

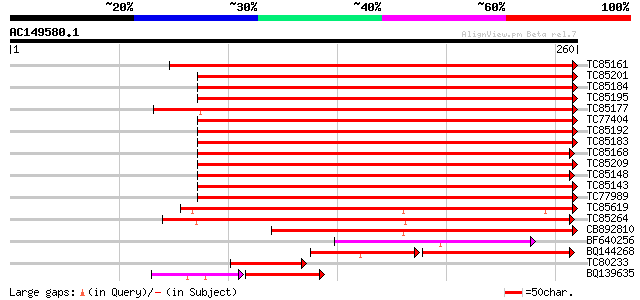
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149580.1 + phase: 0 /pseudo
(260 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85161 similar to GP|4165128|gb|AAD08697.1| lipoxygenase LoxN3 ... 365 e-102
TC85201 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11... 292 8e-80
TC85184 homologue to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.... 287 3e-78
TC85195 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 285 1e-77
TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12... 280 4e-76
TC77404 homologue to GP|493730|emb|CAA55318.1| lipoxygenase {Pis... 261 2e-70
TC85192 homologue to PIR|T12142|T12142 lipoxygenase (EC 1.13.11.... 260 3e-70
TC85183 homologue to SP|P09918|LOX3_PEA Seed lipoxygenase-3 (EC ... 254 3e-68
TC85168 similar to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12... 251 2e-67
TC85209 homologue to PIR|T06454|T06454 probable lipoxygenase (EC... 251 2e-67
TC85148 homologue to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.... 248 2e-66
TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12... 247 3e-66
TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Coryl... 242 9e-65
TC85619 similar to PIR|T11578|T11578 probable lipoxygenase (EC 1... 209 1e-54
TC85264 similar to GP|14589309|emb|CAC43237. lipoxygenase {Sesba... 199 7e-52
CB892810 similar to GP|2826842|emb loxc homologue {Lycopersicon ... 168 2e-42
BF640256 similar to PIR|T06827|T068 lipoxygenase (EC 1.13.11.12)... 108 3e-24
BQ144268 weakly similar to GP|1407701|gb|A lipoxygenase {Solanum... 74 3e-17
TC80233 similar to PIR|T07408|T07408 lipoxygenase (EC 1.13.11.12... 64 6e-11
BQ139635 similar to GP|10505183|gb lipoxygenase {Zantedeschia ae... 46 3e-09
>TC85161 similar to GP|4165128|gb|AAD08697.1| lipoxygenase LoxN3 {Pisum
sativum}, partial (81%)
Length = 1536
Score = 365 bits (938), Expect = e-102
Identities = 177/187 (94%), Positives = 177/187 (94%)
Frame = +2
Query: 74 PPNSTAYHAGTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIH 133
PPN A WLLAKAYAVVNDSC HQLVSHWLKTHAVVEPFVIATNRHLSVLHPIH
Sbjct: 146 PPNKDAKDDEPLIWLLAKAYAVVNDSCWHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIH 325
Query: 134 KLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDL 193
KLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDL
Sbjct: 326 KLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDL 505
Query: 194 KNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEEL 253
KNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEEL
Sbjct: 506 KNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEEL 685
Query: 254 KAFWKEL 260
KAFWKEL
Sbjct: 686 KAFWKEL 706
>TC85201 similar to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11.12).
[Lentil] {Lens culinaris}, partial (49%)
Length = 1275
Score = 292 bits (748), Expect = 8e-80
Identities = 136/174 (78%), Positives = 152/174 (87%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPIHKLL+PHYR TM I
Sbjct: 619 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIHKLLLPHYRDTMNI 798
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R IL+NAGG++E TFL+ Y +EMSAVVYKDW F E+GLP DL RG+AV D +SP
Sbjct: 799 NSLARTILVNAGGVMELTFLWGDYAVEMSAVVYKDWNFTEQGLPNDLIKRGVAVQDPASP 978
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIWAAIKSWVDEYVNFYY+SD DV D EL+AFWKEL
Sbjct: 979 HGVRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYKSDADVVKDSELQAFWKEL 1140
>TC85184 homologue to SP|P38414|LOX1_LENCU Lipoxygenase (EC 1.13.11.12).
