

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149600.13 + phase: 0 /pseudo
(232 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
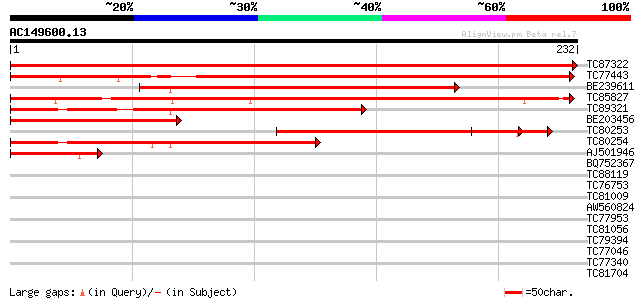
Score E
Sequences producing significant alignments: (bits) Value
TC87322 similar to GP|12322716|gb|AAG51340.1 unknown protein; 28... 464 e-131
TC77443 similar to GP|9758634|dbj|BAB09296.1 RNA-binding protein... 237 3e-63
BE239611 similar to PIR|B84807|B84 probable RNA-binding protein ... 226 7e-60
TC85827 similar to GP|16930473|gb|AAL31922.1 At1g09660/F21M12_5 ... 208 2e-54
TC89321 similar to PIR|B84807|B84807 probable RNA-binding protei... 181 2e-46
BE203456 similar to GP|12322716|gb| unknown protein; 28504-31237... 146 6e-36
TC80253 homologue to PIR|B84807|B84807 probable RNA-binding prot... 142 1e-34
TC80254 similar to PIR|B84807|B84807 probable RNA-binding protei... 129 1e-30
AJ501946 similar to PIR|B84807|B848 probable RNA-binding protein... 65 2e-11
BQ752367 29 1.3
TC88119 similar to GP|10178284|emb|CAC08342. rev interacting pro... 29 1.7
TC76753 SP|O65731|RS5_CICAR 40S ribosomal protein S5 (Fragment).... 28 2.3
TC81009 similar to GP|13605619|gb|AAK32803.1 AT3g49260/F2K15_120... 28 3.0
AW560824 similar to PIR|T04576|T04 hypothetical protein T12H17.2... 28 3.0
TC77953 similar to PIR|D84890|D84890 probable AT-hook DNA-bindin... 28 3.0
TC81056 28 3.9
TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding... 28 3.9
TC77046 similar to PIR|S35942|S35942 probable ATP synthase chain... 27 5.1
TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog ... 27 6.6
TC81704 27 8.7
>TC87322 similar to GP|12322716|gb|AAG51340.1 unknown protein; 28504-31237
{Arabidopsis thaliana}, partial (62%)
Length = 1356
Score = 464 bits (1194), Expect = e-131
Identities = 232/232 (100%), Positives = 232/232 (100%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 60
MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS
Sbjct: 324 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 503
Query: 61 LSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTG 120
LSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTG
Sbjct: 504 LSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVEATTG 683
Query: 121 CRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEEL 180
CRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEEL
Sbjct: 684 CRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEEL 863
Query: 181 LKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNEMIKRAKTD 232
LKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNEMIKRAKTD
Sbjct: 864 LKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSNEMIKRAKTD 1019
>TC77443 similar to GP|9758634|dbj|BAB09296.1 RNA-binding protein-like
{Arabidopsis thaliana}, partial (80%)
Length = 1332
Score = 237 bits (604), Expect = 3e-63
