

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149600.1 - phase: 0
(494 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
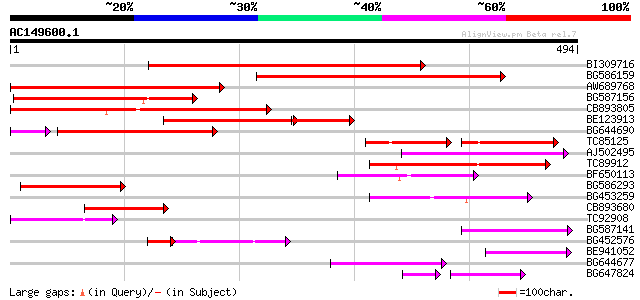
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 232 3e-61
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 196 2e-50
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 189 3e-48
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 186 1e-47
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 172 2e-43
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 113 2e-39
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 125 2e-33
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 73 3e-28
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 114 9e-26
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 112 4e-25
BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vu... 106 2e-23
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 92 3e-19
BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, ... 89 4e-18
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 73 2e-13
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 63 2e-10
BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440... 62 4e-10
BG452576 weakly similar to GP|12005223|gb|A reverse transcriptas... 56 7e-10
BE941052 weakly similar to PIR|B85188|B85 retrotransposon like p... 59 4e-09
BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T... 57 2e-08
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 52 5e-08
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 232 bits (591), Expect = 3e-61
Identities = 108/241 (44%), Positives = 170/241 (69%)
Frame = +2
Query: 122 KVCKLKKALYGLKQSPRAWFGRFTKSMKTFGYKASNSDHTLFLKRGEGKITALIIYVDDM 181
KVC+L+K++YGLKQ+ R W+ + ++S+ +FGY S+SD +LF K + T L++YVDD+
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 182 IVTGNNQDEISSLQKYLTSKFEMKQLGNLKYFMGIEVARSKHGIFLCQRKYTLDLLSETG 241
++ GN+ EI ++ +L +F++K LG+L+YF+G+EVARSK GI L QRKYTL+LL ++G
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 242 LLASKSAETPIEQNHKLFHCLNSNITDKGRYQSLVGKLIYLSHTRPDITYAVNVVSQFMH 301
LA KS TP + + KL + + D+ +Y+ L+GKLIYL+ TRPDI++AV +SQF+
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 302 DPRKPHMDVVERILRYLKSTPGKGILFSNNGHLRVEGYTDADWAGSADDRRSTSGYFTFV 361
P++ H R+L+YLK+ P KG+ +S +L++ + D+DWA R+S +GY+ F+
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
Query: 362 G 362
G
Sbjct: 737 G 739
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 196 bits (498), Expect = 2e-50
Identities = 92/217 (42%), Positives = 138/217 (63%)
Frame = +1
Query: 216 IEVARSKHGIFLCQRKYTLDLLSETGLLASKSAETPIEQNHKLFHCLNSNITDKGRYQSL 275
+EV +++ GI++CQRKY DLL G+ S + PI KL N D +Y+ +