[Lentil] {Lens culinaris}, partial (45%)
Length = 1369
Score = 287 bits (734), Expect = 3e-78
Identities = 134/174 (77%), Positives = 151/174 (86%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ VVNDSC HQLVSHWL THAVVEPF+IATNRHLSV+HPIHKLL+PHYR TM I
Sbjct: 95 WLLAKAFVVVNDSCYHQLVSHWLNTHAVVEPFIIATNRHLSVVHPIHKLLLPHYRDTMNI 274
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +RN+L+NA GI+ESTFL+ Y +EMSAVVYKDW F E+GLP DL RG+AV+D +SP
Sbjct: 275 NALARNVLVNAEGIIESTFLWGNYALEMSAVVYKDWNFIEQGLPNDLIKRGVAVEDPASP 454
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
GLRLLIEDYPYASDGLEIWAAIKSWV EYVNFYY+SD + D EL+AFWKEL
Sbjct: 455 TGLRLLIEDYPYASDGLEIWAAIKSWVGEYVNFYYKSDAAIAQDAELQAFWKEL 616
>TC85195 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (98%)
Length = 2903
Score = 285 bits (729), Expect = 1e-77
Identities = 129/174 (74%), Positives = 151/174 (86%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPF+IATNRHLS +HP+HKLL+PHYR TM I
Sbjct: 1572 WLLAKAYVVVNDSCHHQLVSHWLNTHAVVEPFIIATNRHLSTVHPVHKLLLPHYRDTMNI 1751
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +RN+L+NA GI+ESTFL+ Y +EMSAV Y+DWVF E+GLP DL RG+AV+D +SP
Sbjct: 1752 NSLARNVLVNAEGIIESTFLWGGYALEMSAVAYRDWVFTEQGLPNDLLKRGVAVEDPASP 1931
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HG+RLLIEDYPYASDGLEIWAAIK+WV EYV+FYY+SD + D EL+AFWKEL
Sbjct: 1932 HGIRLLIEDYPYASDGLEIWAAIKTWVGEYVSFYYKSDAAIAQDAELQAFWKEL 2093
>TC85177 similar to PIR|S56655|S56655 lipoxygenase (EC 1.13.11.12) loxG -
garden pea, partial (75%)
Length = 2646
Score = 280 bits (716), Expect = 4e-76
Identities = 133/199 (66%), Positives = 157/199 (78%), Gaps = 5/199 (2%)
Frame = +1
Query: 67 LPYPPPGPPNSTAYHAGTFN-----WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIA 121
+P+P +T+Y T W LAKAYAVVND+C HQL+SHWL THA VEPF+IA
Sbjct: 1288 VPHPDGDGIVTTSYTPATEGVDASIWRLAKAYAVVNDACYHQLISHWLHTHASVEPFIIA 1467
Query: 122 TNRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYKD 181
TNRHLSV+HPIHKLL+PHYR TM INA +R++LI AGGI+EST+LF Y ME+S+ VYKD
Sbjct: 1468 TNRHLSVVHPIHKLLLPHYRNTMNINANARDVLIKAGGIIESTYLFGSYSMELSSDVYKD 1647
Query: 182 WVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYY 241
WVF ++ LP DL RG+AV D PHG+RLLIEDYPYA+DGLEIWAAIKSWV+EYVNFYY
Sbjct: 1648 WVFPDQALPNDLIKRGVAVKDPKFPHGVRLLIEDYPYATDGLEIWAAIKSWVEEYVNFYY 1827
Query: 242 ESDKDVKDDEELKAFWKEL 260
+ D V DD EL+AFWKEL
Sbjct: 1828 KLDAAVADDSELQAFWKEL 1884
>TC77404 homologue to GP|493730|emb|CAA55318.1| lipoxygenase {Pisum sativum},
partial (85%)
Length = 2463
Score = 261 bits (666), Expect = 2e-70
Identities = 121/174 (69%), Positives = 142/174 (81%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQL+SHWL THAVVEPFVIATNR LSV+HPI+KLL PHYR TM I
Sbjct: 1143 WLLAKAYVVVNDSCYHQLMSHWLNTHAVVEPFVIATNRQLSVIHPIYKLLSPHYRDTMNI 1322
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R LINA GI+E TFL KY +EMS+ VYK+WVF ++ LP DL R MAV+D SSP
