

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.7 + phase: 1 /pseudo
(576 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
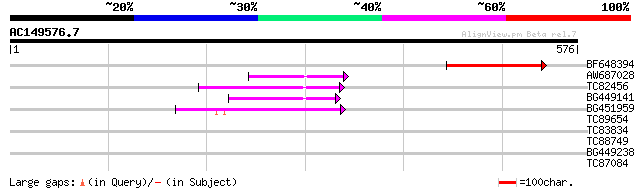
Score E
Sequences producing significant alignments: (bits) Value
BF648394 122 4e-28
AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~... 66 3e-11
TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far... 64 2e-10
BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidop... 54 2e-07
BG451959 homologue to GP|6175165|gb| Mutator-like transposase {A... 50 3e-06
TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein... 40 0.003
TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabid... 37 0.016
TC88749 34 0.13
BG449238 weakly similar to GP|19071826|dbj PnC401 homologue {Ara... 29 4.3
TC87084 similar to GP|2529683|gb|AAC62866.1| unknown protein {Ar... 28 7.4
>BF648394
Length = 650
Score = 122 bits (306), Expect = 4e-28
Identities = 61/102 (59%), Positives = 77/102 (74%)
Frame = -3
Query: 444 VEGAHGVVKEYLSTSKGDLGTCSQKIDEMLANQFGEIQSSFGRSVTVLEHRYKDVTLYSG 503
VE A ++K+YL S GDLGTC +KI +ML QF IQ++FG+SV+VLEHR+K VTLYSG
Sbjct: 648 VESAXDLLKKYLDNSVGDLGTCWEKIHDMLLLQFIAIQTTFGQSVSVLEHRFKYVTLYSG 469
Query: 504 LGGHMSRQAMNFIFVEEARSRKTLCIKKKTCGCVQRMSYGLP 545
+GGH+SR A++ I +EE RKTLC+ GCVQR SYGLP
Sbjct: 468 IGGHVSRYALDNIALEETHCRKTLCMYNDI*GCVQRTSYGLP 343
>AW687028 weakly similar to GP|9909176|dbj| hypothetical protein~similar to
Arabidopsis thaliana F20D10.300, partial (11%)
Length = 662
Score = 66.2 bits (160), Expect = 3e-11
Identities = 37/102 (36%), Positives = 52/102 (50%)
Frame = +1
Query: 243 VLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNS 302
VL D+TYK N Y PL G T G A + + +++ W L L+ ++ +
Sbjct: 4 VLAFDTTYKKNKYNYPLCIFSGCNHHSQTIIFGVALLEDETIESYKWVLNRFLECME--N 177
Query: 303 DMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHVGKNIRSRI 344
PK VVTD D SM A+ V PD+S LC +H+ KN + I
Sbjct: 178 KFPKAVVTDGDGSMREAIKQVFPDASHRLCAWHLHKNAQENI 303
>TC82456 weakly similar to GP|18057159|gb|AAL58182.1 putative far-red
impaired response protein {Oryza sativa}, partial (7%)
Length = 1244
Score = 63.5 bits (153), Expect = 2e-10
Identities = 35/150 (23%), Positives = 68/150 (45%), Gaps = 1/150 (0%)
Frame = +2
Query: 192 RGDLTEMQYLISKLE-ENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTY 250
RGD+ + +++ +N + + + +Q++FW + F V+ +D+TY
Sbjct: 710 RGDVDAIHSFFHRMQKQNSQFYCAMDMDDKRNIQNLFWADARCRAAYEYFGEVITLDTTY 889
Query: 251 KTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVT 310
T+ Y +PL VGV T +G A + + TW L+ + + P ++T
