

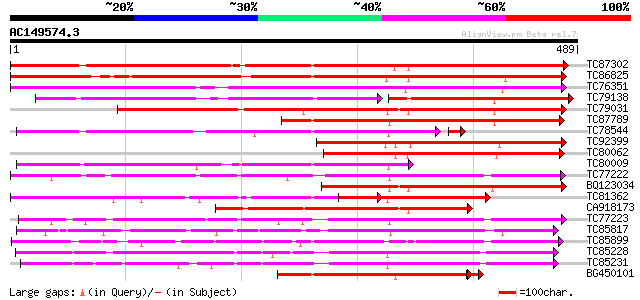
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.3 + phase: 0
(489 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase... 544 e-155
TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransfe... 372 e-103
TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycos... 362 e-100
TC79138 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 209 1e-99
TC79031 weakly similar to GP|13492674|gb|AAK28303.1 phenylpropan... 339 2e-93
TC87789 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 249 1e-66
TC78544 weakly similar to PIR|T03747|T03747 glucosyltransferase ... 242 9e-66
TC92399 weakly similar to GP|5763524|dbj|BAA83484.1 UDP-glucose:... 241 4e-64
TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:... 230 8e-61
TC80009 weakly similar to PIR|T03747|T03747 glucosyltransferase ... 213 1e-55
TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 213 1e-55
BQ123034 weakly similar to GP|14334982|gb putative glucosyltrans... 213 2e-55
TC81362 similar to GP|19911201|dbj|BAB86927. glucosyltransferase... 201 7e-52
CA918173 weakly similar to GP|11034537|dbj putative glucosyl tra... 200 1e-51
TC77223 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 197 6e-51
TC85817 weakly similar to PIR|B85014|B85014 probable flavonol gl... 195 3e-50
TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:... 194 6e-50
TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltran... 194 8e-50
TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {R... 193 1e-49
BG450101 similar to GP|4115534|dbj UDP-glycose:flavonoid glycosy... 189 2e-48
>TC87302 similar to GP|19911199|dbj|BAB86926. glucosyltransferase-8 {Vigna
angularis}, partial (53%)
Length = 1787
Score = 544 bits (1401), Expect = e-155
Identities = 275/489 (56%), Positives = 367/489 (74%), Gaps = 7/489 (1%)
Frame = +1
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M +++ LHI+ FPF+ +GH IP +D+A++F+S+G++VTIVTT LN P IS+ + ++KI+
Sbjct: 34 MTNENQELHIIFFPFLANGHIIPCVDLARVFSSRGLKVTIVTTHLNVPLISRTIGKAKIN 213
Query: 61 FNNIDIQTIKFPCVE-AGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHC 119
I+TIKFP E GLPEGCEN +S + F + LL++P E +L Q+KP C
Sbjct: 214 -----IKTIKFPSPEETGLPEGCENSESALAPDKFIKFMKSTLLLREPLEHVLEQEKPDC 378
Query: 120 VVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLP 179
+VADMFFPW+TDSAAKF IPRIVFHG FF LC C ++Y+P VSS T+ F + +LP
Sbjct: 379 LVADMFFPWSTDSAAKFNIPRIVFHGLGFFPLCVLACTRQYKPQDKVSSYTEPFVVPNLP 558
Query: 180 GNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREV 239
G I +T++QLP L ++D + F KL EE +SEV+S+GVI N+FYELE VYAD+YR
Sbjct: 559 GEITLTKMQLPQ-LPQHDKV---FTKLLEESNESEVKSFGVIANTFYELEPVYADHYRNE 726
Query: 240 LGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHF 299
LG K WH+GP S+ NR+ EE+ + RG+EASID+HECLKWL +K NSV+Y+CFGSMT F
Sbjct: 727 LGRKAWHLGPVSLCNRDTEEK--ACRGREASIDEHECLKWLQSKEPNSVIYVCFGSMTVF 900
Query: 300 LNSQLKEIAMGLEASGHNFIWVVRTQTE-DGD--EWLPEGFEERTE--GKGLIIRGWSPQ 354