Sbjct: 1323 NALARESLINANGIIERTFLPSKYAVEMSSAVYKNWVFPDQALPADLIKRNMAVEDPSSP 1502
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
+GLRLLIEDYPYA DGLEIW IK+WV +YV+ YY +D D+K+D EL+ +WKE+
Sbjct: 1503 YGLRLLIEDYPYAVDGLEIWTTIKTWVQDYVSVYYATDNDIKNDSELQHWWKEV 1664
>TC85192 homologue to PIR|T12142|T12142 lipoxygenase (EC 1.13.11.12) 1 - fava
bean, complete
Length = 2918
Score = 260 bits (665), Expect = 3e-70
Identities = 120/174 (68%), Positives = 144/174 (81%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY +VNDSC HQLVSHWL THAVVEPFVIATNR LSVLHPI+KLL PHYR TM I
Sbjct: 1510 WLLAKAYVIVNDSCFHQLVSHWLNTHAVVEPFVIATNRQLSVLHPIYKLLHPHYRDTMNI 1689
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R+ L+NA GI+E TFL+ Y ME+S+ VYKDWVF ++ LP DL RG+AV D SSP
Sbjct: 1690 NALARSSLVNADGIIEKTFLWGGYAMEISSKVYKDWVFTDQALPADLIKRGIAVADSSSP 1869
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HGLRLLIEDYPYA DGL+IW AIK+WV +YV+ Y+ SD+ ++ D EL+++WKE+
Sbjct: 1870 HGLRLLIEDYPYAVDGLDIWDAIKTWVQDYVSIYFTSDEKIQQDSELQSWWKEV 2031
>TC85183 homologue to SP|P09918|LOX3_PEA Seed lipoxygenase-3 (EC
1.13.11.12). [Garden pea] {Pisum sativum}, partial (57%)
Length = 1623
Score = 254 bits (648), Expect = 3e-68
Identities = 116/174 (66%), Positives = 141/174 (80%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKAY VVNDSC HQLVSHWL THAVVEPFVIATNRHLS LHPI+KLL PHYR TM I
Sbjct: 411 WLLAKAYVVVNDSCYHQLVSHWLNTHAVVEPFVIATNRHLSYLHPIYKLLYPHYRDTMNI 590
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N+ +R L+N GGI+E TFL+ +Y MEMS+ VYK+W + LP DL RGMA+++ SSP
Sbjct: 591 NSLARQSLVNDGGIIEKTFLWGRYSMEMSSKVYKNWTLPGQALPADLIKRGMAIEEPSSP 770
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
G++L++EDYPYA DGLEIWAAIK+WV +YV+ YY +D ++ D EL+A+WKEL
Sbjct: 771 CGVKLVVEDYPYAHDGLEIWAAIKTWVQDYVSLYYTTDDILRQDSELQAWWKEL 932
>TC85168 similar to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12) - garden
pea, partial (98%)
Length = 2890
Score = 251 bits (642), Expect = 2e-67
Identities = 114/173 (65%), Positives = 141/173 (80%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDSC HQL+SHWL THAVVEPF+IATNRHLSVLHPI+KLL PH+R T+ I
Sbjct: 1577 WLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTINI 1756
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINAGGI+E TFL +E+S++VY+DWVF ++ LP DL RG+AV+D SSP
Sbjct: 1757 NGLARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQALPADLIKRGLAVEDPSSP 1936
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HGLRL +EDYPYA DGLEIW AIK+WV +YV+ YY +D+ V+ D EL+ +WKE
Sbjct: 1937 HGLRLALEDYPYAVDGLEIWDAIKAWVQDYVSLYYPTDEVVQKDTELQTWWKE 2095
>TC85209 homologue to PIR|T06454|T06454 probable lipoxygenase (EC
1.13.11.12) - garden pea, partial (68%)
Length = 2002
Score = 251 bits (642), Expect = 2e-67
Identities = 116/174 (66%), Positives = 141/174 (80%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
WLLAKA+ +VNDSC HQL+SHWL THAVVEPF+IATNRHLSVLHPI+KLL PH+R T+ I
