEMDWDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYPSTCVCEKPFIDEL 427
KKL LFM + W+ FS KV++ QRGFMGEP E++AGRGEFHE PSTC+CE +
Sbjct: 435 KKLYRLFMNKENTTWEEFSSKVRSFQRGFMGEPKEVRAGRGEFHENPSTCICED--TEAK 492
Query: 428 SEDGIRPPKLAFRNLTIGTDAEINRGNEESFELPKPN 464
++ + KL +N + DA + N++ E P+
Sbjct: 493 AKAQLESRKLGKKNKSTNKDAAVTVTNDDQTEEDDPD 529
>At4g38390 putative growth regulator protein
Length = 551
Score = 528 bits (1359), Expect = e-150
Identities = 251/411 (61%), Positives = 316/411 (76%), Gaps = 1/411 (0%)
Query: 10 IWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILNA 69
+W S+ S YY C F +N YLLIA SGGLNQQRTGI DAVV A ILNA
Sbjct: 95 LWSSRLSNLYYACSNATDTFQVTDTRSQTNRYLLIATSGGLNQQRTGIIDAVVAAYILNA 154
Query: 70 TLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDKVMRSMDKPPYTMRVP 129
TLVVP+LD S+WKD S+F +IFDV+WFI++L+KDV I+K +P + + +MRVP
Sbjct: 155 TLVVPKLDQKSYWKDTSNFEDIFDVDWFISHLSKDVKIIKELPKEEQSRISTSLQSMRVP 214
Query: 130 RKSDPEYYLDQVLPILLRRRVLQLTKFDYRLANDLDDELQKLRCRVNYHALRFTKPIRQL 189
RK P YL +VLPIL ++ V+QL+KFDYRL+N LD ELQKLRCRVNYHA+R+T+ I ++
Sbjct: 215 RKCTPSCYLQRVLPILTKKHVVQLSKFDYRLSNALDTELQKLRCRVNYHAVRYTESINRM 274
Query: 190 GQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIRKRWTTLPDLSPD 249
GQ +V RMRK A ++A+HLRFEPDMLAFSGCY+GGG+KER ELG +R+RW TL +P+
Sbjct: 275 GQLLVDRMRKKAKHFVALHLRFEPDMLAFSGCYYGGGQKERLELGAMRRRWKTLHAANPE 334
Query: 250 GERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEML 309
R+ G+CPLTP E+GLMLR LG+ + +LYVASGE+YGG++T+ PLR LFPN++TKE L
Sbjct: 335 KVRRHGRCPLTPEEIGLMLRGLGFGKEVHLYVASGEVYGGEDTLAPLRALFPNLHTKETL 394
Query: 310 -AEEELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAK 368
+++EL PF +FSSR+AA+D+IVCD+S+ FV NNNGNMARILAG+RRY+GHK TIRPNAK
Sbjct: 395 TSKKELAPFANFSSRMAALDFIVCDKSDAFVTNNNGNMARILAGRRRYLGHKVTIRPNAK 454
Query: 369 KLSALFMARHEMDWDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYPSTCVC 419
KL +F RH M W FS KV+ Q GFMGEP EMK G GEFHE P++C+C
Sbjct: 455 KLYEIFKNRHNMTWGEFSSKVRRYQTGFMGEPDEMKPGEGEFHENPASCIC 505
>At1g20550 hypothetical protein
Length = 529
Score = 463 bits (1192), Expect = e-131
Identities = 227/420 (54%), Positives = 302/420 (71%), Gaps = 41/420 (9%)
Query: 2 GNDHAPIDIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAV 61
G + DIW S ++++YGC F A ++ YL IA SGGLNQQRTGI DAV
Sbjct: 76 GGGRSDRDIWRSWNAEFFYGCCNASSKFPNAKAITRNDRYLAIATSGGLNQQRTGIVDAV 135
Query: 62 VVARILNATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDKVMRSMDK 121
V ARILNATLV+P+LD S+WKD SDF+NIFDV+WF+++L+KDV I++++P K ++
Sbjct: 136 VAARILNATLVIPKLDQKSYWKDASDFSNIFDVDWFMSFLSKDVKIIEKLPQKGGQTWS- 194
Query: 122 PPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLANDLDDELQKLRCRVNYHALR 181
P MRVPRK + + Y+++VLP+L +R +QL KFDYRL+N L D+LQKLR
Sbjct: 195 -PRRMRVPRKCNEKCYINRVLPVLQKRHAVQLNKFDYRLSNKLRDDLQKLR--------- 244
Query: 182 FTKPIRQLGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIRKRWT 241
FEPDMLAFSGCY+GGGEKE++ELG IR+RW
Sbjct: 245 -----------------------------FEPDMLAFSGCYYGGGEKEKKELGTIRRRWK 275
Query: 242 TLPDLSPDGERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFP 301
TL +P+ +R++G+CPLTP EVGLMLRALGY +D ++YVASGE+YGG++++ PL+ LFP
Sbjct: 276 TLHVNNPEKQRRQGRCPLTPEEVGLMLRALGYGSDVHIYVASGEVYGGEKSLAPLKALFP 335
Query: 302 NIYTKEMLAEE-ELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHK 360
+ Y+K+ +A + ELKPF S+SSR+AA+D++VCDES+VFV NNNGNMARILAG+RRY GHK
Sbjct: 336 HFYSKDTIATKMELKPFSSYSSRMAALDFLVCDESDVFVTNNNGNMARILAGRRRYFGHK 395
Query: 361 RTIRPNAKKLSALFMARHEMDWDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYPSTCVCE 420
TIRPNAKKL LFM++ W+ F+ +V+ Q+GFMGEP E++AG+GEFHE P+ C+CE
Sbjct: 396 PTIRPNAKKLYKLFMSKENTTWEEFASRVRTFQKGFMGEPKEVRAGKGEFHENPAACICE 455
>At2g01480 similar to axi 1 protein from Nicotiana tabacum