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 276 VGKLIYLSHTRPDITYAVNVVSQFMHDPRKPHMDVVERILRYLKSTPGKGILFSNNGHLR 335
VG L+YL+ TRPD+ Y ++++S+FM+ P + HM V+R+LRYL T GI++ NG +
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 336 VEGYTDADWAGSADDRRSTSGYFTFVGGNLVTWRSKKQPVVARSSAEVEFRGMALGMCEL 395
+E YTD+D+AG DDR+STSGY + V+W SKKQPVV S+ + EF A C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 396 LWVKSVLSDLGFEPKEAMSLYCDNTSAIEIAHNPVQH 432
+W++ VL LG+ ++++YCDN S I+++ NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 189 bits (479), Expect = 3e-48
Identities = 86/187 (45%), Positives = 123/187 (64%)
Frame = +1
Query: 1 MVEEMTTLKKNNTWDLVTLPRGKKTVGCRWVFTIKHKADGTIDRYKARLVAKGYTQSYGV 60
M E L N TWDLV LP KK +GC+WV+ +K DG+++++KARLVAKG++Q+ G
Sbjct: 109 MKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQTLGC 288
Query: 61 DYQETFAPVAKLNTVRILLSLAANQDWPLLQFDVKNAFLHGEISEDIYMDTPPGMVYSNG 120
DY ETF+PV K T+R++L++A W + Q D+ NAFL+G + E++YM P G +N
Sbjct: 289 DYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFEAANK 468
Query: 121 LKVCKLKKALYGLKQSPRAWFGRFTKSMKTFGYKASNSDHTLFLKRGEGKITALIIYVDD 180
VCKL K+LYGLKQ+PRAW+ T + FG+ S D +L + G L IYVDD
Sbjct: 469 SLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXIYVDD 648
Query: 181 MIVTGNN 187
+++TG++
Sbjct: 649 ILITGSS 669
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 186 bits (473), Expect = 1e-47
Identities = 91/162 (56%), Positives = 119/162 (73%), Gaps = 2/162 (1%)
Frame = -1
Query: 4 EMTTLKKNNTWDLVTLPRGKKTVGCRWVFTIKHKADGTIDRYKARLVAKGYTQSYGVDYQ 63
E + KN+TW LP+GKK V RW+FTIK+KADG+I+R K RLVA+G+T +YG DY
Sbjct: 501 EAGAMIKNDTWYESELPKGKKAVSSRWIFTIKYKADGSIERKKTRLVARGFTLTYGEDYI 322
Query: 64 ETFAPVAKLNTVRILLSLAANQDWPLLQFDVKNAFLHGEISEDIYMDTPPGM--VYSNGL 121
ETFAPVAKL+T+RI+LSLA N W L Q DVKNAFL GE+ +++YM PPG+ + G
Sbjct: 321 ETFAPVAKLHTIRIVLSLAVNLGWGLWQMDVKNAFLQGELEDEVYMYPPPGLEHLVKRG- 145
Query: 122 KVCKLKKALYGLKQSPRAWFGRFTKSMKTFGYKASNSDHTLF 163
V +LKKA+YGLKQSPRAW+ + + ++ G++ S DHTLF
Sbjct: 144 NVLRLKKAIYGLKQSPRAWYNKLSTTLNGRGFRKSELDHTLF 19
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 172 bits (437), Expect = 2e-43
Identities = 91/232 (39%), Positives = 145/232 (62%), Gaps = 4/232 (1%)
Frame = +3
Query: 1 MVEEMTTLKKNNTWDLVTLPRGKKTVGCRWVFTIKHKADGTIDRYKARLVAKGYTQSYGV 60
M EM ++NNTW+L L G KT+G +W+F K +G I++YKARLVAKGY+Q YGV
Sbjct: 90 MNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNENGEIEKYKARLVAKGYSQQYGV 269
Query: 61 DYQETFAPVAKLNTVRILLSLAA--NQDWPLLQFDVK-NAFLHGEISEDIYMDTPPGMVY 117
DY E FAPVA+ +T+R++++LAA +D + K ++ + + + + ++ V
Sbjct: 270 DYTEVFAPVARWDTIRMVIALAAQIKRDGVCIS*M*KAHSCMEN*MRKFLLIN---HRVM 440
Query: 118 SNGLKVCKLKKALYGLKQSPRAWFGRFTKSMKTFGYKASNSDHTLFLKRGE-GKITALII 176
+ ++K+ALYGLKQ+PRAW+ R G++ +HTLF+K E GKI + +