Sbjct: 723 WLLAKAHVIVNDSCYHQLMSHWLNTHAVVEPFIIATNRHLSVLHPINKLLDPHFRDTINI 902
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +RN LINA GI+E TFL +EMS+ YK+WVF ++ LP DL RG+AV+D SSP
Sbjct: 903 NGLARNALINADGIIEETFLPGPNSVEMSSAAYKNWVFTDQALPADLIKRGLAVEDPSSP 1082
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HGLRL+IEDYPYA DGLEIW AIKSWV +YV+ YY +D+ V+ D EL+ +WKE+
Sbjct: 1083HGLRLVIEDYPYAVDGLEIWDAIKSWVQDYVSLYYPNDEAVQKDTELQTWWKEV 1244
>TC85148 homologue to PIR|T06827|T06827 lipoxygenase (EC 1.13.11.12) - garden
pea, complete
Length = 2962
Score = 248 bits (632), Expect = 2e-66
Identities = 112/173 (64%), Positives = 140/173 (80%)
Frame = +2
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W+LAKA+ +VNDSC HQL+SHWL THAV+EPF+IATNRHLSVLHPI+KLL PHYR T+ I
Sbjct: 1577 WMLAKAHVIVNDSCYHQLMSHWLNTHAVMEPFIIATNRHLSVLHPINKLLFPHYRDTINI 1756
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
N +R LINAGGI+E TF +E+S+ VYK+WVF ++ LP DL RG+AV+D SSP
Sbjct: 1757 NGLARQALINAGGIIEQTFCPGPNSIEISSAVYKNWVFTDQALPADLIKRGLAVEDPSSP 1936
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKE 259
HGLRL+IEDYPYA DGLEIW AIK+WV +YV+ YY +D+ V+ D E++ +WKE
Sbjct: 1937 HGLRLVIEDYPYAVDGLEIWDAIKAWVQDYVSLYYPTDEAVQKDTEIQKWWKE 2095
>TC85143 similar to PIR|T11852|T11852 lipoxygenase (EC 1.13.11.12) - kidney
bean, partial (93%)
Length = 3009
Score = 247 bits (631), Expect = 3e-66
Identities = 113/174 (64%), Positives = 139/174 (78%)
Frame = +1
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAY VVND+C HQL+SHWL TH V+EPF+IATNR LSV+HPIHKLL PHYR TM I
Sbjct: 1537 WQLAKAYVVVNDACFHQLMSHWLNTHCVIEPFIIATNRCLSVVHPIHKLLQPHYRDTMNI 1716
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R+ LI+ GGI+E FL Y +EMS+ VYKDWVF ++ LP DL RGMAV D S+P
Sbjct: 1717 NALARSSLISGGGIIEQAFLPGPYAVEMSSAVYKDWVFPDQALPADLIKRGMAVQDASAP 1896
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HGLRL+I+DYP+A DGLEIW AIK+WV +YV+ YY++D ++ D EL+ FWKE+
Sbjct: 1897 HGLRLVIKDYPFAVDGLEIWDAIKAWVKDYVSLYYKTDVAIQRDAELQEFWKEV 2058
>TC77989 similar to GP|16904543|emb|CAD10740. lipoxygenase {Corylus
avellana}, partial (54%)
Length = 1763
Score = 242 bits (618), Expect = 9e-65
Identities = 113/174 (64%), Positives = 140/174 (79%)
Frame = +3
Query: 87 WLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTMTI 146
W LAKAYA VNDS HQLVSHWL THAV+EPF+IATNR LS+LHPIHKLL PH++ TM I
Sbjct: 357 WQLAKAYAAVNDSGYHQLVSHWLFTHAVIEPFIIATNRQLSLLHPIHKLLKPHFKDTMHI 536
Query: 147 NARSRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNRGMAVDDDSSP 206
NA +R+ LINAGG++E T K+ +EMSAVVYK+WVF E+ LP +L RG+A D +SP
Sbjct: 537 NALARHTLINAGGVLEKTVFPGKFALEMSAVVYKNWVFTEQALPANLLKRGIAGPDSNSP 716
Query: 207 HGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKDDEELKAFWKEL 260
HGL+LLIEDYP+A DGLEIW AI++WV EY +FYY SD +++D EL+ +WKE+
Sbjct: 717 HGLKLLIEDYPFAVDGLEIWDAIETWVSEYCSFYYTSDDMIENDYELQFWWKEV 878