Identities = 133/248 (53%), Positives = 165/248 (65%), Gaps = 17/248 (6%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRAS---------GKSGLMQNQGFSDYDRVQFGSTKP-------- 43
M VLP C RLLNQEILR + G+SGL + G P
Sbjct: 237 MAVLPHCYRLLNQEILRVTTILGNASVLGQSGLEHGSPLAAGGMFSKGGLDPNGWVSRFQ 416
Query: 44 SLMPSLDTTSSFTGWNSLSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGR 103
S MPSL +S W LS +G S+ +VKK +R+DIP D+ P FNFVGR
Sbjct: 417 SEMPSLIQSSPTPSW--LSPQGS----------SSGLLVKKTIRVDIPVDSFPNFNFVGR 560
Query: 104 LLGPRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPV 163
LLGPRGNSLKRVEA T CRV IRG+GSIKD +EE++RG+PG+EHLNEPLHIL+EAELP
Sbjct: 561 LLGPRGNSLKRVEANTECRVLIRGRGSIKDTAREEMMRGKPGYEHLNEPLHILVEAELPA 740
Query: 164 NVVDLRLRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSFREESPQLSGSLSPFTSN 223
++D RL QA+EI+E+LL+PV+ES D YK+QQLRELAM+N + REE +SGS+SPF ++
Sbjct: 741 EIIDARLMQAREILEDLLRPVEESHDFYKKQQLRELAMINGTLREEGSPMSGSVSPFHNS 920
Query: 224 EMIKRAKT 231
+KRAKT
Sbjct: 921 LGMKRAKT 944
>BE239611 similar to PIR|B84807|B84 probable RNA-binding protein [imported] -
Arabidopsis thaliana, partial (40%)
Length = 656
Score = 226 bits (575), Expect = 7e-60
Identities = 109/137 (79%), Positives = 121/137 (87%), Gaps = 6/137 (4%)
Frame = +1
Query: 54 SFTGWNSLSHE------GLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGP 107
+ TGWNSLSH+ GLN+DWQRAP + NSHIVKK+LRLDIP D +P FNFVGRLLGP
Sbjct: 7 NLTGWNSLSHDMLAEVNGLNMDWQRAPVVPNSHIVKKILRLDIPKDGYPNFNFVGRLLGP 186
Query: 108 RGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVD 167
RGNSLKRVEATTGCRV+IRGKGSIKD DKEELLRGRPG+EHL++ LHILIEAELP N+VD
Sbjct: 187 RGNSLKRVEATTGCRVYIRGKGSIKDLDKEELLRGRPGYEHLSDELHILIEAELPANIVD 366
Query: 168 LRLRQAQEIIEELLKPV 184
+RLR AQEIIEELLKPV
Sbjct: 367 VRLRHAQEIIEELLKPV 417
>TC85827 similar to GP|16930473|gb|AAL31922.1 At1g09660/F21M12_5
{Arabidopsis thaliana}, partial (65%)
Length = 1499
Score = 208 bits (529), Expect = 2e-54
Identities = 123/247 (49%), Positives = 153/247 (61%), Gaps = 16/247 (6%)
Frame = +1
Query: 1 MQVLPLCTRLLNQEILR-ASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWN 59
+QVLP TRLL QEI R +S SG + D F +P L T W
Sbjct: 319 LQVLPQSTRLLTQEIRRISSAGSGFIMEHDHPDSSTTPF---RPPLPQHPITRPMDFDWP 489
Query: 60 SLSHEG----------LNVDWQRAPAISNSHIVKKMLRLDIPHDNHPT-FNFVGRLLGPR 108
G V W I + IVK+++RLD+P D +P +NFVGR+LGPR
Sbjct: 490 HREDNGNIQRMGSFQASPVGWHGPQGIPTTPIVKRVIRLDVPVDKYPNQYNFVGRILGPR 669
Query: 109 GNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDL 168
GNSLKRVEA T CRV+IRG GS+KD KEE L+ +PG+EHL EPLH+L+EAE P ++++
Sbjct: 670 GNSLKRVEAMTECRVYIRGCGSVKDSIKEEKLKDKPGYEHLKEPLHLLVEAEFPEDIINS 849
Query: 169 RLRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSFREE----SPQLSGSLSPFTSNE 224
RL A ++E LLKPVDES D YK+QQLRELAM+N + REE SP +S S+SPF SN
Sbjct: 850 RLDHAVAVLENLLKPVDESLDHYKKQQLRELAMINGTLREESPSMSPSMSPSMSPFNSNG 1029
