

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149576.2 + phase: 0
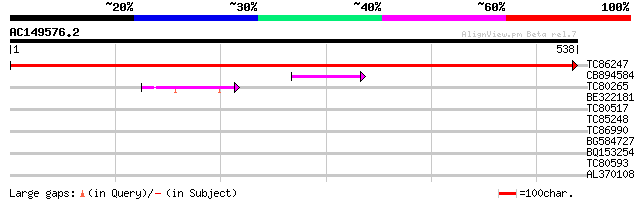
(538 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86247 similar to PIR|T04767|T04767 hypothetical protein T16H5.... 1092 0.0
CB894584 66 4e-11
TC80265 similar to GP|15450695|gb|AAK96619.1 AT5g13640/MSH12_10 ... 47 2e-05
BE322181 similar to GP|22137198|gb At1g27480/F17L21_28 {Arabidop... 35 0.056
TC80517 similar to PIR|S46306|S46306 cytochrome b5 - common toba... 31 1.4
TC85248 similar to GP|20975622|emb|CAD31716. putative ripening r... 30 3.1
TC86990 similar to GP|22136840|gb|AAM91764.1 unknown protein {Ar... 30 3.1
BG584727 weakly similar to PIR|T08399|T083 hypothetical protein ... 29 5.2
BQ153254 similar to GP|7300898|gb| CG6954 gene product {Drosophi... 29 5.2
TC80593 similar to GP|1633130|pdb|1SCH|A Chain A Peanut Peroxid... 28 6.8
AL370108 28 8.9
>TC86247 similar to PIR|T04767|T04767 hypothetical protein T16H5.220 -
Arabidopsis thaliana, partial (63%)
Length = 2073
Score = 1092 bits (2824), Expect = 0.0
Identities = 531/539 (98%), Positives = 533/539 (98%), Gaps = 1/539 (0%)
Frame = +3
Query: 1 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 60
MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW
Sbjct: 201 MTMLLEDVLRSVELWLRLIKKPQPQAYVNPNLDPVLLVPGVGGSILNAVNESDGSQERVW 380
Query: 61 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAV 120
VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAV
Sbjct: 381 VRFLSAEYKLKTKLWSRYDPSTGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAV 560
Query: 121 YYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLI 180
YYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLI
Sbjct: 561 YYFHDMIVQMQKWGYQEGKTLFGFGYDFRQSNRLQETMDRFAEKLELIYNAAGGKKIDLI 740
Query: 181 SHSMGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFF 240
SHSMGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFF
Sbjct: 741 SHSMGGLLVKCFMTLHSDIFEKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFF 920
Query: 241 ISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPPR-DSI 299
ISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYP ++
Sbjct: 921 ISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILESYPATVIAL 1100
Query: 300 EIFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATP 359
FKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATP
Sbjct: 1101RFFKQALVNNKVNHEGEELPLPFNSHIFEWANKTREILSSAKLPSGVKFYNIYGTNLATP 1280
Query: 360 HSICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGIL 419
HSICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGIL
Sbjct: 1281HSICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGIL 1460
Query: 420 CEPHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKD 479
CEPHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKD
Sbjct: 1461CEPHLFRILKHWLKAGDPDPFYNPLNDYVILPTAFEMERHKEKGLEVASLKEEWEIISKD 1640
Query: 480 QDGQSNTGDNKMTLSSISVSQEGANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVDAL 538
QDGQSNTGDNKMTLSSISVSQEGANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVDAL
Sbjct: 1641QDGQSNTGDNKMTLSSISVSQEGANKSHSEAHATVFVHTDNDGKQHIELNAVAVSVDAL 1817
>CB894584
Length = 211
Score = 65.9 bits (159), Expect = 4e-11
Identities = 38/70 (54%), Positives = 41/70 (58%)
Frame = +1
Query: 268 KHVPLLELWRERLHEDGKSHVILESYPPRDSIEIFKQALVNNKVNHEGEELPLPFNSHIF 327
KH P E RER E+GK ES PPR+S E KQA NNK EGEE P P N+
Sbjct: 1 KHDPQPEQRRERQQEEGKPQENRES*PPRESTESVKQAHENNKAKQEGEEAPQPCNAQRV 180
Query: 328 EWANKTREIL 337
E ANKTRE L
Sbjct: 181 EGANKTRENL 210
>TC80265 similar to GP|15450695|gb|AAK96619.1 AT5g13640/MSH12_10
{Arabidopsis thaliana}, partial (38%)
Length = 855
Score = 46.6 bits (109), Expect = 2e-05
Identities = 35/108 (32%), Positives = 50/108 (45%), Gaps = 15/108 (13%)
Frame = +3
Query: 126 MIVQMQKWGYQEGKTLFGFGYDFRQSNRLQE----TMDRFAEKLELIYNAAGGKKIDLIS 181
+I + + GY+E KT++ YD+R S + E T+ R +EL+ + GG K +I
Sbjct: 186 LIANLARIGYEE-KTMYMAAYDWRISFQNTEVRDQTLSRIKSNIELMVSTNGGNKAVIIP 362
Query: 182 HSMGGLLVKCFMTLHS-----------DIFEKYVKNWIAICAPFQGAP 218
HSMG L FM D KY+K + I PF G P
Sbjct: 363 HSMGVLYFLHFMKWVEAPAPMGGGGGPDWCSKYIKAIVNIGGPFLGVP 506
>BE322181 similar to GP|22137198|gb At1g27480/F17L21_28 {Arabidopsis
thaliana}, partial (29%)
Length = 673
Score = 35.4 bits (80), Expect = 0.056
Identities = 36/145 (24%), Positives = 59/145 (39%), Gaps = 24/145 (16%)
Frame = +1
Query: 29 NPNLDPVLLVPGVGGSILNAVNESDGSQERVWVRFLSAEYKLKT---KLWSRYDPS---- 81
N NL+PV+L+PG GG+ L A + + +K K +LW +D S
Sbjct: 166 NNNLNPVILIPGNGGNQLEAKLTTKYKPSTLICDPWYPPFKKKNGWFRLW--FDSSVLLA 339
Query: 82 -------TGKTVTLDQKSRIVVPEDRHGLHAIDVLDPDLVIGSEAVY----------YFH 124
+ T+ DQ +D + ++ P S +Y Y
Sbjct: 340 PFTKCFASRMTLYYDQDL-----DDYFNVPGVETRVPSFGSTSSLLYLNPRLKLVTGYMA 504
Query: 125 DMIVQMQKWGYQEGKTLFGFGYDFR 149
++ +++ GY +G+ LFG YDFR
Sbjct: 505 PLVESLEQLGYIDGQNLFGAPYDFR 579
>TC80517 similar to PIR|S46306|S46306 cytochrome b5 - common tobacco,
partial (94%)
Length = 741
Score = 30.8 bits (68), Expect = 1.4
Identities = 17/56 (30%), Positives = 25/56 (44%), Gaps = 5/56 (8%)
Frame = -1
Query: 244 WSMHQLLIE--CPSIYELMACPNFHWKHVPLLELWRE---RLHEDGKSHVILESYP 294
W M+Q L PS+Y+ + C +H H L LW++ LH G + P
Sbjct: 327 WLMNQCLQHNTLPSLYKHLYCDQYHQNH*THLSLWKKLELHLHRGGPPRTLSHRKP 160
>TC85248 similar to GP|20975622|emb|CAD31716. putative ripening related
protein {Cicer arietinum}, complete
Length = 919
Score = 29.6 bits (65), Expect = 3.1
Identities = 16/55 (29%), Positives = 26/55 (47%)