++QLKEIAMGLEAS FIWVVR + +G+ EWLPEGFEER E GKGLIIRGW+PQ
Sbjct: 901 SDAQLKEIAMGLEASEVPFIWVVRKSAKSEGENLEWLPEGFEERIEGSGKGLIIRGWAPQ 1080
Query: 355 VMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK 414
VMIL+HE++G FVTHCGWNS LEGV AG+PM+TWP+ EQFYN K +++++K GV VGV+
Sbjct: 1081VMILDHESVGGFVTHCGWNSTLEGVSAGLPMVTWPMYGEQFYNAKFLSDIVKIGVGVGVQ 1260
Query: 415 KWV-MKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNAL 473
W+ M G+ V+ D +EKAV+R+M G+EA EMR++AK +MA++AVE GSSY+ + L
Sbjct: 1261TWIGMGGGEPVKKDVIEKAVRRIMVGDEAEEMRSRAKEFGKMARRAVEVGGSSYNDFSNL 1440
Query: 474 IEELRSLSH 482
IE+L+S ++
Sbjct: 1441IEDLKSRAY 1467
>TC86825 similar to GP|18151384|dbj|BAB83692. ABA-glucosyltransferase {Vigna
angularis}, partial (87%)
Length = 1704
Score = 372 bits (954), Expect = e-103
Identities = 211/489 (43%), Positives = 297/489 (60%), Gaps = 9/489 (1%)
Frame = +3
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M+ +++ + I FPF+G GH IP +D A++FA G TI+TTP N ++ + +
Sbjct: 72 MNHETDAVKIYFFPFVGGGHQIPMVDTARVFAKHGATSTIITTPSNALQFQNSITRDQKS 251
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCV 120
+ I IQ + PE E D+ +S P +I L +P ++ L+Q +P +
Sbjct: 252 NHPITIQILT-------TPENTEVTDT--DMSAGPMIDTSILL--EPLKQFLVQHRPDGI 398
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPG 180
V DMF WA D + IPRIVF+G F C + +K+ +N+SSD++ F + LP
Sbjct: 399 VVDMFHRWAGDIIDELKIPRIVFNGNGCFPRCVIENTRKHVVLENLSSDSEPFIVPGLPD 578
Query: 181 NIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVL 240
I+MTR Q P + S + IK E S G ++NSFY+LE YADY R L
Sbjct: 579 IIEMTRSQTPIFMRNPSQFS-------DRIKQLEENSLGTLINSFYDLEPAYADYVRNKL 737
Query: 241 GIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFL 300
G K W +GP S+ NR+ E++ RGK+ +ID+ CL WL++K NSV+Y+ FGS+
Sbjct: 738 GKKAWLVGPVSLCNRSVEDK--KERGKQPTIDEQSCLNWLNSKKPNSVLYISFGSVARVP 911
Query: 301 NSQLKEIAMGLEASGHNFIWVVR---TQTEDGDEWLPEGFEERTE--GKGLIIRGWSPQV 355
QLKEIA GLEAS +FIWVV +++ ++W+ + FE R + KGLI RGW+PQ+
Sbjct: 912 MKQLKEIAYGLEASDQSFIWVVGKILNSSKNEEDWVLDKFERRMKEMDKGLIFRGWAPQL 1091
Query: 356 MILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKK 415
+ILEHEA+G F+THCGWNS LEGV AGVPM TWP++AEQF NEKL+T+VL+ GV VG ++
Sbjct: 1092LILEHEAVGGFMTHCGWNSTLEGVCAGVPMATWPLSAEQFINEKLITDVLRIGVQVGSRE 1271
Query: 416 WVMKVGDNVEW---DAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLN 471
W + E + VE AVK++M E E+ EMR + K + E AK+AVEE GSSY ++
Sbjct: 1272WGSWDEERKELVGREKVELAVKKLMAESEDTEEMRRRVKSIFENAKRAVEEGGSSYDDIH 1451
Query: 472 ALIEELRSL 480
ALI+ SL
Sbjct: 1452ALIQCKTSL 1478
>TC76351 similar to GP|7635494|emb|CAB88666.1 putative UDP-glycose {Cicer
arietinum}, partial (92%)
Length = 1645
Score = 362 bits (928), Expect = e-100
Identities = 197/488 (40%), Positives = 288/488 (58%), Gaps = 8/488 (1%)
Frame = +2
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M ++S PL I + PF GH IP +++A+L ASK VTI+TTP N K +E+ K
Sbjct: 74 MGTESKPLKIYMLPFFAQGHLIPLVNLARLVASKNQHVTIITTPSNAQLFDKTIEEEKAA 253
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCV 120