Length = 567
Score = 326 bits (835), Expect = 2e-89
Identities = 175/393 (44%), Positives = 248/393 (62%), Gaps = 17/393 (4%)
Query: 38 SNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWF 97
SNGY+ + A+GGLNQQRT I +AV VA LNATLV+P ++S W+D S F +I+D +F
Sbjct: 154 SNGYIYVEANGGLNQQRTSICNAVAVAGYLNATLVIPNFHYHSIWRDPSKFGDIYDEEFF 213
Query: 98 INYLAKDVTIVKRVPDKVMRSMDKPP---YTMRVPRKSDPEYYLDQVLPILLRRRVLQLT 154
++ L+ DV +V +P+ +M D Y RV S +YY D +LP LL ++++++
Sbjct: 214 VSTLSNDVRVVDTIPEYLMERFDHNMTNVYNFRVKAWSPIQYYRDSILPKLLEEKIIRIS 273
Query: 155 KFDYRLANDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMAN----RYIAVHLR 210
F RL+ D +Q+LRC NY AL+F+K I LG+ +V RM++ + +Y++VHLR
Sbjct: 274 PFANRLSFDAPQAVQRLRCLANYEALKFSKTILTLGETLVKRMKEQSANHGAKYVSVHLR 333
Query: 211 FEPDMLAFSGCYFGGGEKERQELGEIRKR-WT---TLPD--LSPDGERKRGKCPLTPHEV 264
FE DM+AFS C F GG +E+Q++ R+R W T P + P R+ GKCPLTP EV
Sbjct: 334 FEEDMVAFSCCIFDGGNQEKQDMIAARERGWKGKFTKPGRVIRPGAIRQNGKCPLTPLEV 393
Query: 265 GLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEELKPFLSFSSR 323
GLMLR +G+ TY+++ASGEIY + TM PL ++FPN+ TKEMLA EEEL P+ +FSSR
Sbjct: 394 GLMLRGMGFNKSTYIFLASGEIYDANRTMAPLLEMFPNLQTKEMLASEEELAPYKNFSSR 453
Query: 324 LAAVDYIVCDESNVFVANNNGNMARILAGQRRYM--GHKRTIRPNAKKLSALFMARHEMD 381
+AA+DY VC S VFV GN L G RRYM GH +TIRP+ +KL+ LF +
Sbjct: 454 MAAIDYTVCLHSEVFVTTQGGNFPHFLMGHRRYMFGGHSKTIRPDKRKLAILF-DNPNIG 512
Query: 382 WDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYP 414
W +F R++ + + E+K + +P
Sbjct: 513 WRSFKRQMLNMRSHSDSKGFELKRPNDSIYTFP 545
>At1g14970 auxin-independent growth promoter, putative
Length = 458
Score = 324 bits (831), Expect = 6e-89
Identities = 178/401 (44%), Positives = 250/401 (61%), Gaps = 18/401 (4%)
Query: 38 SNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWF 97
SNG++ I A+GGLNQQRT I +AV VA LNATLV+P ++S WKD S F +I+D +F
Sbjct: 51 SNGFIFIEANGGLNQQRTSICNAVAVAGYLNATLVIPNFHYHSIWKDPSKFGDIYDEEYF 110
Query: 98 INYLAKDVTIVKRVPDKVMRSMD---KPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLT 154
I+ LA DV +V VP+ +M D Y RV + YY D VLP LL +V++++
Sbjct: 111 IDTLANDVRVVDTVPEYLMERFDYNLTNVYNFRVKAWAPTSYYRDSVLPKLLEEKVIRIS 170
Query: 155 KFDYRLANDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKM----ANRYIAVHLR 210
F RL+ D +Q+ RC N ALRF+KPI G+ +V +M+ + A +Y++VHLR
Sbjct: 171 PFANRLSFDAPRAVQRFRCLANNVALRFSKPILTQGETLVNKMKGLSANNAGKYVSVHLR 230
Query: 211 FEPDMLAFSGCYFGGGEKERQELGEIRKR-WT---TLPD--LSPDGERKRGKCPLTPHEV 264
FE DM+AFS C F GG++E+Q++ R+R W T P + P R GKCPLTP EV
Sbjct: 231 FEEDMVAFSCCVFDGGDQEKQDMIAARERGWKGKFTKPGRVIRPGANRLNGKCPLTPLEV 290
Query: 265 GLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEELKPFLSFSSR 323
GLMLR +G+ TY+++A+G IY + TM PL ++FPN+ TKEMLA EE+L PF +FSSR
Sbjct: 291 GLMLRGMGFNKSTYIFLAAGPIYSANRTMAPLLEMFPNLQTKEMLASEEDLAPFKNFSSR 350
Query: 324 LAAVDYIVCDESNVFVANNNGNMARILAGQRRYM--GHKRTIRPNAKKLSALFMARHEMD 381
+AA+DY VC S VFV GN L G RRY+ GH +TI+P+ +KL+ LF ++
Sbjct: 351 MAAIDYTVCLHSEVFVTTQGGNFPHFLMGHRRYLFGGHSKTIQPDKRKLAVLF-DNPKLG 409
Query: 382 WDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYP-STCVCEK 421
W +F R++ + + + E+K + +P C+C K
Sbjct: 410 WKSFKRQMLSMRSHSDSKGFELKRSSDSIYIFPCPDCMCRK 450
>At1g14020 growth regulator like protein
Length = 499
Score = 308 bits (788), Expect = 5e-84
Identities = 171/402 (42%), Positives = 242/402 (59%), Gaps = 13/402 (3%)
Query: 31 PAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFAN 90
P R SNG LL++ +GGLNQ R+ I D V VAR+LN TLVVPELD SFW D S F +
Sbjct: 84 PPPRNYTSNGILLVSCNGGLNQMRSAICDMVTVARLLNLTLVVPELDKTSFWADPSGFED 143
Query: 91 IFDVNWFINYLAKDVTIVKRVPDKVMRSMDKPPYTMRVPRKSDPEYYLDQVLPILLRRRV 150
IFDV FI+ L +V I++R+P + R + M SD +YYL QVLP+ + +V