Frame = +1
Query: 238 NFFISKWSMHQLLIECPSIYELMACPNFHWKHVPLLELWRERLHEDGKSHVILES 292
NFF ++ +H++ I C ++E W H ++ W + DGK H ES
Sbjct: 94 NFFATQ--LHEMQIHCERVHETKLHQGDDWHHTDTVKHWTYVI--DGKVHTCHES 246
>TC86990 similar to GP|22136840|gb|AAM91764.1 unknown protein {Arabidopsis
thaliana}, partial (64%)
Length = 2354
Score = 29.6 bits (65), Expect = 3.1
Identities = 13/33 (39%), Positives = 18/33 (54%)
Frame = +3
Query: 201 EKYVKNWIAICAPFQGAPGCTNSTFLNGMSFVE 233
E+ K W +C P+ G CT TFL ++VE
Sbjct: 531 EESFKAWGKVCVPYLG*ISCTRYTFLKVSTWVE 629
>BG584727 weakly similar to PIR|T08399|T083 hypothetical protein F18B3.60 -
Arabidopsis thaliana, partial (7%)
Length = 854
Score = 28.9 bits (63), Expect = 5.2
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 6/80 (7%)
Frame = +3
Query: 202 KYVKNWIAICAPFQGAPGCTNSTFLNGMSFVEGWEQNFFISKWSMHQLLIECPSIYELMA 261
KY+ + +C + G N TFL +F E F W L+E S ++ MA
Sbjct: 576 KYMLKLLVLCTAKK*NSGL*NRTFL---AFFEFSS*LNF*KAW-----LVESCSFFQSMA 731
Query: 262 ------CPNFHWKHVPLLEL 275
C +F W H+P L++
Sbjct: 732 VNAFLGCASFSWPHIPPLDV 791
>BQ153254 similar to GP|7300898|gb| CG6954 gene product {Drosophila
melanogaster}, partial (0%)
Length = 491
Score = 28.9 bits (63), Expect = 5.2
Identities = 12/33 (36%), Positives = 20/33 (60%)
Frame = -2
Query: 427 ILKHWLKAGDPDPFYNPLNDYVILPTAFEMERH 459
I ++W+K P +Y+ LND + L TA ++H
Sbjct: 109 IFENWVKPYIPFIYYDSLNDILFLNTACRTQKH 11
>TC80593 similar to GP|1633130|pdb|1SCH|A Chain A Peanut Peroxidase,
partial (68%)
Length = 1137
Score = 28.5 bits (62), Expect = 6.8
Identities = 19/66 (28%), Positives = 29/66 (43%)
Frame = +2
Query: 361 SICYGNADKPVSDLQELRYLQARYVCVDGDGTVPVESAKADGFNAEERVGIPGEHRGILC 420
++ G D ++L E L A ++ +DG + + GF AEE V + G H L
Sbjct: 335 NVLLGRRDSTTANLSEANTLPAPFLNLDG----LITAFAKKGFTAEEMVTLSGAHTIGLV 502
Query: 421 EPHLFR 426
FR
Sbjct: 503 RCRFFR 520
>AL370108
Length = 455
Score = 28.1 bits (61), Expect = 8.9
Identities = 13/47 (27%), Positives = 23/47 (48%), Gaps = 3/47 (6%)
Frame = -2
Query: 245 SMHQLLIECPSIYELMA---CPNFHWKHVPLLELWRERLHEDGKSHV 288
S + +++C + ++ CPN H +P+ +W L ED S V
Sbjct: 361 SRYHRIVDCQAFAAVIGIGTCPNGHGLRIPITSIWTRLLLEDCHSVV 221
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,255,651
Number of Sequences: 36976
Number of extensions: 281843
Number of successful extensions: 1408
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 1352
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1407
length of query: 538
length of database: 9,014,727
effective HSP length: 101
effective length of query: 437
effective length of database: 5,280,151
effective search space: 2307425987
effective search space used: 2307425987
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149576.2