Sbjct: 890 LTSKYDLPLVPFVGVNHHGQTILLGCAILSNLDAKTLTWLFTRWLECM--HGHAPNGIIT 1063
Query: 311 DRDPSMMNAVANVLPDSSAILCYFHVGKNI 340
+ D +M NA+ P + C +H+ K +
Sbjct: 1064EEDKAMKNAIEVAFPKARHRWCLWHIMKKV 1153
>BG449141 similar to GP|15983442|gb At1g10240/F14N23_12 {Arabidopsis
thaliana}, partial (33%)
Length = 690
Score = 53.9 bits (128), Expect = 2e-07
Identities = 24/114 (21%), Positives = 62/114 (54%)
Frame = +3
Query: 223 VQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSK 282
+++I W++ +S++L++ F ++ D+T++ + MPL VG+ + + G + +
Sbjct: 321 LENIAWSYASSIQLYDIFGDAVVFDTTHRLTAFDMPLGLWVGINNYGMPCFFGCVLLRDE 500
Query: 283 KEDNFTWALQMLLKLLKPNSDMPKVVVTDRDPSMMNAVANVLPDSSAILCYFHV 336
+F+WA++ L + N P+ ++TD++ + A++ +P + C + +
Sbjct: 501 TVRSFSWAIKAFLGFM--NGKAPQTILTDQNICLKEALSAEMPMTKHAFCIWMI 656
>BG451959 homologue to GP|6175165|gb| Mutator-like transposase {Arabidopsis
thaliana}, partial (28%)
Length = 658
Score = 49.7 bits (117), Expect = 3e-06
Identities = 46/192 (23%), Positives = 79/192 (40%), Gaps = 19/192 (9%)
Frame = +2
Query: 169 KKKESITNIKRVYNECHKFKKAKRGDLTEMQYLISKLEEN-----EYVNYVRE------- 216
K KE + I RV+ +K+A RG M + EE +Y +V+
Sbjct: 77 KPKEILEEIHRVHGITLSYKQAWRGKEHIMAAMRGSFEEGYRLLPQYCAHVKRTNPGSIA 256
Query: 217 ----KPESQIVQDIFWTHPTSVK-LFNTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLT 271
P Q +F + S+ L N +L +D Y + Y L G
Sbjct: 257 SVYGNPSDNCFQRLFISFQASIYGLLNACRPLLGLDRIYLKSKYLGTLLLATGFDGDGAL 436
Query: 272 YSVGFAFMMSKKEDNFTWALQMLLKLLKPNSD-MPKV-VVTDRDPSMMNAVANVLPDSSA 329
+ + F + + +DN+ W L L LL+ N++ MP++ +++DR +++ V P +
Sbjct: 437 FPLAFGVVDEENDDNWMWFLSKLHNLLEINTENMPRLTILSDRQQGIVDGVEANFPTAFH 616
Query: 330 ILCYFHVGKNIR 341
C H+ N R
Sbjct: 617 GFCMRHLSDNFR 652
>TC89654 weakly similar to PIR|T05644|T05644 hypothetical protein F20D10.290
- Arabidopsis thaliana, partial (65%)
Length = 1358
Score = 39.7 bits (91), Expect = 0.003
Identities = 21/67 (31%), Positives = 29/67 (42%)
Frame = +2
Query: 198 MQYLISKLEENEYVNYVREKPESQIVQDIFWTHPTSVKLFNTFPTVLIMDSTYKTNLYRM 257
+ YL EN Y + +IFW T ++ F +I D+TYKTN YR+
Sbjct: 134 LDYLKRMRAENHAFFYAVQSDVDNAGGNIFWADETCRTNYSYFGDTVIFDTTYKTNQYRV 313
Query: 258 PLFEIVG 264
P G
Sbjct: 314 PFASFTG 334
Score = 34.7 bits (78), Expect = 0.10
Identities = 21/90 (23%), Positives = 43/90 (47%)
Frame = +3
Query: 255 YRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKLLKPNSDMPKVVVTDRDP 314
Y +PL +G+ + + ++++ E ++ W + L+ + P + TD DP
Sbjct: 312 YHLPLS--LGLIIMVNLFYLAVLLILNESEPSYIWLFKTWLRAVSGRP--PVSITTDLDP 479
Query: 315 SMMNAVANVLPDSSAILCYFHVGKNIRSRI 344