Sbjct: 144 IFDVRHFIDSLRDEVRILRRLPKRFSRKYGYQMFEMPPVSWSDEKYYLKQVLPLFSKHKV 203
Query: 151 LQLTKFDYRLAND-LDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVHL 209
+ + D RLAN+ L LQ LRCRVN+ L+FT + LG ++V R+ + ++A+HL
Sbjct: 204 VHFNRTDTRLANNGLPLSLQWLRCRVNFQGLKFTPQLEALGSKLV-RILQQRGPFVALHL 262
Query: 210 RFEPDMLAFSGCYFGGGEKERQELGEIRKR--WTTLPDLSPDGERKRGKCPLTPHEVGLM 267
R+E DMLAFSGC G E+E +EL ++R W ++ + R +G CPLTP EV L+
Sbjct: 263 RYEMDMLAFSGCTHGCTEEEAEELKKMRYTYPWWREKEIVSEERRAQGLCPLTPEEVALV 322
Query: 268 LRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLAEE-ELKPFLSFSSRLAA 326
L+ALG+ +T +Y+A+GEIYG + + LR+ FP I KEML E EL+ F + SS++AA
Sbjct: 323 LKALGFEKNTQIYIAAGEIYGSEHRLSVLREAFPRIVKKEMLLESAELQQFQNHSSQMAA 382
Query: 327 VDYIVCDESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSALFMARHE--MDWDT 384
+D++V SN F+ +GNMA+++ G RRY+G+K+TI + K+L L H + WD
Sbjct: 383 LDFMVSVASNTFIPTYDGNMAKVVEGHRRYLGYKKTILLDRKRLVELLDLHHNKTLTWDQ 442
Query: 385 FSRKVKACQRGFMGEP------GEMKAGRGEFHEYPSTCVCE 420
F+ VK G P + F+ P C+CE
Sbjct: 443 FAVAVKEAHERRAGAPTHRRVISDKPKEEDYFYANPQECLCE 484
>At5g15740 unknown protein
Length = 508
Score = 303 bits (777), Expect = 1e-82
Identities = 167/377 (44%), Positives = 238/377 (62%), Gaps = 8/377 (2%)
Query: 31 PAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFAN 90
P R ++NGYL+++ +GGLNQ R I D V VAR +N TL+VPELD SFW D S+F +
Sbjct: 99 PPKRVYVNNGYLMVSCNGGLNQMRAAICDMVTVARYMNVTLIVPELDKTSFWNDPSEFKD 158
Query: 91 IFDVNWFINYLAKDVTIVKRVPDKVMRSMDKPPYTMRVPRK-SDPEYYLDQVLPILLRRR 149
IFDV+ FI+ L +V I+K +P ++ + ++ Y P S+ YY +Q+LP++ + +
Sbjct: 159 IFDVDHFISSLRDEVRILKELPPRLKKRVELGVYHEMPPISWSNMSYYQNQILPLVKKHK 218
Query: 150 VLQLTKFDYRLAND-LDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVH 208
VL L + D RLAN+ L E+QKLRCRVN++ L+FT I +LG+R+V +R+ ++ +H
Sbjct: 219 VLHLNRTDTRLANNGLPVEVQKLRCRVNFNGLKFTPQIEELGRRVVKILRE-KGPFLVLH 277
Query: 209 LRFEPDMLAFSGCYFGGGEKERQELGEIRKR--WTTLPDLSPDGERKRGKCPLTPHEVGL 266
LR+E DMLAFSGC G +E +EL +R W ++ + +RK G CPLTP E L
Sbjct: 278 LRYEMDMLAFSGCSHGCNPEEEEELTRMRYAYPWWKEKVINSELKRKDGLCPLTPEETAL 337
Query: 267 MLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLAEEELKPFL-SFSSRLA 325
L ALG + +Y+A+GEIYGG M+ L D FPN+ KE L E F + SS++A
Sbjct: 338 TLTALGIDRNVQIYIAAGEIYGGQRRMKALTDAFPNVVRKETLLESSDLDFCRNHSSQMA 397
Query: 326 AVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSALFMARHE--MDWD 383
A+DY+V ES++FV N+GNMAR++ G RR++G K+TI+ N + L L E + WD
Sbjct: 398 ALDYLVALESDIFVPTNDGNMARVVEGHRRFLGFKKTIQLNRRFLVKLIDEYTEGLLSWD 457
Query: 384 TFSRKVKACQRGFMGEP 400
FS VKA MG P
Sbjct: 458 VFSSTVKAFHSTRMGSP 474
>At1g35510 putative growth regulator protein (At1g35510)
Length = 568
Score = 303 bits (775), Expect = 2e-82
Identities = 173/405 (42%), Positives = 243/405 (59%), Gaps = 24/405 (5%)
Query: 38 SNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWF 97
SNGY +I A+GGLNQQR I DAV VA +LNATLV+P NS W+D S F +IFD ++F
Sbjct: 159 SNGYFIIEANGGLNQQRLSICDAVAVAGLLNATLVIPIFHLNSVWRDSSKFGDIFDEDFF 218
Query: 98 INYLAKDVTIVKRVPDKVMRSMD---KPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLT 154
I L+K+V +VK +P V+ + +R+ S P YYL +VLP LLR +++
Sbjct: 219 IYALSKNVNVVKELPKDVLERYNYNISSIVNLRLKAWSSPAYYLQKVLPQLLRLGAVRVA 278
Query: 155 KFDYRLANDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRM----RKMANRYIAVHLR 210
F RLA+ + +Q LRC N+ ALRF +PIR L +++V RM + +Y++VHLR
Sbjct: 279 PFSNRLAHAVPAHIQGLRCLANFEALRFAEPIRLLAEKMVDRMVTKSVESGGKYVSVHLR 338
Query: 211 FEPDMLAFSGCYFGGGEKERQEL---------GEIRKRWTTLPDLSPDGERKRGKCPLTP 261
FE DM+AFS C + G+ E+ E+ G+ R+R + P R GKCPLTP
Sbjct: 339 FEMDMVAFSCCEYDFGQAEKLEMDMARERGWKGKFRRRGRV---IRPGANRIDGKCPLTP 395
Query: 262 HEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEELKPFLSF 320
EVG+MLR +G+ N T +YVA+G IY D+ M PLR +FP + TK+ LA EEL PF