Sbjct: 441 *RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTKEGFEKCPYEHTLFVKLSEGGKILIISL 620
Query: 177 YVDDMIVTGNNQDEISSLQKYLTSKFEMKQLGNLKYFMGIEVARSKHGIFLC 228
YVDD+I GN+++ +K + +F M LG + YF+G+EV +++ GI++C
Sbjct: 621 YVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGKMHYFLGVEVTQNEKGIYIC 776
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 113 bits (283), Expect(2) = 2e-39
Identities = 58/118 (49%), Positives = 76/118 (64%), Gaps = 1/118 (0%)
Frame = +1
Query: 135 QSPRAWFGRFTKSMKTFGYKASNSDHTLFLKRGEG-KITALIIYVDDMIVTGNNQDEISS 193
QSPR WF RFT +K FGY +DH +F+K K LI+YVDD+ +TG++ I
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 194 LQKYLTSKFEMKQLGNLKYFMGIEVARSKHGIFLCQRKYTLDLLSETGLLASKSAETP 251
L+ L +FE+K LGNLKYF+G+EVAR K G + QRKY LDLL ET ++ K+ P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 67.8 bits (164), Expect(2) = 2e-39
Identities = 33/55 (60%), Positives = 40/55 (72%)
Frame = +2
Query: 246 KSAETPIEQNHKLFHCLNSNITDKGRYQSLVGKLIYLSHTRPDITYAVNVVSQFM 300
K +ETP++ KL N + DKGRYQ LVGKLIYLSHTRPDI++ V +SQFM
Sbjct: 338 KPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 125 bits (313), Expect(2) = 2e-33
Identities = 60/141 (42%), Positives = 94/141 (66%), Gaps = 1/141 (0%)
Frame = -2
Query: 42 IDRYKARLVAKGYTQSYGVDYQETFAPVAKLNTVRILLSLAANQDWPLLQFDVKNAFLHG 101
I R K++LV +GY Q G+DY E F+PVA++ +RIL++ AA + L Q DVK+AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 102 EISEDIYMDTPPGMVYSN-GLKVCKLKKALYGLKQSPRAWFGRFTKSMKTFGYKASNSDH 160
++ E++++ PPG + V +L K LYGLKQ+PRAW+ R +K + G+K D+
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 161 TLFLKRGEGKITALIIYVDDM 181
TLFL + E ++ + +YVDD+
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 35.8 bits (81), Expect(2) = 2e-33
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = -3
Query: 1 MVEEMTTLKKNNTWDLVTLPRGKKTVGCRWVFTIK 35
M EE+ +++ W LV P GK +G RWVF K
Sbjct: 537 MQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNK 433
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 72.8 bits (177), Expect(2) = 3e-28
Identities = 30/85 (35%), Positives = 56/85 (65%)
Frame = +3
Query: 394 ELLWVKSVLSDLGFEPKEAMSLYCDNTSAIEIAHNPVQHDRTKHVEIDRHFIKEKLDAGT 453
E +W++ ++ +LG + +E +++YCD+ SA+ IA NP H RTKH+ I HF++E ++ G+
Sbjct: 249 EAIWMQRLMEELGHK-QEQITVYCDSQSALHIARNPAFHSRTKHIGIQYHFVREVVEEGS 425
Query: 454 IVFPFVRSEQQLADMLTKGVSSKVF 478
+ + + LAD +TK +++ F
Sbjct: 426 VDMQKIHTNDNLADAMTKSINTDKF 500
Score = 70.5 bits (171), Expect(2) = 3e-28
Identities = 35/75 (46%), Positives = 48/75 (63%)
Frame = +1
Query: 311 VERILRYLKSTPGKGILFSNNGHLRVEGYTDADWAGSADDRRSTSGYFTFVGGNLVTWRS 370
V+RI+RY+K T G + F + L V GY D+D+AG D R+ST+GY + G V+W S
Sbjct: 1 VKRIMRYIKGTSGVAVCFGGS-ELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLS 177