>TC85619 similar to PIR|T11578|T11578 probable lipoxygenase (EC 1.13.11.12)
CPRD46 drought-inducible - cowpea, partial (75%)
Length = 2452
Score = 209 bits (531), Expect = 1e-54
Identities = 99/185 (53%), Positives = 139/185 (74%), Gaps = 3/185 (1%)
Frame = +3
Query: 79 AYHA-GTFNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLLV 137
A+H+ G + W LAKA+ + +DS HQLVSHWL+TH EP++IATNR LS +HPI+KLL+
Sbjct: 1086 AWHSTGVWLWRLAKAHVLAHDSGYHQLVSHWLRTHCATEPYIIATNRQLSAMHPIYKLLL 1265
Query: 138 PHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVY-KDWVFAEEGLPTDLKNR 196
PH+R TM INA +R LINA GI+ES+F ++ + +S++ Y K W F + LP DL +R
Sbjct: 1266 PHFRYTMEINALAREALINADGIIESSFTPKQLSLLVSSIAYDKHWQFDLQALPNDLIHR 1445
Query: 197 GMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESD-KDVKDDEELKA 255
G+A D ++PHGL+L IEDYPYA+DGL +W IKSWV +YVN YY ++ ++V+ D+EL+A
Sbjct: 1446 GLAEKDPNAPHGLKLAIEDYPYANDGLVLWDTIKSWVTDYVNHYYNNESRNVESDKELQA 1625
Query: 256 FWKEL 260
+W E+
Sbjct: 1626 WWDEI 1640
>TC85264 similar to GP|14589309|emb|CAC43237. lipoxygenase {Sesbania
rostrata}, partial (51%)
Length = 1774
Score = 199 bits (507), Expect = 7e-52
Identities = 96/198 (48%), Positives = 134/198 (67%), Gaps = 9/198 (4%)
Frame = +2
Query: 71 PPGPPNSTAYHAGT--------FNWLLAKAYAVVNDSCCHQLVSHWLKTHAVVEPFVIAT 122
PP PN+ + T + W+LAKA+ ND+ HQL HWL+THA +EPF+++
Sbjct: 311 PPSGPNTRSKRVVTPALDATTNWMWMLAKAHVCSNDAGVHQLAHHWLRTHACMEPFILSA 490
Query: 123 NRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVYK-D 181
+R LS +HPI KLL PH R T+ INA +R LINA G++ES F +Y ME+S+ YK +
Sbjct: 491 HRQLSAMHPIFKLLDPHMRYTLEINALARQSLINADGVIESCFTPGRYAMEISSAAYKTN 670
Query: 182 WVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYY 241
W F ++ LP DL RGMAV D + PHGL+L+++DYPYA DGL IW+AI++WV YVN+YY
Sbjct: 671 WRFDQDSLPQDLIRRGMAVPDPTQPHGLKLIMKDYPYAEDGLLIWSAIENWVRTYVNYYY 850
Query: 242 ESDKDVKDDEELKAFWKE 259
+ + +D EL+A++ E
Sbjct: 851 PNPSLIINDRELQAWYSE 904
>CB892810 similar to GP|2826842|emb loxc homologue {Lycopersicon
pimpinellifolium}, partial (34%)
Length = 812
Score = 168 bits (426), Expect = 2e-42
Identities = 80/141 (56%), Positives = 106/141 (74%), Gaps = 1/141 (0%)
Frame = +3
Query: 121 ATNRHLSVLHPIHKLLVPHYRGTMTINARSRNILINAGGIVESTFLFEKYCMEMSAVVY- 179
ATNR LS +HP++KLL PH R T+ INA R ILI++ G++ESTF +KY ME+S+V Y
Sbjct: 3 ATNRQLSTMHPVYKLLHPHLRYTLQINALGREILISSYGVIESTFFTKKYSMELSSVAYD 182
Query: 180 KDWVFAEEGLPTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNF 239
K W F +GLP DL +RGMAV+D S+ HGL+L IEDYP+A+DGL IW AIK WV +YVN
Sbjct: 183 KLWQFDLQGLPNDLLHRGMAVEDPSAQHGLKLAIEDYPFANDGLLIWDAIKQWVTDYVNH 362
Query: 240 YYESDKDVKDDEELKAFWKEL 260
YY + ++ D+EL+A+W E+
Sbjct: 363 YYPNPSIIESDQELQAWWTEV 425
>BF640256 similar to PIR|T06827|T068 lipoxygenase (EC 1.13.11.12) - garden
pea, partial (10%)
Length = 374
Score = 108 bits (269), Expect = 3e-24
Identities = 55/122 (45%), Positives = 71/122 (58%), Gaps = 30/122 (24%)