Sbjct: 396 LEVGMMLRGMGFNNSTLVYVAAGNIYKADKYMAPLRQMFPLLQTKDTLATPEELAPFKGH 455
Query: 321 SSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYM--GHKRTIRPNAKKLSALFMARH 378
SSRLAA+DY VC S VFV+ GN L G RRY+ GH TI+P+ +KL L + +
Sbjct: 456 SSRLAALDYTVCLHSEVFVSTQGGNFPHFLIGHRRYLYKGHAETIKPDKRKLVQL-LDKP 514
Query: 379 EMDWDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYP-STCVCEKP 422
+ WD F ++++ R + E++ + +P C+C++P
Sbjct: 515 SIRWDYFKKQMQDMLRHNDAKGVELRKPAASLYTFPMPDCMCKEP 559
>At2g37980 similar to axi 1 protein from Nicotiana tabacum
Length = 638
Score = 301 bits (772), Expect = 4e-82
Identities = 178/434 (41%), Positives = 253/434 (58%), Gaps = 24/434 (5%)
Query: 9 DIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILN 68
+IW+ S Y C R ++ + R +NGYLL+ A+GGLNQ RTGI D V A+I+N
Sbjct: 199 EIWQKPESGNYRQCASRPKN--RSRLRRKTNGYLLVHANGGLNQMRTGICDMVAAAKIMN 256
Query: 69 ATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDK--VMRSMDKPPYTM 126
ATLV+P LDH SFW D S F +IFD F+N L DV IV+ +P + MR + K P +
Sbjct: 257 ATLVLPLLDHESFWTDPSTFKDIFDWRHFMNVLKDDVDIVEYLPPRYAAMRPLLKAPVSW 316
Query: 127 RVPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLAND-LDDELQKLRCRVNYHALRFTKP 185
S YY ++LP+L + +V++ T D RLAN+ L +Q+LRCR NY AL ++K
Sbjct: 317 -----SKASYYRSEMLPLLKKHKVIKFTHTDSRLANNGLPPSIQRLRCRANYQALGYSKE 371
Query: 186 IRQLGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIR---KRWTT 242
I G+ +V R+R + +IA+HLR+E DMLAF+GC E +EL +R K W
Sbjct: 372 IEDFGKVLVNRLRNNSEPFIALHLRYEKDMLAFTGCSHNLTAGEAEELRIMRYNVKHWKE 431
Query: 243 LPDLSPDGERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPN 302
++ R +G CP++P E + L+A+GY + T +Y+ +GEIYGG+ +M R+ +PN
Sbjct: 432 -KEIDSRERRIQGGCPMSPREAAIFLKAMGYPSSTTVYIVAGEIYGGN-SMDAFREEYPN 489
Query: 303 IYTKEMLA-EEELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKR 361
++ LA EEEL+PF + +RLAA+DYIV ES+VFV +GNMA+ + G RR+ G K+
Sbjct: 490 VFAHSYLATEEELEPFKPYQNRLAALDYIVALESDVFVYTYDGNMAKAVQGHRRFEGFKK 549
Query: 362 TIRPNAKKLSALFMARHE--MDWDTFSRKVKACQRGFMGE-----PGEMKAGRGEFHEYP 414
TI P+ L E M WD FS +VK +G PGE F+ P
Sbjct: 550 TINPDRLNFVRLIDHLDEGVMSWDEFSSEVKRLHNNRIGAPYARLPGEFPRLEENFYANP 609
Query: 415 S-TCVCEKPFIDEL 427
C+C K ++L
Sbjct: 610 QPDCICNKSQPEQL 623
>At2g03280 axi 1 -like protein
Length = 481
Score = 301 bits (770), Expect = 7e-82
Identities = 170/398 (42%), Positives = 243/398 (60%), Gaps = 14/398 (3%)
Query: 34 RERMSNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFD 93
R SNG LL++ +GGLNQ R I D V VAR+LN TLVVPELD SFW D SDF +IFD
Sbjct: 84 RNYTSNGILLVSCNGGLNQMRAAICDMVTVARLLNLTLVVPELDKKSFWADTSDFEDIFD 143
Query: 94 VNWFINYLAKDVTIVKRVPDKVMRSMDKPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQL 153
+ FI+ L +V I++R+P + + + M S+ +YYL QVLP +R+V+
Sbjct: 144 IKHFIDSLRDEVRIIRRLPKRYSKKYGFKLFEMPPVSWSNDKYYLQQVLPRFSKRKVIHF 203
Query: 154 TKFDYRLAND-LDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVHLRFE 212
+ D RLAN+ L +LQ+LRCRVN+ LRFT I LG ++V R+ + ++A+HLR+E
Sbjct: 204 VRSDTRLANNGLSLDLQRLRCRVNFQGLRFTPRIEALGSKLV-RILQQRGSFVALHLRYE 262
Query: 213 PDMLAFSGCYFGGGEKERQELGEIRKR--WTTLPDLSPDGERKRGKCPLTPHEVGLMLRA 270
DMLAFSGC G ++E +EL ++R W ++ + R +G CPLTP E L+L+A
Sbjct: 263 MDMLAFSGCTHGCTDEEAEELKKMRYAYPWWREKEIVSEERRVQGLCPLTPEEAVLVLKA 322
Query: 271 LGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLAE-EELKPFLSFSSRLAAVDY 329
LG+ DT +Y+A+GEI+GG + + L++ FP I KEML + EL+ F + SS++AA+D+
Sbjct: 323 LGFQKDTQIYIAAGEIFGGAKRLALLKESFPRIVKKEMLLDPTELQQFQNHSSQMAALDF 382
Query: 330 IVCDESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSALFMARHE--MDWDTFSR 387
IV SN F+ GNMA+++ G RRY+G K+TI + K+L L + + WD F+
Sbjct: 383 IVSVASNTFIPTYYGNMAKVVEGHRRYLGFKKTILLDRKRLVELLDLHNNKTLSWDQFAV 442