Query: 371 KKQPVVARSSAEVEF 385
K Q VVA S+ E E+
Sbjct: 178 KLQTVVALSTTEAEY 222
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 114 bits (285), Expect = 9e-26
Identities = 55/146 (37%), Positives = 87/146 (58%)
Frame = +2
Query: 342 ADWAGSADDRRSTSGYFTFVGGNLVTWRSKKQPVVARSSAEVEFRGMALGMCELLWVKSV 401
+DWAG + R+STSGY +G ++W SKKQPVVA S+AE E+ + +W++ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 402 LSDLGFEPKEAMSLYCDNTSAIEIAHNPVQHDRTKHVEIDRHFIKEKLDAGTIVFPFVRS 461
L + E +YCDN SAI ++ NPV H R+KH++I H I+E + +V + +
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 462 EQQLADMLTKGVSSKVFNESLLKLGM 487
E+++AD+ TK + + F + LGM
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGM 439
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 112 bits (279), Expect = 4e-25
Identities = 53/160 (33%), Positives = 99/160 (61%), Gaps = 2/160 (1%)
Frame = +1
Query: 314 ILRYLKSTPGKGILFSNNGHLR--VEGYTDADWAGSADDRRSTSGYFTFVGGNLVTWRSK 371
+L+YL + + ++ +EGY DAD+AG+ D R+S SG+ + G ++W++
Sbjct: 16 VLKYLNESLKSSLKYTKAAQEEDALEGYVDADYAGNVDTRKSLSGFVFTLYGTTISWKAN 195
Query: 372 KQPVVARSSAEVEFRGMALGMCELLWVKSVLSDLGFEPKEAMSLYCDNTSAIEIAHNPVQ 431
+Q VV S+ + E+ G+ + +W+K ++ +LG +E + ++CD+ SAI +A++ V
Sbjct: 196 QQSVVTLSTTQAEYIAFVEGVKDAIWLKGMIGELGI-TQEYVKIHCDSQSAIHLANHQVY 372
Query: 432 HDRTKHVEIDRHFIKEKLDAGTIVFPFVRSEQQLADMLTK 471
H+RTKH++I HFI++ +++ IV + SE+ AD+ TK
Sbjct: 373 HERTKHIDIRLHFIRDMIESKEIVVEKMASEENPADVFTK 492
>BF650113 weakly similar to GP|4753889|emb| Tpv2-1c {Phaseolus vulgaris},
partial (13%)
Length = 494
Score = 106 bits (265), Expect = 2e-23
Identities = 56/126 (44%), Positives = 75/126 (59%), Gaps = 3/126 (2%)
Frame = +1
Query: 286 RPDITYAVNVVSQFMHDPRKPHMDVVERILRYLKSTPGKGILFSNNGHLRVEG---YTDA 342
RPDI Y+V+V+S+FMHDPRKPH+ RILRY++ T G+LF V Y+D+
Sbjct: 121 RPDICYSVSVISKFMHDPRKPHLIAANRILRYVRGTMEYGLLFPYGAKSEVYELICYSDS 300
Query: 343 DWAGSADDRRSTSGYFTFVGGNLVTWRSKKQPVVARSSAEVEFRGMALGMCELLWVKSVL 402
DW G DRRSTSGY ++W +KKQP+ A SS E E+ + LW+ SV+
Sbjct: 301 DWCG---DRRSTSGYVFKFNDAAISWCTKKQPITALSSYEAEYIAGTFATFQALWLDSVI 471
Query: 403 SDLGFE 408
+L E
Sbjct: 472 KELKCE 489
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 92.4 bits (228), Expect = 3e-19
Identities = 44/92 (47%), Positives = 60/92 (64%)
Frame = +2
Query: 10 KNNTWDLVTLPRGKKTVGCRWVFTIKHKADGTIDRYKARLVAKGYTQSYGVDYQETFAPV 69
+ T LV P G K +G RW++ IK DGT+ +YKARLVAKGY + G+D+ E FAPV
Sbjct: 47 QKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPV 226
Query: 70 AKLNTVRILLSLAANQDWPLLQFDVKNAFLHG 101
++ T+ +LL+LAA + DVK AFL+G
Sbjct: 227 VRIETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG453259 homologue to GP|21434|emb|CA ORF4 {Solanum tuberosum}, partial (5%)
Length = 657