Query: 225 MIKRAKT 231
M KRAKT
Sbjct: 1030M-KRAKT 1047
>TC89321 similar to PIR|B84807|B84807 probable RNA-binding protein
[imported] - Arabidopsis thaliana, partial (62%)
Length = 907
Score = 181 bits (460), Expect = 2e-46
Identities = 93/152 (61%), Positives = 116/152 (76%), Gaps = 6/152 (3%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 60
+++LP +RLLNQEILR SG ++ NQGF+D+DR++ S P ++S+ TGWN+
Sbjct: 477 IKILPNSSRLLNQEILRVSG---MLSNQGFADFDRLRHRSPSPL------SSSNLTGWNN 629
Query: 61 LSHE------GLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKR 114
L HE G+ +DWQ APA +S+ VK++LRL+IP D +P FNFVGRLLGPRGNSLKR
Sbjct: 630 LQHERLCGTPGMTMDWQGAPASPSSYTVKRILRLEIPVDTYPNFNFVGRLLGPRGNSLKR 809
Query: 115 VEATTGCRVFIRGKGSIKDFDKEELLRGRPGF 146
VEATTGCRVFIRGKGSIKD DKEE LRGRPG+
Sbjct: 810 VEATTGCRVFIRGKGSIKDPDKEEKLRGRPGY 905
>BE203456 similar to GP|12322716|gb| unknown protein; 28504-31237
{Arabidopsis thaliana}, partial (10%)
Length = 539
Score = 146 bits (369), Expect = 6e-36
Identities = 70/70 (100%), Positives = 70/70 (100%)
Frame = +2
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 60
MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS
Sbjct: 329 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNS 508
Query: 61 LSHEGLNVDW 70
LSHEGLNVDW
Sbjct: 509 LSHEGLNVDW 538
>TC80253 homologue to PIR|B84807|B84807 probable RNA-binding protein
[imported] - Arabidopsis thaliana, partial (39%)
Length = 1063
Score = 142 bits (358), Expect = 1e-34
Identities = 74/102 (72%), Positives = 84/102 (81%), Gaps = 1/102 (0%)
Frame = +3
Query: 110 NSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLR 169
+SLKRVEATTGCRV+IRGKGSIKD +KE+ LRGRPG+EHLNE LHILIEA+LP NVVD+R
Sbjct: 12 HSLKRVEATTGCRVYIRGKGSIKDPEKEDKLRGRPGYEHLNENLHILIEADLPANVVDIR 191
Query: 170 LRQAQEIIEELLKPVDESQDIYKRQQLRELAMLNSSF-REES 210
LRQAQEIIEELLKPVDESQD R + ++ F R ES
Sbjct: 192 LRQAQEIIEELLKPVDESQDFI*RAAVARISSAEFKFQRRES 317
Score = 49.7 bits (117), Expect = 1e-06
Identities = 23/33 (69%), Positives = 29/33 (87%)
Frame = +1
Query: 190 IYKRQQLRELAMLNSSFREESPQLSGSLSPFTS 222
+++ QQLRELA+LNS+FREESP SGS+SPF S
Sbjct: 253 LFRGQQLRELALLNSNFREESPGPSGSVSPFNS 351
>TC80254 similar to PIR|B84807|B84807 probable RNA-binding protein
[imported] - Arabidopsis thaliana, partial (60%)
Length = 637
Score = 129 bits (323), Expect = 1e-30
Identities = 71/136 (52%), Positives = 88/136 (64%), Gaps = 9/136 (6%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTG--- 57
+QVLP+C+RLLNQEILR SG ++ NQGF D+DR+Q S P +L + S TG
Sbjct: 237 LQVLPICSRLLNQEILRVSG---MLSNQGFGDFDRLQHRSPSPMASSNLMSNVSGTGMGA 407
Query: 58 WNSLSHE------GLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNS 111
WNSL E G+N+DWQ APA +S VK++LRL+IP D P FNF + G
Sbjct: 408 WNSLQQERLCGPPGMNMDWQSAPASPSSFTVKRILRLEIPVDTFPNFNFCWKTSGT*RQF 587