Query: 388 KVKACQRG-FMGEPGEMKA------GRGEFHEYPSTCV 418
VK +G MGEP K F+ P C+
Sbjct: 443 AVKDAHQGRRMGEPTHRKVISVRPKEEDYFYANPQECI 480
>At3g54100 unknown protein
Length = 638
Score = 298 bits (763), Expect = 4e-81
Identities = 176/434 (40%), Positives = 255/434 (58%), Gaps = 24/434 (5%)
Query: 9 DIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILN 68
+IW+ S Y C R +++ +R +NGYL++ A+GGLNQ RTGI D V VA+I+N
Sbjct: 195 EIWQKPESGNYRQCVTRPKNYTRL--QRQTNGYLVVHANGGLNQMRTGICDMVAVAKIMN 252
Query: 69 ATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDK--VMRSMDKPPYTM 126
ATLV+P LDH SFW D S F +IFD F+N L DV IV+ +P + M+ + K P +
Sbjct: 253 ATLVLPLLDHESFWTDPSTFKDIFDWRNFMNVLKHDVDIVEYLPPQYAAMKPLLKAPVSW 312
Query: 127 RVPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLAND-LDDELQKLRCRVNYHALRFTKP 185
S YY ++LP+L R +VL+ T D RLAN+ L +Q+LRCR NY AL +TK
Sbjct: 313 -----SKASYYRSEMLPLLKRHKVLKFTLTDSRLANNGLPPSIQRLRCRANYQALLYTKE 367
Query: 186 IRQLGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIR---KRWTT 242
I LG+ +V R+R YIA+HLR+E DMLAF+GC +E +EL +R K W
Sbjct: 368 IEDLGKILVNRLRNNTEPYIALHLRYEKDMLAFTGCNHNLTTEEAEELRIMRYSVKHWKE 427
Query: 243 LPDLSPDGERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPN 302
++ R +G CP++P E + L+A+GY + T +Y+ +GEIY G E+M R +PN
Sbjct: 428 -KEIDSRERRIQGGCPMSPREAAIFLKAMGYPSSTTVYIVAGEIY-GSESMDAFRAEYPN 485
Query: 303 IYTKEMLA-EEELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKR 361
+++ LA EEEL+PF + +RLAA+DYIV ES+VFV +GNMA+ + G R++ G ++
Sbjct: 486 VFSHSTLATEEELEPFSQYQNRLAALDYIVALESDVFVYTYDGNMAKAVQGHRKFEGFRK 545
Query: 362 TIRPNAKKLSALFMARHE--MDWDTFSRKVKACQRGFMGE-----PGEMKAGRGEFHEYP 414
+I P+ L E + W+ FS +VK R +G P + F+ P
Sbjct: 546 SINPDRLNFVRLIDHFDEGIISWEEFSSEVKRLNRDRIGAAYGRLPAALPRLEENFYANP 605
Query: 415 S-TCVCEKPFIDEL 427
C+C K ++L
Sbjct: 606 QPDCICNKSHPEQL 619
>At3g02250 putative auxin-independent growth promoter
Length = 512
Score = 296 bits (758), Expect = 2e-80
Identities = 165/377 (43%), Positives = 234/377 (61%), Gaps = 8/377 (2%)
Query: 31 PAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFAN 90
P R +NGYL+++ +GGLNQ R I D V +AR +N TL+VPELD SFW D S+F +
Sbjct: 99 PPKRIYQNNGYLMVSCNGGLNQMRAAICDMVTIARYMNVTLIVPELDKTSFWNDPSEFKD 158
Query: 91 IFDVNWFINYLAKDVTIVKRVPDKVMRSMDKPPY-TMRVPRKSDPEYYLDQVLPILLRRR 149
IFDV+ FI+ L +V I+K +P ++ R + Y TM S+ YY DQ+LP++ + +
Sbjct: 159 IFDVDHFISSLRDEVRILKELPPRLKRRVRLGLYHTMPPISWSNMSYYQDQILPLVKKYK 218
Query: 150 VLQLTKFDYRLA-NDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVH 208
V+ L K D RLA N+L E+QKLRCR N++ LRFT I +LG+R+V +R+ ++ +H
Sbjct: 219 VVHLNKTDTRLANNELPVEIQKLRCRANFNGLRFTPKIEELGRRVVKILRE-KGPFLVLH 277
Query: 209 LRFEPDMLAFSGCYFGGGEKERQELGEIRKRWTTLPDLSPDGE--RKRGKCPLTPHEVGL 266
LR+E DMLAFSGC G E +EL +R + + D E RK G CPLTP E L
Sbjct: 278 LRYEMDMLAFSGCSHGCNRYEEEELTRMRYAYPWWKEKVIDSELKRKEGLCPLTPEETAL 337
Query: 267 MLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKE-MLAEEELKPFLSFSSRLA 325
L ALG + +Y+A+GEIYGG ++ L D+FPN+ KE +L +L + SS++A
Sbjct: 338 TLSALGIDRNVQIYIAAGEIYGGKRRLKALTDVFPNVVRKETLLDSSDLSFCKNHSSQMA 397
Query: 326 AVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSALFMARHE--MDWD 383
A+DY++ ES++FV GNMA+++ G RR++G K+TI N K L L +E + W+
Sbjct: 398 ALDYLISLESDIFVPTYYGNMAKVVEGHRRFLGFKKTIELNRKLLVKLIDEYYEGLLSWE 457
Query: 384 TFSRKVKACQRGFMGEP 400
FS VKA MG P
Sbjct: 458 VFSTTVKAFHATRMGGP 474
>At1g22460 Similar to auxin-independent growth promoter
Length = 565
Score = 296 bits (757), Expect = 2e-80
Identities = 161/340 (47%), Positives = 227/340 (66%), Gaps = 8/340 (2%)
Query: 38 SNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWF 97
S GYLL+ +GGLNQ R GI D V +ARI+NATLVVPELD SFW+D S F+++FD + F
Sbjct: 160 SRGYLLVHTNGGLNQMRAGICDMVAIARIINATLVVPELDKRSFWQDTSKFSDVFDEDHF 219