Frame = +2
Query: 150 SRNILINAGGIVESTFLFEKYCMEMSAVVYKDWVFAEEGLPTDLKNR------------- 196
+R LINAGGI+E TFL +E+S++VY+DWVF ++ LP DL R
Sbjct: 8 ARQSLINAGGIIEQTFLPGPNSVEISSIVYRDWVFTDQALPADLIKRYFLTTIWTIYIKI 187
Query: 197 -----------------GMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNF 239
G+AV+D SSPHGLRL + DYPYA DGL+IW A K+WV +YV+
Sbjct: 188 RSNFIIIIDNKLSA*FRGLAVEDPSSPHGLRLALXDYPYAVDGLQIWDAXKAWVQDYVSL 367
Query: 240 YY 241
YY
Sbjct: 368 YY 373
>BQ144268 weakly similar to GP|1407701|gb|A lipoxygenase {Solanum tuberosum},
partial (9%)
Length = 751
Score = 74.3 bits (181), Expect(2) = 3e-17
Identities = 35/70 (50%), Positives = 47/70 (67%)
Frame = +2
Query: 190 PTDLKNRGMAVDDDSSPHGLRLLIEDYPYASDGLEIWAAIKSWVDEYVNFYYESDKDVKD 249
P L RG+ + D + PHGL LLIE+YP DGLEI AI++WV+EY FY SD +++
Sbjct: 314 PALLLKRGITISDSNHPHGLILLIENYPITVDGLEICDAIETWVNEYCTFYSTSDDMIEN 493
Query: 250 DEELKAFWKE 259
D EL+ WK+
Sbjct: 494 DYELQF*WKK 523
Score = 30.8 bits (68), Expect(2) = 3e-17
Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Frame = +3
Query: 139 HYRGTMTINARSRNILINAGG-IVESTFLFEKYCMEMSAVVYKDWVFAEEG 188
H + T +A + + LINAGG + E+T + +EMSAV Y + V +E+G
Sbjct: 156 HIKDTEDNSALAMHTLINAGGELHENTESIA*FALEMSAVKY*NRV*SEQG 308
>TC80233 similar to PIR|T07408|T07408 lipoxygenase (EC 1.13.11.12) loxC
chloroplast - tomato, partial (4%)
Length = 1077
Score = 63.9 bits (154), Expect = 6e-11
Identities = 26/35 (74%), Positives = 32/35 (91%)
Frame = +1
Query: 102 HQLVSHWLKTHAVVEPFVIATNRHLSVLHPIHKLL 136
H+L+SHWL+TH VVEPFVIATN+ LS +HPI+KLL
Sbjct: 973 HELISHWLRTHCVVEPFVIATNKQLSTMHPIYKLL 1077
>BQ139635 similar to GP|10505183|gb lipoxygenase {Zantedeschia aethiopica},
partial (20%)
Length = 554
Score = 45.8 bits (107), Expect(2) = 3e-09
Identities = 20/36 (55%), Positives = 27/36 (74%)
Frame = +3
Query: 109 LKTHAVVEPFVIATNRHLSVLHPIHKLLVPHYRGTM 144
L+THA +EP++IAT+R L +HPI+ LL P R TM
Sbjct: 444 LRTHACMEPYIIATHRQLXSMHPIYXLLXPXTRYTM 551
Score = 32.3 bits (72), Expect(2) = 3e-09
Identities = 19/47 (40%), Positives = 25/47 (52%), Gaps = 5/47 (10%)
Frame = +1
Query: 66 TLPYPPPGPPNSTAY---HAGTFNWL--LAKAYAVVNDSCCHQLVSH 107
+LP P N Y H T +W+ LAKA+ ND+ HQLV+H
Sbjct: 280 SLPPTLTSPQNKKVYTQGHDATAHWIWKLAKAHVSSNDAGIHQLVNH 420
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.316 0.137 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,667,193
Number of Sequences: 36976
Number of extensions: 163201
Number of successful extensions: 2452
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 1954
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2296
length of query: 260
length of database: 9,014,727
effective HSP length: 94
effective length of query: 166
effective length of database: 5,538,983
effective search space: 919471178
effective search space used: 919471178
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149580.1


