

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.9 + phase: 2 /pseudo
(88 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
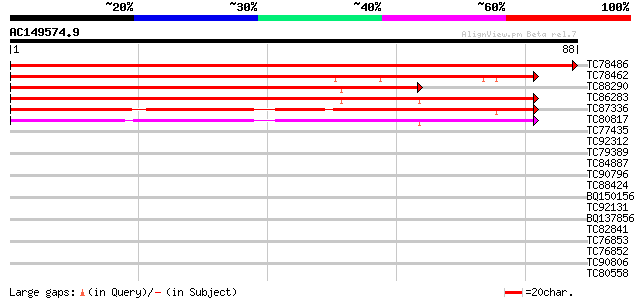
Score E
Sequences producing significant alignments: (bits) Value
TC78486 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 162 2e-41
TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 91 8e-20
TC88290 similar to GP|21592780|gb|AAM64729.1 nucleic acid bindin... 70 1e-13
TC86283 EGAD|127388|135851 Alfin-1 {Medicago sativa}, partial (97%) 69 4e-13
TC87336 similar to GP|10178138|dbj|BAB11550. nucleic acid bindin... 58 7e-10
TC80817 similar to GP|10178138|dbj|BAB11550. nucleic acid bindin... 53 2e-08
TC77435 similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna... 32 0.043
TC92312 weakly similar to GP|4567275|gb|AAD23688.1| expressed pr... 32 0.056
TC79389 similar to GP|14189890|dbj|BAB55874. response regulator ... 31 0.095
TC84887 similar to SP|Q92HF5|IF2_RICCN Translation initiation fa... 30 0.12
TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Ar... 30 0.16
TC88424 similar to GP|22136724|gb|AAM91681.1 unknown protein {Ar... 30 0.21
BQ150156 similar to GP|4096175|gb|A early embryogenesis protein ... 29 0.28
TC92131 similar to GP|4115937|gb|AAD03447.1| contains similarity... 29 0.28
BQ137856 29 0.36
TC82841 similar to GP|3152572|gb|AAC17053.1| Contains homology t... 28 0.47
TC76853 similar to PIR|T09581|T09581 probable high mobility grou... 28 0.61
TC76852 similar to PIR|T09581|T09581 probable high mobility grou... 28 0.61
TC90806 similar to GP|15217295|gb|AAK92639.1 Putative phosphoino... 28 0.61
TC80558 similar to GP|17933427|gb|AAL48287.1 aminoimidazole ribo... 28 0.80
>TC78486 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (89%)
Length = 1089
Score = 162 bits (410), Expect = 2e-41
Identities = 82/88 (93%), Positives = 83/88 (94%)
Frame = +3
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE 60
KRLFT INDLPTIFEVVTGSAKKQTKEKPSVSSHNS KSKSGSKARGSELAKYSKPPAKE
Sbjct: 432 KRLFTLINDLPTIFEVVTGSAKKQTKEKPSVSSHNSIKSKSGSKARGSELAKYSKPPAKE 611
Query: 61 DDEEVDDEEEYQGECTACGENYVSASDE 88
DDE VDDEEE QGEC ACGE+YVSASDE
Sbjct: 612 DDEGVDDEEEDQGECAACGESYVSASDE 695
>TC78462 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (85%)
Length = 1281
Score = 90.9 bits (224), Expect = 8e-20
Identities = 52/89 (58%), Positives = 66/89 (73%), Gaps = 7/89 (7%)
Frame = +3
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSE-LAKYSKP--- 56
KRLF INDLP+I+EVVTG+AKKQ KEK SVS+H+ +KSKS SKAR E K +KP
Sbjct: 564 KRLFNMINDLPSIYEVVTGAAKKQVKEKSSVSNHSGSKSKSSSKARAPEPQVKQTKPLEL 743
Query: 57 PAKEDDEEVDDEEEYQ-GE--CTACGENY 82
P ++ EE+D+E+E + GE C ACGE+Y
Sbjct: 744 PKDDEVEELDEEDEDEHGETLCGACGEHY 830
>TC88290 similar to GP|21592780|gb|AAM64729.1 nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (60%)
Length = 774
Score = 70.1 bits (170), Expect = 1e-13
Identities = 38/65 (58%), Positives = 47/65 (71%), Gaps = 1/65 (1%)
Frame = +1
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSEL-AKYSKPPAK 59
KRLF IN+LPT+FEVVTG+AKKQ KEK SVS+++ +KSKS SKAR E ++ K