Query: 98 INYLAKDVTIVKRVPDKVMRSMDKPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLTKFD 157
IN L+KD+ ++K++P K + + K S YY +++ + +V++ K D
Sbjct: 220 INALSKDIRVIKKLP-KGIDGLTK--VVKHFKSYSGLSYYQNEIASMWDEYKVIRAAKSD 276
Query: 158 YRLA-NDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVHLRFEPDML 216
RLA N+L ++QKLRCR Y ALRF+ IR +G+ +V RMR YIA+HLRFE +ML
Sbjct: 277 SRLANNNLPPDIQKLRCRACYEALRFSTKIRSMGELLVDRMRSY-GLYIALHLRFEKEML 335
Query: 217 AFSGCYFGGGEKERQELGEIRKR--WTTLPDLSPDGERKRGKCPLTPHEVGLMLRALGYT 274
AFSGC G E EL IRK + + D+ +R +G CPLTP EVG++L ALGY+
Sbjct: 336 AFSGCNHGLSASEAAELRRIRKNTAYWKVKDIDGRVQRLKGYCPLTPKEVGILLTALGYS 395
Query: 275 NDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEELKPFLSFSSRLAAVDYIVCD 333
+DT +Y+A+GEIYGG+ + LR F + +KE LA EELK F++ S+++AA+DYIV
Sbjct: 396 SDTPVYIAAGEIYGGESRLADLRSRFSMLMSKEKLATREELKTFMNHSTQMAALDYIVSI 455
Query: 334 ESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSAL 373
ES+VF+ + +GNMAR + G RR++GH++TI P+ K + L
Sbjct: 456 ESDVFIPSYSGNMARAVEGHRRFLGHRKTISPDRKAIVRL 495
>At5g64600 auxin-independent growth promoter-like protein
Length = 539
Score = 295 bits (756), Expect = 3e-80
Identities = 169/404 (41%), Positives = 245/404 (59%), Gaps = 28/404 (6%)
Query: 38 SNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWF 97
S+ Y+ + ++GGLNQ RTGI D V VA I+NATLV+PELD SFW+D S F++IFD F
Sbjct: 134 SDHYITVKSNGGLNQMRTGIADIVAVAHIMNATLVIPELDKRSFWQDSSVFSDIFDEEQF 193
Query: 98 INYLAKDVTIVKRVPDKVMRSMDKPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLTKFD 157
I L +DV ++K++P +V + P + Y +++ + +V+ + K D
Sbjct: 194 IKSLRRDVKVIKKLPKEV----ESLPRARKHFTSWSSVGYYEEMTHLWKEYKVIHVAKSD 249
Query: 158 YRLA-NDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVHLRFEPDML 216
RLA NDL ++Q+LRCRV Y L F+ I LGQ++V R++ A RYIA+HLR+E DML
Sbjct: 250 SRLANNDLPIDVQRLRCRVLYRGLCFSPAIESLGQKLVERLKSRAGRYIALHLRYEKDML 309
Query: 217 AFSGCYFGGGEKERQELGEIRK---RWTTLPDLSPDGERKRGKCPLTPHEVGLMLRALGY 273
AF+GC +G + E +EL +R+ W + ++ +R+ G CPLTP EVG+ L+ LGY
Sbjct: 310 AFTGCTYGLTDAESEELRVMRESTSHW-KIKSINSTEQREEGLCPLTPKEVGIFLKGLGY 368
Query: 274 TNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEELKPFLSFSSRLAAVDYIVC 332
+ T +Y+A+GEIYGGD+ + L+ FPN+ KE LA EELK F +++ AA+DYI+
Sbjct: 369 SQSTVIYIAAGEIYGGDDRLSELKSRFPNLVFKETLAGNEELKGFTGHATKTAALDYIIS 428
Query: 333 DESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSALF--MARHEM-DWDTFSRKV 389
ES+VFV +++GNMAR + G RR++GH+RTI P+ K L LF M R ++ + S V
Sbjct: 429 VESDVFVPSHSGNMARAVEGHRRFLGHRRTITPDRKGLVKLFVKMERGQLKEGPKLSNFV 488
Query: 390 KACQRGFMGEPGEMKA------GRGEF--------HEYPSTCVC 419
+ G P K GR F + YP C+C
Sbjct: 489 NQMHKDRQGAPRRRKGPTQGIKGRARFRTEEAFYENPYPE-CIC 531
>At3g26370 unknown protein
Length = 557
Score = 288 bits (738), Expect = 3e-78
Identities = 166/408 (40%), Positives = 244/408 (59%), Gaps = 28/408 (6%)
Query: 35 ERMSNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDV 94
E +NGY+ I A GGLNQQR I +AV VA+I+NATL++P L + WKD + F +IFDV
Sbjct: 155 ENETNGYVFIHAEGGLNQQRIAICNAVAVAKIMNATLILPVLKQDQIWKDTTKFEDIFDV 214
Query: 95 NWFINYLAKDVTIVKRVPD------KVMRSMDKPPYTMRVPRKSDPEYYLDQVLPILLRR 148
+ FI+YL DV IV+ +PD ++ S+ + +P+ + ++Y+D VLP + +
Sbjct: 215 DHFIDYLKDDVRIVRDIPDWFTDKAELFSSIRRT--VKNIPKYAAAQFYIDNVLPRIKEK 272
Query: 149 RVLQLTKFDYRLAND-LDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMA---NRY 204
+++ L F RL D + E+ +LRCRVNYHAL+F I Q+ +V RMR N Y
Sbjct: 273 KIMALKPFVDRLGYDNVPQEINRLRCRVNYHALKFLPEIEQMADSLVSRMRNRTGNPNPY 332
Query: 205 IAVHLRFEPDMLAFSGCYFGGGEKERQELGEIR-KRWTTLPDLSPDG---------ERKR 254
+A+HLRFE M+ S C F G +E+ ++ E R K W P +G +RK
Sbjct: 333 MALHLRFEKGMVGLSFCDFVGTREEKVKMAEYRQKEW---PRRFKNGSHLWQLALQKRKE 389