Query: 112 LKRVEATTGCRVFIRG 127
+RVEATTGCRVFIRG
Sbjct: 588 FERVEATTGCRVFIRG 635
>AJ501946 similar to PIR|B84807|B848 probable RNA-binding protein [imported]
- Arabidopsis thaliana, partial (12%)
Length = 421
Score = 65.5 bits (158), Expect = 2e-11
Identities = 31/39 (79%), Positives = 37/39 (94%), Gaps = 1/39 (2%)
Frame = +3
Query: 1 MQVLPLCTRLLNQEILRASGKSGLMQN-QGFSDYDRVQF 38
MQVLPLC+RLLNQEILR SGK+GL+QN QGF+D+DR+QF
Sbjct: 285 MQVLPLCSRLLNQEILRVSGKNGLLQNHQGFNDFDRMQF 401
>BQ752367
Length = 959
Score = 29.3 bits (64), Expect = 1.3
Identities = 14/44 (31%), Positives = 21/44 (46%)
Frame = -2
Query: 73 APAISNSHIVKKMLRLDIPHDNHPTFNFVGRLLGPRGNSLKRVE 116
AP N H+ + + + NHP + +G LG RG + VE
Sbjct: 526 APCRDNRHVKETITTATLWRSNHPGRHAIGTELGKRGKHVAAVE 395
>TC88119 similar to GP|10178284|emb|CAC08342. rev interacting protein
mis3-like {Arabidopsis thaliana}, partial (79%)
Length = 1369
Score = 28.9 bits (63), Expect = 1.7
Identities = 10/24 (41%), Positives = 17/24 (70%)
Frame = +1
Query: 104 LLGPRGNSLKRVEATTGCRVFIRG 127
L+GP ++LK +E TGC + ++G
Sbjct: 613 LVGPNSSTLKALEILTGCYILVQG 684
>TC76753 SP|O65731|RS5_CICAR 40S ribosomal protein S5 (Fragment). [Chickpea
Garbanzo] {Cicer arietinum}, partial (66%)
Length = 1221
Score = 28.5 bits (62), Expect = 2.3
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +3
Query: 36 VQFGSTKPSLMPSLDTTSSFTGWNSLSHE 64
+Q G KPSL L T+ + WN SH+
Sbjct: 357 IQLGKKKPSLTTMLVTSYNLWHWNPFSHQ 443
>TC81009 similar to GP|13605619|gb|AAK32803.1 AT3g49260/F2K15_120
{Arabidopsis thaliana}, partial (19%)
Length = 935
Score = 28.1 bits (61), Expect = 3.0
Identities = 18/56 (32%), Positives = 29/56 (51%)
Frame = +1
Query: 44 SLMPSLDTTSSFTGWNSLSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFN 99
S M L TTS+ T +++S + + +D P+ N ++ L HD+ PTFN
Sbjct: 247 SYMTLLSTTSTTT--DNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTFN 408
>AW560824 similar to PIR|T04576|T04 hypothetical protein T12H17.200 -
Arabidopsis thaliana, partial (38%)
Length = 531
Score = 28.1 bits (61), Expect = 3.0
Identities = 12/38 (31%), Positives = 27/38 (70%), Gaps = 1/38 (2%)
Frame = -2
Query: 162 PVNVVDLRLRQAQEIIEELLKPVDESQDIYKR-QQLRE 198
P+N+ + ++A++++EE ++P ES+ ++ QQL+E
Sbjct: 287 PLNIWSNQKQRAEQVVEETIQPAKESRTFHEV*QQLQE 174
>TC77953 similar to PIR|D84890|D84890 probable AT-hook DNA-binding protein
[imported] - Arabidopsis thaliana, partial (57%)
Length = 1695
Score = 28.1 bits (61), Expect = 3.0
Identities = 12/38 (31%), Positives = 27/38 (70%), Gaps = 1/38 (2%)
Frame = -3
Query: 162 PVNVVDLRLRQAQEIIEELLKPVDESQDIYKR-QQLRE 198
P+N+ + ++A++++EE ++P ES+ ++ QQL+E
Sbjct: 1045 PLNIWSNQKQRAEQVVEETIQPAKESRTFHEV*QQLQE 932
>TC81056
Length = 951
Score = 27.7 bits (60), Expect = 3.9
Identities = 23/87 (26%), Positives = 38/87 (43%)
Frame = +1
Query: 18 ASGKSGLMQNQGFSDYDRVQFGSTKPSLMPSLDTTSSFTGWNSLSHEGLNVDWQRAPAIS 77