Score = 89.0 bits (219), Expect = 4e-18
Identities = 60/144 (41%), Positives = 82/144 (56%), Gaps = 2/144 (1%)
Frame = -2
Query: 314 ILRYLKSTPGKGILFSNNGHLRVEGYTDADWAGSADDRRSTSGYFTFVGGNLVTWRSKKQ 373
++ L S+ + I+F L +E Y + AG DR STSGY F+GGN+V KQ
Sbjct: 566 LIEGLXSSGTQMIVFKRE*KLSMEVYXNTVCAGWIVDRGSTSGY*MFLGGNMV---E*KQ 396
Query: 374 PVVARSSAEVEFRGMALGMCELL--WVKSVLSDLGFEPKEAMSLYCDNTSAIEIAHNPVQ 431
VVAR F + G+ ++ +K L DL K+ M+L+ +N IAHNPVQ
Sbjct: 395 NVVAR*VQRHNFELCSQGL*RVMDEELKIKLDDLIINYKDPMTLF*NNNFVSRIAHNPVQ 216
Query: 432 HDRTKHVEIDRHFIKEKLDAGTIV 455
H RTKH+EID+HFI EKL +G I+
Sbjct: 215 HYRTKHIEIDQHFIIEKLYSGLIL 144
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 73.2 bits (178), Expect = 2e-13
Identities = 38/73 (52%), Positives = 47/73 (64%)
Frame = -2
Query: 66 FAPVAKLNTVRILLSLAANQDWPLLQFDVKNAFLHGEISEDIYMDTPPGMVYSNGLKVCK 125
F P+ KLNT+ LLS+ A ++ L DVK AFL G++ EDIYM P G G V K
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGFS*EVGKMVGK 375
Query: 126 LKKALYGLKQSPR 138
LKK++YGLKQ PR
Sbjct: 374 LKKSMYGLKQGPR 336
Score = 33.1 bits (74), Expect = 0.25
Identities = 14/42 (33%), Positives = 26/42 (61%)
Frame = -3
Query: 181 MIVTGNNQDEISSLQKYLTSKFEMKQLGNLKYFMGIEVARSK 222
++V G+N DEI +L+ + + +MK LG K +G+++ K
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDK 146
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 63.2 bits (152), Expect = 2e-10
Identities = 40/94 (42%), Positives = 56/94 (59%)
Frame = +2
Query: 1 MVEEMTTLKKNNTWDLVTLPRGKKTVGCRWVFTIKHKADGTIDRYKARLVAKGYTQSYGV 60
M +E+ L++NNT L L GKK V CR V+ I HKA+G I++YKA+LVAK + Q G
Sbjct: 329 MEQEIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANGEIEKYKAQLVAKDFVQVEGE 508
Query: 61 DYQETFAPVAKLNTVRILLSLAANQDWPLLQFDV 94
D+ + K + R LL++AA + L DV
Sbjct: 509 DF*D-LCLSNKDDNCRCLLTIAAAKG*QLHLMDV 607
>BG587141 similar to PIR|H86461|H86 hypothetical protein AAF32440.1
[imported] - Arabidopsis thaliana, partial (20%)
Length = 731
Score = 62.4 bits (150), Expect = 4e-10
Identities = 31/97 (31%), Positives = 53/97 (53%)
Frame = +3
Query: 394 ELLWVKSVLSDLGFEPKEAMSLYCDNTSAIEIAHNPVQHDRTKHVEIDRHFIKEKLDAGT 453
+ +W++ +LS++ +EP E + + DN S I + NPV H R H+ HFI+E ++ G
Sbjct: 135 QAMWLQDLLSEVTWEPCEEVVIRIDNQSVIALTRNPVFHGRGNHIHKRYHFIRECVENGQ 314
Query: 454 IVFPFVRSEQQLADMLTKGVSSKVFNESLLKLGMCDI 490
+ V E+ A + TK + +F E +GM D+
Sbjct: 315 VEVEHVPGEKHRAYI*TKALGRIIFREIRYYIGMIDL 425
>BG452576 weakly similar to GP|12005223|gb|A reverse transcriptase-like
protein {Spiranthes hongkongensis}, partial (25%)
Length = 676
Score = 56.2 bits (134), Expect(2) = 7e-10
Identities = 32/102 (31%), Positives = 59/102 (57%)
Frame = +2
Query: 143 RFTKSMKTFGYKASNSDHTLFLKRGEGKITALIIYVDDMIVTGNNQDEISSLQKYLTSKF 202
+ ++ + GYK S +DH++F + T ++Y+DD++ ++ E ++ +L F