Sbjct: 526 KRLFNLINELPTVFEVVTGAAKKQVKEKSSVSNNSGSKSKSSSKARAPEAQSRQPKAALL 705
Query: 60 EDDEE 64
DEE
Sbjct: 706 PKDEE 720
>TC86283 EGAD|127388|135851 Alfin-1 {Medicago sativa}, partial (97%)
Length = 2105
Score = 68.6 bits (166), Expect = 4e-13
Identities = 38/88 (43%), Positives = 55/88 (62%), Gaps = 6/88 (6%)
Frame = +3
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSEL----AKYSKP 56
KRLF INDLPT+FE+ TG+AK+ + + ++ +++K KS K+R SE K S P
Sbjct: 1473 KRLFQMINDLPTVFELATGTAKQSKDQLTAHNNGSNSKYKSSGKSRQSESQTKGVKMSAP 1652
Query: 57 PAKEDD--EEVDDEEEYQGECTACGENY 82
+E D EE +D++E C ACG+NY
Sbjct: 1653 VKEEVDSGEEEEDDDEQGATCGACGDNY 1736
>TC87336 similar to GP|10178138|dbj|BAB11550. nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (95%)
Length = 1223
Score = 57.8 bits (138), Expect = 7e-10
Identities = 35/84 (41%), Positives = 51/84 (60%), Gaps = 2/84 (2%)
Frame = +3
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE 60
KRLF+ IN+LPT+FEVVT +K K+KP+V S + ++ GS R S+ + P
Sbjct: 555 KRLFSLINELPTVFEVVTD--RKPIKDKPTVDSGSKSR---GSTKRSSD-GQVKSNPKLV 716
Query: 61 DDEEVDDEEEYQGE--CTACGENY 82
DD+ ++EE+ E C +CG NY
Sbjct: 717 DDQGYEEEEDEHSETLCGSCGGNY 788
>TC80817 similar to GP|10178138|dbj|BAB11550. nucleic acid binding
protein-like {Arabidopsis thaliana}, partial (67%)
Length = 573
Score = 52.8 bits (125), Expect = 2e-08
Identities = 33/84 (39%), Positives = 47/84 (55%), Gaps = 2/84 (2%)
Frame = +2
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE 60
KRLF+ IN+LPT+FE+VT K KP+ S + ++ GS R S+ S P
Sbjct: 290 KRLFSLINELPTVFEIVT-DRKPIKDNKPAADSGSKSR---GSTKRSSDGQVKSNPKFPV 457
Query: 61 DD--EEVDDEEEYQGECTACGENY 82
DD EE +++E + C +CG NY
Sbjct: 458 DDGYEEEEEDEHSETLCGSCGGNY 529
>TC77435 similar to GP|7211427|gb|AAF40306.1| RNA helicase {Vigna radiata},
partial (67%)
Length = 1828
Score = 32.0 bits (71), Expect = 0.043
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 2/65 (3%)
Frame = +1
Query: 8 NDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKE--DDEEV 65
ND+ + E V+ K+ +K++ S + S GS K K K +DEE+
Sbjct: 199 NDVVSEVEAVSAKKKESSKKRKKSSDDDEETKSDISSEEGSRKVKKEKKKKKSKVEDEEI 378
Query: 66 DDEEE 70
+EEE
Sbjct: 379 VEEEE 393
>TC92312 weakly similar to GP|4567275|gb|AAD23688.1| expressed protein
{Arabidopsis thaliana}, partial (29%)
Length = 1319
Score = 31.6 bits (70), Expect = 0.056
Identities = 18/47 (38%), Positives = 26/47 (55%)
Frame = +1
Query: 16 VVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDD 62
++T SAK ++ SVSS NK KS +G++ +K SK K D
Sbjct: 739 LITSSAKDESVNNDSVSSEEKNKPKSKETVKGAD-SKTSKESDKVSD 876
>TC79389 similar to GP|14189890|dbj|BAB55874. response regulator 9 {Zea
mays}, partial (33%)
Length = 1532
Score = 30.8 bits (68), Expect = 0.095
Identities = 20/69 (28%), Positives = 32/69 (45%), Gaps = 6/69 (8%)
Frame = +1
Query: 7 INDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGS------ELAKYSKPPAKE 60
+ +L I++ V K KEK S + S SGS R + E K +K +
Sbjct: 520 LEELKNIWQHVIRKKKSDPKEKNKTSKPDKTTSDSGSGLRSAGAENSDENGKLTKKRKDQ 699
Query: 61 DDEEVDDEE 69
D++E +D+E
Sbjct: 700 DEDEDEDKE 726
>TC84887 similar to SP|Q92HF5|IF2_RICCN Translation initiation factor IF-2.