Query: 255 GKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEE 313
G+CPL P EV ++LRA+GY +T +YVASG++YGG M PLR++FPN+ TKE LA +EE
Sbjct: 390 GRCPLEPGEVAVILRAMGYPKETQIYVASGQVYGGQNRMAPLRNMFPNLVTKEDLAGKEE 449
Query: 314 LKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHK-RTIRPNAKKLSA 372
L F + LAA+D++VC +S+VFV + GN A+++ G RRYMGH+ ++I+P+ +S
Sbjct: 450 LTTFRKHVTSLAALDFLVCLKSDVFVMTHGGNFAKLIIGARRYMGHRQKSIKPDKGLMSK 509
Query: 373 LFMARHEMDWDTFSRKVKACQRGFMGEPGEMKAGRGEFHEYPSTCVCE 420
F + M W TF V + G P E + + C+C+
Sbjct: 510 SFGDPY-MGWATFVEDVVVTHQTRTGLPEETFPNYDLWENPLTPCMCK 556
>At5g35570 unknown protein
Length = 652
Score = 285 bits (730), Expect = 3e-77
Identities = 169/395 (42%), Positives = 235/395 (58%), Gaps = 16/395 (4%)
Query: 38 SNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHNSFWKDDSDFANIFDVNWF 97
+NGYLLI A+GGLNQ R GI D V VA+I+ ATLV+P LDH+S+W DDS F ++FD F
Sbjct: 251 TNGYLLINANGGLNQMRFGICDMVAVAKIMKATLVLPSLDHSSYWADDSGFKDLFDWQHF 310
Query: 98 INYLAKDVTIVKRVPDKVMRSMDKPPYTMRVPRKSDPEYYLDQVLPILLRRRVLQLTKFD 157
I L D+ IV+ +P ++ P+ S YY +VLP+L + V+ LT D
Sbjct: 311 IEELKDDIHIVEMLPSEL---AGIEPFVKTPISWSKVGYYKKEVLPLLKQHIVMYLTHTD 367
Query: 158 YRLA-NDLDDELQKLRCRVNYHALRFTKPIRQLGQRIVMRMRKMANRYIAVHLRFEPDML 216
RLA NDL D +QKLRCRVNY AL+++ PI +LG +V RMR+ Y+A+HLR+E DML
Sbjct: 368 SRLANNDLPDSVQKLRCRVNYRALKYSAPIEELGNVLVSRMRQDRGPYLALHLRYEKDML 427
Query: 217 AFSGCYFGGGEKERQELGEIRKRWTTLPDLSPDGERKR--GKCPLTPHEVGLMLRALGYT 274
AF+GC +E +EL ++R + + +G +R G CPLTP E L+LRAL +
Sbjct: 428 AFTGCSHSLTAEEDEELRQMRYEVSHWKEKEINGTERRLQGGCPLTPRETSLLLRALEFP 487
Query: 275 NDTYLYVASGEIYGGDETMQPLRDLFPNIYTKEMLA-EEELKPFLSFSSRLAAVDYIVCD 333
+ + +Y+ +GE Y G+ +M PL FPNI++ +LA +EEL PF + + LA +DYIV
Sbjct: 488 SSSRIYLVAGEAY-GNGSMDPLNTDFPNIFSHSILATKEELSPFNNHQNMLAGLDYIVAL 546
Query: 334 ESNVFVANNNGNMARILAGQRRYMGHKRTIRPNAKKLSALFMARHE--MDWDTFSRKVKA 391
+S VF+ +GNMA+ + G RR+ K+TI P+ L A E + W FS KVK
Sbjct: 547 QSEVFLYTYDGNMAKAVQGHRRFEDFKKTINPDKMNFVKLVDALDEGRISWKKFSSKVKK 606
Query: 392 CQRGFMGEP-----GEMKAGRGEFHEYP-STCVCE 420
+ G P GE F+ P C+CE
Sbjct: 607 LHKDRNGAPYNRESGEFPKLEESFYANPLPGCICE 641
>At1g29200 hypothetical protein
Length = 698
Score = 276 bits (707), Expect = 1e-74
Identities = 161/422 (38%), Positives = 245/422 (57%), Gaps = 45/422 (10%)
Query: 13 SQYSKYYYGCKERGRHFYPAIRER-MSNGYLLIAASGGLNQQRTGITDAVVVARILNATL 71
S S + C + + R R +SNGY++++A+GGLNQQR I +AV VA +LNATL
Sbjct: 247 SSKSSQWKPCADNNKAAVALERSRELSNGYIMVSANGGLNQQRVAICNAVAVAALLNATL 306
Query: 72 VVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDKVMRSMDKPPYTM----R 127
V+P +++ WKD S F +I+ + FI YL +V IVK +P ++S D ++
Sbjct: 307 VLPRFLYSNVWKDPSQFGDIYQEDHFIEYLKDEVNIVKNLPQH-LKSTDNKNLSLVTDTE 365
Query: 128 VPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLAND-LDDELQKLRCRVNYHALRFTKPI 186
+ +++ P Y++ VLP+L + ++ L + RL D L ++Q+LRC+ N+HAL+F I
Sbjct: 366 LVKEATPVDYIEHVLPLLKKYGMVHLFGYGNRLGFDPLPFDVQRLRCKCNFHALKFAPKI 425
Query: 187 RQLGQRIVMRMRKMAN--------------------------RYIAVHLRFEPDMLAFSG 220
++ G +V R+R+ +Y+A+HLRFE DM+A+S
Sbjct: 426 QEAGSLLVKRIRRFKTSRSRLEEALLGESMVKSTVKGEEEPLKYLALHLRFEEDMVAYSL 485
Query: 221 CYFGGGEKERQELGEIR--------KRWTTLPDLSPDGERKRGKCPLTPHEVGLMLRALG 272
C FGGGE ER+EL R KR +SP+ RK GKCPLTP E L+L LG
Sbjct: 486 CDFGGGEAERKELQAYREDHFPLLLKRLKKSKPVSPEELRKTGKCPLTPEEATLVLAGLG 545
Query: 273 YTNDTYLYVASGEIYGGDETMQPLRDLFPNIYTKE-MLAEEELKPFLSFSSRLAAVDYIV 331
+ TY+Y+A +IYGG M PL L+PNI TKE +L +EL PF +FSS+LAA+D+I
Sbjct: 546 FKRKTYIYLAGSQIYGGSSRMLPLTRLYPNIATKETLLTPQELAPFKNFSSQLAALDFIA 605
Query: 332 CDESNVFVANNNGN-MARILAGQRRYMGHKR--TIRPNAKKLSALFMARHEMDWDTFSRK 388
C S+VF ++G+ ++ +++G R Y G+ + T+RPN K+L+A+ + W F +
Sbjct: 606 CIASDVFAMTDSGSQLSSLVSGFRNYYGNGQAPTLRPNKKRLAAILSDSETIKWKIFEDR 665
Query: 389 VK 390
V+
Sbjct: 666 VR 667