Sbjct: 251 KLLSTLISLGYKQSPNDHSIF--SFGRRFTIFLVYLDDIVFARDDHSETQLVKSHLDKNF 424
Query: 203 EMKQLGNLKYFMGIEVARSKHGIFLCQRKYTLDLLSETGLLA 244
+ LG L Y +G+++A S+ I Q KYT++LL E+G LA
Sbjct: 425 IIIGLGTLHY-VGVKIA*SES*IIDDQCKYTIELLEESGQLA 547
Score = 25.0 bits (53), Expect(2) = 7e-10
Identities = 9/24 (37%), Positives = 15/24 (62%)
Frame = +3
Query: 121 LKVCKLKKALYGLKQSPRAWFGRF 144
+ VCK ++YGLK + R + +F
Sbjct: 159 IAVCKFHNSIYGLKHAHRQ*YSKF 230
>BE941052 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 480
Score = 58.9 bits (141), Expect = 4e-09
Identities = 30/75 (40%), Positives = 41/75 (54%)
Frame = +2
Query: 415 LYCDNTSAIEIAHNPVQHDRTKHVEIDRHFIKEKLDAGTIVFPFVRSEQQLADMLTKGVS 474
L CD SA + HNPV H R KH+ ID HF+++ + G + V + QLAD LTK +S
Sbjct: 23 LRCDYLSATYLTHNPVYHSRMKHISIDIHFVRDLVQQGKLKVQHVCTVDQLADCLTKPLS 202
Query: 475 SKVFNESLLKLGMCD 489
K+G+ D
Sbjct: 203 KSRHQLLRNKIGVTD 247
>BG644677 weakly similar to GP|7682800|gb| Hypothetical protein T15F17.l
{Arabidopsis thaliana}, partial (3%)
Length = 539
Score = 57.0 bits (136), Expect = 2e-08
Identities = 30/101 (29%), Positives = 54/101 (52%)
Frame = -3
Query: 280 IYLSHTRPDITYAVNVVSQFMHDPRKPHMDVVERILRYLKSTPGKGILFSNNGHLRVEGY 339
I L+ P+IT+++N++S++ P H + ++ I +YLK G+ +S + + GY
Sbjct: 531 ILLTLQGPNITFSINLLSRYSSAPTMRH*NGIKHICKYLKGIIDMGLFYSKDCSPDLIGY 352
Query: 340 TDADWAGSADDRRSTSGYFTFVGGNLVTWRSKKQPVVARSS 380
+A + RS +GY G +++WRS K +A SS
Sbjct: 351 VNA*YLSDPHKARS*TGYIFTCGNTVISWRSTK*STIATSS 229
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 51.6 bits (122), Expect(2) = 5e-08
Identities = 27/66 (40%), Positives = 40/66 (59%), Gaps = 1/66 (1%)
Frame = -3
Query: 385 FRGMALGMCELLWVKSVLSDLGFEPKEAMSLYCDNTSAI-EIAHNPVQHDRTKHVEIDRH 443
+R + +CE+ W+ +L+DL F + LYCDN SA IA N +RTKH+E+D H
Sbjct: 374 YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQSAARHIAANSSFLERTKHIELDCH 195
Query: 444 FIKEKL 449
++ KL
Sbjct: 194 IVRVKL 177
Score = 23.5 bits (49), Expect(2) = 5e-08
Identities = 11/33 (33%), Positives = 19/33 (57%)
Frame = -2
Query: 343 DWAGSADDRRSTSGYFTFVGGNLVTWRSKKQPV 375
D + D +S S + F+G +L+ W+S K+ V
Sbjct: 486 D*SSCLDT*KSISYFCIFLGDSLICWKS*KKKV 388
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,530,711
Number of Sequences: 36976
Number of extensions: 192049
Number of successful extensions: 834
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 821
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 824
length of query: 494
length of database: 9,014,727
effective HSP length: 100
effective length of query: 394
effective length of database: 5,317,127
effective search space: 2094948038
effective search space used: 2094948038
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149600.1