{Rickettsia conorii}, partial (1%)
Length = 742
Score = 30.4 bits (67), Expect = 0.12
Identities = 21/63 (33%), Positives = 31/63 (48%)
Frame = -1
Query: 19 GSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTAC 78
GS K+ +E ++ S+ S S GSE S+PPA DEE ++E+ +G
Sbjct: 424 GSDPKENEESDPKAAEESDPKASSSL--GSEGNLSSEPPAASVDEESGEKEKRKGNEDDA 251
Query: 79 GEN 81
EN
Sbjct: 250 REN 242
>TC90796 similar to GP|3395431|gb|AAC28763.1| unknown protein {Arabidopsis
thaliana}, partial (20%)
Length = 965
Score = 30.0 bits (66), Expect = 0.16
Identities = 20/54 (37%), Positives = 23/54 (42%), Gaps = 6/54 (11%)
Frame = +2
Query: 21 AKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKP------PAKEDDEEVDDE 68
A T SS N N+ KS S A K S P P E+DEE +DE
Sbjct: 272 ASSSTMSSEVQSSFNQNEVKSSSPAESVSTPKASPPAEVYSEPKGENDEEEEDE 433
>TC88424 similar to GP|22136724|gb|AAM91681.1 unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1635
Score = 29.6 bits (65), Expect = 0.21
Identities = 18/69 (26%), Positives = 31/69 (44%)
Frame = +2
Query: 20 SAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACG 79
+A+++TK++P H S+ S++ A DD YQ EC ACG
Sbjct: 1394 TAEEKTKKEPDEKQHMQMTDARCSRCTDSKVGLREIIHAGHDDR-------YQLECVACG 1552
Query: 80 ENYVSASDE 88
++ + +E
Sbjct: 1553 NSWYPSRNE 1579
>BQ150156 similar to GP|4096175|gb|A early embryogenesis protein {Oryza
sativa}, partial (4%)
Length = 1059
Score = 29.3 bits (64), Expect = 0.28
Identities = 15/65 (23%), Positives = 29/65 (44%)
Frame = -1
Query: 18 TGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGECTA 77
T S + P S+ + + GS+ G L + +PP + + + + +G C
Sbjct: 276 TTSGRPSRALSPPTSTLSLSLRGVGSECHGVMLRREGRPPEQPPQDPIPQPKRQEGGC-G 100
Query: 78 CGENY 82
CG++Y
Sbjct: 99 CGQSY 85
>TC92131 similar to GP|4115937|gb|AAD03447.1| contains similarity to human
PCF11p homolog (GB:AF046935) {Arabidopsis thaliana},
partial (24%)
Length = 1003
Score = 29.3 bits (64), Expect = 0.28
Identities = 21/75 (28%), Positives = 33/75 (44%), Gaps = 6/75 (8%)
Frame = +3
Query: 14 FEVVTGSAKKQTKEKPSVSSHNSNKS-KSGSKARGSELAKYSKPPAKEDDEEVDDE---- 68
+ V K K+KPS S G++A G+E A P ++++ D+E
Sbjct: 573 WHVTKNRMSKNRKQKPSRKWFVSETMWLXGAEALGTESAPGFLPTETTEEKKEDEELAVP 752
Query: 69 -EEYQGECTACGENY 82
+E Q C CGE +
Sbjct: 753 ADEDQNTCALCGEPF 797
>BQ137856
Length = 975
Score = 28.9 bits (63), Expect = 0.36
Identities = 12/43 (27%), Positives = 24/43 (54%)
Frame = +2
Query: 32 SSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEEYQGE 74
+ HN + S A +E + ++PP +E DE +D ++Y+ +
Sbjct: 512 TQHNERRYGRISSAHEAEERENARPPHRE*DETIDKRKKYRAD 640
>TC82841 similar to GP|3152572|gb|AAC17053.1| Contains homology to DNAJ
heatshock protein gb|U32803 from Haemophilus