>At1g04910 Similar to auxin-independent growth promoter (axi 1)
Length = 519
Score = 276 bits (706), Expect = 2e-74
Identities = 159/376 (42%), Positives = 229/376 (60%), Gaps = 8/376 (2%)
Query: 9 DIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILN 68
++WES S + + P +E +NGYL + +GGLNQQR+ I +AV+ ARI+N
Sbjct: 61 ELWESAKSGGWRPSSAPRSDWPPPTKE--TNGYLRVRCNGGLNQQRSAICNAVLAARIMN 118
Query: 69 ATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPD--KVMRSMDKPPYTM 126
ATLV+PELD NSFW DDS F I+DV FI L DV IV ++PD K ++ + +
Sbjct: 119 ATLVLPELDANSFWHDDSGFQGIYDVEHFIETLKYDVKIVGKIPDVHKNGKTKKIKAFQI 178
Query: 127 RVPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLANDLDD-ELQKLRCRVNYHALRFTKP 185
R PR + E+YL L + + LT F +RLA ++D+ E Q+LRCRVNYHALRF
Sbjct: 179 RPPRDAPIEWYLTTALKAMREHSAIYLTPFSHRLAEEIDNPEYQRLRCRVNYHALRFKPH 238
Query: 186 IRQLGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIRKRWTTLPD 245
I +L + IV ++R + ++++HLRFE DMLAF+GC+ +E++ L + RK
Sbjct: 239 IMKLSESIVDKLRSQGH-FMSIHLRFEMDMLAFAGCFDIFNPEEQKILRKYRKENFADKR 297
Query: 246 LSPDGERKRGKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNIYT 305
L + R GKCPLTP EVGL+LRA+ + N T +Y+A+GE++GG++ M+P R LFP +
Sbjct: 298 LIYNERRAIGKCPLTPEEVGLILRAMRFDNSTRIYLAAGELFGGEQFMKPFRTLFPRLDN 357
Query: 306 KEMLAEEELKPFLSFSSRLAAVDYIVCDESNVFVANNNG--NMARILAGQRRYMGHKRTI 363
+ E S +AVDY+VC S++F+ +G N A L G R Y G + TI
Sbjct: 358 HSSVDPSEELSATSQGLIGSAVDYMVCLLSDIFMPTYDGPSNFANNLLGHRLYYGFRTTI 417
Query: 364 RPNAKKLSALFMARHE 379
RP+ K L+ +F+AR +
Sbjct: 418 RPDRKALAPIFIAREK 433
>At5g01100 unknown protein
Length = 631
Score = 265 bits (677), Expect = 4e-71
Identities = 156/429 (36%), Positives = 242/429 (56%), Gaps = 26/429 (6%)
Query: 9 DIWESQYSKYYYGCKERGRHFYPAIRERMSNGYLLIAASGGLNQQRTGITDAVVVARILN 68
+IW Y C R ++ P + +NGYLL+ A+GGLNQ RTGI D V +A+I+N
Sbjct: 190 EIWNQPEVGNYQKCVARPKNQRPI---KQTNGYLLVHANGGLNQMRTGICDMVAIAKIMN 246
Query: 69 ATLVVPELDHNSFWKDDSDFANIFDVNWFINYLAKDVTIVKRVPDKV--MRSMDKPPYTM 126
ATLV+P LDH+SFW D S F +IFD FI LA+DV IV+ +P + ++ ++K P +
Sbjct: 247 ATLVLPFLDHSSFWSDPSSFKDIFDWKHFIKVLAEDVNIVEYLPQEFASIKPLEKNPVSW 306
Query: 127 RVPRKSDPEYYLDQVLPILLRRRVLQLTKFDYRLANDL-DDELQKLRCRVNYHALRFTKP 185
S YY + + +L + +V+ D RLAN+ +Q+LRCR NY ALR+++
Sbjct: 307 -----SKSSYYRNSISKLLKKHKVIVFNHTDSRLANNSPPPSIQRLRCRANYEALRYSED 361
Query: 186 IRQLGQRIVMRMRKMANRYIAVHLRFEPDMLAFSGCYFGGGEKERQELGEIRKRWTTLPD 245
I L + R+R+ Y+A+HLR+E DMLAF+GC +E +L ++R +
Sbjct: 362 IENLSNVLSSRLRENNEPYLALHLRYEKDMLAFTGCNHSLSNEESIDLEKMRFSIPHWKE 421
Query: 246 LSPDGERKR--GKCPLTPHEVGLMLRALGYTNDTYLYVASGEIYGGDETMQPLRDLFPNI 303
+G +R G CP+TP E + L+A+G+ + T +Y+ +G+IY G +M + FPN+
Sbjct: 422 KVINGTERRLEGNCPMTPREAAVFLKAMGFPSTTNIYIVAGKIY-GQNSMTAFHEEFPNV 480
Query: 304 YTKEMLA-EEELKPFLSFSSRLAAVDYIVCDESNVFVANNNGNMARILAGQRRYMGHKRT 362
+ LA EEEL + +RLAA+DY + ES++F +GNMA+ + G RR+ G ++T
Sbjct: 481 FFHNTLATEEELSTIKPYQNRLAALDYNLALESDIFAYTYDGNMAKAVQGHRRFEGFRKT 540
Query: 363 IRPNAKKLSALF--MARHEMDWDTFSRKVKACQRGFMGEPGEMKAGRG--------EFHE 412
I P+ ++ L + + W+ FS KVK + +G P + G+ F+
Sbjct: 541 INPDRQRFVRLIDRLDAGLISWEDFSSKVKKMHQHRIGAPYLRQPGKAGMSPKLEENFYA 600
Query: 413 YP-STCVCE 420
P CVC+
Sbjct: 601 NPLPGCVCD 609
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,243,605
Number of Sequences: 26719
Number of extensions: 499423
Number of successful extensions: 1169
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 992
Number of HSP's gapped (non-prelim): 46
length of query: 467
length of database: 11,318,596
effective HSP length: 103
effective length of query: 364
effective length of database: 8,566,539
effective search space: 3118220196
effective search space used: 3118220196
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC119408.12