+ AVA VLP + C + + + RS++
Sbjct: 480 VIQVAVAQVLPPTRHRFCKWSIFRENRSKL 569
>TC83834 similar to PIR|G96565|G96565 F6D8.26 [imported] - Arabidopsis
thaliana, partial (24%)
Length = 1032
Score = 37.4 bits (85), Expect = 0.016
Identities = 21/121 (17%), Positives = 52/121 (42%), Gaps = 1/121 (0%)
Frame = +3
Query: 179 RVYNECHKFKKAKRGDLTEMQYLISKLE-ENEYVNYVREKPESQIVQDIFWTHPTSVKLF 237
R +++ ++GD + + +++ N Y+ + + +++ W S
Sbjct: 645 RAFSKYSNKLNLRKGDTQAIYNFLCRMQLTNPNFFYLMDFNDEGHLRNALWVDAKSRAAC 824
Query: 238 NTFPTVLIMDSTYKTNLYRMPLFEIVGVTSTYLTYSVGFAFMMSKKEDNFTWALQMLLKL 297
F V+ D+TY N Y +PL +VG+ + +G + + +++ W + +K
Sbjct: 825 GYFSDVIYFDNTYLVNKYEIPLVALVGINHHGQSVLLGCGLLAGEIIESYKWLFRTWIKC 1004
Query: 298 L 298
+
Sbjct: 1005I 1007
>TC88749
Length = 1221
Score = 34.3 bits (77), Expect = 0.13
Identities = 34/78 (43%), Positives = 41/78 (51%), Gaps = 2/78 (2%)
Frame = +1
Query: 23 QRSGHDSFL--FDTRYMER*RGIARLSPSTRK*GWIYGYH*KIL*R*KCYVEVGW*TERR 80
QR G D L YMER*R A +T K*G IY ++ KI + V V * E
Sbjct: 502 QRKGIDRRLATIQPSYMER*RRHA*FDTATSK*GRIYSWYKKIQ-HTESNVGVDV*KEW* 678
Query: 81 S*ITEDKSEA*SNGIKKM 98
S ++K EA*SN IKK+
Sbjct: 679 SQSIQEKIEA*SNMIKKL 732
>BG449238 weakly similar to GP|19071826|dbj PnC401 homologue {Arabidopsis
thaliana}, partial (11%)
Length = 617
Score = 29.3 bits (64), Expect = 4.3
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Frame = +3
Query: 500 LYSGLGGHM-SRQAMNFIFVEEARSRK---TLCIKKKTCGCV 537
L +G G H S+ MN I +E K T CI KKTCG V
Sbjct: 135 LVTGKGSHSYSKNQMNRIAKDEYSGSKLTCTFCICKKTCGVV 260
>TC87084 similar to GP|2529683|gb|AAC62866.1| unknown protein {Arabidopsis
thaliana}, partial (81%)
Length = 2109
Score = 28.5 bits (62), Expect = 7.4
Identities = 12/28 (42%), Positives = 17/28 (59%)
Frame = -1
Query: 160 KKYLTNLKKKKKESITNIKRVYNECHKF 187
KK +TN+K KKKE +T + N + F
Sbjct: 2064 KKKITNIKYKKKEKVTKFSGILNFFYPF 1981
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.334 0.144 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,964,350
Number of Sequences: 36976
Number of extensions: 229632
Number of successful extensions: 1698
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1269
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 421
Number of HSP's gapped (non-prelim): 1347
length of query: 576
length of database: 9,014,727
effective HSP length: 101
effective length of query: 475
effective length of database: 5,280,151
effective search space: 2508071725
effective search space used: 2508071725
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149576.7