AS + M N+ F R+ + S+ + S +G + L H + P I
Sbjct: 586 ASSSAARMGNRAFLPNARIGLAEIEGQQFHSVGSESP-SGQSPLRHRSPS-----PPNID 747
Query: 78 NSHIVKKMLRLDIPHDNHPTFNFVGRL 104
+SH++K + D PH T +F+G L
Sbjct: 748 HSHLMKSLAMQDHPH-TRKTSHFLGGL 825
>TC79394 weakly similar to PIR|T49019|T49019 probable RNA binding protein -
Arabidopsis thaliana, partial (74%)
Length = 1909
Score = 27.7 bits (60), Expect = 3.9
Identities = 25/95 (26%), Positives = 40/95 (41%)
Frame = +2
Query: 107 PRGNSLKRVEATTGCRVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVV 166
P GNS +G +V + G D D+EE +E E E+ V
Sbjct: 188 PGGNSEPEKPTDSGEQVDLDG-----DNDQEESSEEEVEYE----------EVEVEEEVE 322
Query: 167 DLRLRQAQEIIEELLKPVDESQDIYKRQQLRELAM 201
+ + +E +EE KP+DE + K++ LA+
Sbjct: 323 EEEEEEEEEEVEEESKPLDEEDEADKKKHAELLAL 427
>TC77046 similar to PIR|S35942|S35942 probable ATP synthase chain - soybean,
partial (98%)
Length = 1146
Score = 27.3 bits (59), Expect = 5.1
Identities = 22/71 (30%), Positives = 34/71 (46%), Gaps = 7/71 (9%)
Frame = +2
Query: 132 KDFDKEELLRGRPG-------FEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELLKPV 184
KD K L + G F+ +N L I E ELP + L L+ A+ +EEL K
Sbjct: 557 KDLKKPLLRNDKKGMDLLLAEFDKINNKLGIRKE-ELPKHEEQLELKIAKAQLEELKKDA 733
Query: 185 DESQDIYKRQQ 195
E+ + K+++
Sbjct: 734 VEAMETQKKRE 766
>TC77340 SP|Q9BAE0|FTSH_MEDSA Cell division protein ftsH homolog chloroplast
precursor (EC 3.4.24.-). [Alfalfa] {Medicago sativa},
complete
Length = 2473
Score = 26.9 bits (58), Expect = 6.6
Identities = 25/100 (25%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Frame = +3
Query: 122 RVFIRGKGSIKDFDKEELLRGRPGFEHLNEPLHILIEAELPVNVVDLRLRQAQEIIEELL 181
+V RGK KD D +++ R PGF + +++ EA + DL+ EI + L
Sbjct: 1548 QVHSRGKALAKDVDFDKIARRTPGFTGA-DLQNLMNEAAILAARRDLKEISKDEIADALE 1724
Query: 182 KPV--DESQDIYKRQQLRELAMLNSSFREESPQLSGSLSP 219
+ + E ++ ++ ++L ++ E L G+L P
Sbjct: 1725 RIIAGPEKKNAVVSEEKKKLV----AYHEAGHALVGALMP 1832
>TC81704
Length = 571
Score = 26.6 bits (57), Expect = 8.7
Identities = 20/70 (28%), Positives = 31/70 (43%)
Frame = -2
Query: 43 PSLMPSLDTTSSFTGWNSLSHEGLNVDWQRAPAISNSHIVKKMLRLDIPHDNHPTFNFVG 102
P+ + SL SF W+ L H +++ PA+ VK + IP++ FNF+
Sbjct: 204 PNGISSLHVPFSFAIWSRLRHNLVSI*LYI*PAVHVKFFVKH*ILFPIPYN---IFNFIQ 34
Query: 103 RLLGPRGNSL 112
P N L
Sbjct: 33 TENNPFRNRL 4
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,198,598
Number of Sequences: 36976
Number of extensions: 78741
Number of successful extensions: 393
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 383
length of query: 232
length of database: 9,014,727
effective HSP length: 93
effective length of query: 139
effective length of database: 5,575,959
effective search space: 775058301
effective search space used: 775058301
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149600.13