influenzae., partial (38%)
Length = 1131
Score = 28.5 bits (62), Expect = 0.47
Identities = 18/74 (24%), Positives = 34/74 (45%)
Frame = +2
Query: 10 LPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEE 69
L +EV++ S KK+ ++ + KS + + S + +D+ E EE
Sbjct: 17 LQCAYEVLSDSVKKRDYDEQLRKEESMAKS----------VCQKSHSSSHQDNTEYRSEE 166
Query: 70 EYQGECTACGENYV 83
+ +CT CG ++V
Sbjct: 167SRRIQCTKCGNSHV 208
>TC76853 similar to PIR|T09581|T09581 probable high mobility group protein
HMG1 - sword bean, partial (97%)
Length = 907
Score = 28.1 bits (61), Expect = 0.61
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = +3
Query: 26 KEKPSVSSHNSNKSKSGSKAR-GSELAKYSKPPAKEDDEEVDDEEEYQGEC 75
KE+ V++ NK G GS+ +K ED+E+ DD+E+ + C
Sbjct: 408 KEEYEVATQAYNKKLEGKDEEDGSDKSKSEVNDEDEDEEDEDDDEDDE*NC 560
>TC76852 similar to PIR|T09581|T09581 probable high mobility group protein
HMG1 - sword bean, partial (93%)
Length = 731
Score = 28.1 bits (61), Expect = 0.61
Identities = 17/48 (35%), Positives = 25/48 (51%)
Frame = +2
Query: 23 KQTKEKPSVSSHNSNKSKSGSKARGSELAKYSKPPAKEDDEEVDDEEE 70
K+ EK + + K+ S+ GSE SK +DDE+ DDEE+
Sbjct: 350 KEEYEKTMRAYNMGITEKNASEEEGSEK---SKSEVNDDDEDGDDEED 484
>TC90806 similar to GP|15217295|gb|AAK92639.1 Putative phosphoinositide
phosphatase {Oryza sativa} [Oryza sativa (japonica
cultivar-group)], partial (21%)
Length = 1285
Score = 28.1 bits (61), Expect = 0.61
Identities = 17/43 (39%), Positives = 21/43 (48%)
Frame = +2
Query: 1 KRLFTPINDLPTIFEVVTGSAKKQTKEKPSVSSHNSNKSKSGS 43
K LF P + + TI + TG KKQ +SSHN GS
Sbjct: 563 KSLFEPYSVITTI-HIRTGLNKKQLTYSWGISSHNKENQYCGS 688
>TC80558 similar to GP|17933427|gb|AAL48287.1 aminoimidazole ribonucleotide
carboxylase {Vigna unguiculata}, partial (15%)
Length = 691
Score = 27.7 bits (60), Expect = 0.80
Identities = 15/45 (33%), Positives = 25/45 (55%)
Frame = -1
Query: 44 KARGSELAKYSKPPAKEDDEEVDDEEEYQGECTACGENYVSASDE 88
K G + K S+ +E++EE ++EEE +G G+N V+ E
Sbjct: 334 KRLGCSM*KASEEEEEEEEEEEEEEEEERG*VDLKGKNEVTVCRE 200
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.299 0.121 0.324
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,264,914
Number of Sequences: 36976
Number of extensions: 21735
Number of successful extensions: 276
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 245
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 269
length of query: 88
length of database: 9,014,727
effective HSP length: 64
effective length of query: 24
effective length of database: 6,648,263
effective search space: 159558312
effective search space used: 159558312
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.6 bits)
S2: 51 (24.3 bits)
Medicago: description of AC149574.9