++I + IKFP + GLP G EN+ + A ++ EE + + P
Sbjct: 254 GHHIRVHIIKFPSAQLGLPTGVENLFAASDNQTAGKIHMAAHFVKADIEEFMKENPPDVF 433
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPG 180
++D+ F W+ +A IPR+VF+ S F +C Q ++ + ++ SD+ ++I
Sbjct: 434 ISDIIFTWSESTAKNLQIPRLVFNPISIFDVCMIQAIQSHP--ESFVSDSGPYQIHG--- 598
Query: 181 NIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVL 240
LP+ LT S FA+L E + ++E S+GVIVNSF EL+ Y +YY +
Sbjct: 599 --------LPHPLTLPIKPSPGFARLTESLIEAENDSHGVIVNSFAELDEGYTEYYENLT 754
Query: 241 GIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFL 300
G K WH+GP S+ +++ ++SI KH+ L WLDTK +SV+Y+ FGS+
Sbjct: 755 GRKVWHVGPTSLMVEIPKKKKVVSTENDSSITKHQSLTWLDTKEPSSVLYISFGSLCRLS 934
Query: 301 NSQLKEIAMGLEASGHNFIWVVR-TQTEDGDEWLPEGFEER--TEGKGLIIRGWSPQVMI 357
N QLKE+A G+EAS H F+WVV + ED D WLP+GF ER E KG++I+GW PQ +I
Sbjct: 935 NEQLKEMANGIEASKHQFLWVVHGKEGEDEDNWLPKGFVERMKEEKKGMLIKGWVPQALI 1114
Query: 358 LEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWV 417
L+H +IG F+THCGWN+ +E + +GVPM+T P +Q+YNEKLVTEV + GV VG +W
Sbjct: 1115LDHPSIGGFLTHCGWNATVEAISSGVPMVTMPGFGDQYYNEKLVTEVHRIGVEVGAAEWS 1294
Query: 418 MKVGDN----VEWDAVEKAVKRVMEGE-EAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNA 472
M D V + +EKAVK++M+ E E+R +AK + E A KAV+E GSS + L
Sbjct: 1295MSPYDAKKTVVRAERIEKAVKKLMDSNGEGGEIRKRAKEMKEKAWKAVQEGGSSQNCLTK 1474
Query: 473 LIEELRSL 480
L++ L S+
Sbjct: 1475LVDYLHSV 1498
>TC79138 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (36%)
Length = 1575
Score = 209 bits (531), Expect(2) = 1e-99
Identities = 115/300 (38%), Positives = 170/300 (56%), Gaps = 1/300 (0%)
Frame = +2
Query: 23 PTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQTIKFPCVEAGLPEGC 82
P D+A LFAS G VTI+TTP N I K++ N++ + T+ FP + GLP G
Sbjct: 59 PLCDIATLFASCGHHVTIITTPSNAQIILKSIPSH----NHLRLHTVPFPSHQVGLPLGV 226
Query: 83 ENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFFPWATDSAAKFGIPRIV 142
EN+ + +V A LL+ P + Q P C+VAD F W + A + IPR+
Sbjct: 227 ENLAFVNNVDNSCKIHHATMLLRSPINHFVEQDSPDCIVADFMFLWVDELANRLHIPRLA 406
Query: 143 FHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTRLQLPNTLTENDPISQS 202
F+G S F++CA + +K FE + + G LP+ +T N ++
Sbjct: 407 FNGFSLFAICAMESLKARD-----------FESSIIQG--------LPHCITLNAMPPKA 529
Query: 203 FAKLFEEIKDSEVRSYGVIVNSFYELENV-YADYYREVLGIKEWHIGPFSIHNRNKEEEI 261
K E + ++E++SYG+IVN+F EL+ Y ++Y + +G + WH+GP S+ R +E+
Sbjct: 530 LTKFMEPLLETELKSYGLIVNNFTELDGEEYIEHYEKTIGHRAWHLGPSSLICRTTQEKA 709
Query: 262 PSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWV 321
RG+ + +D HECL WL++K NSV+Y+CFGS+ HF N QL EIA +EASGH FIWV
Sbjct: 710 D--RGQTSVVDVHECLSWLNSKQPNSVLYICFGSLCHFTNKQLYEIASAIEASGHQFIWV 883
Score = 173 bits (438), Expect(2) = 1e-99
Identities = 89/165 (53%), Positives = 120/165 (71%), Gaps = 5/165 (3%)
Frame = +1
Query: 327 EDGDEWLPEGFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMI 386
++ ++W+P+GFEER G +IIRGW+PQV+IL H AIGAF+THCGWNS +E V AGVPMI
Sbjct: 919 DENEKWMPKGFEERNIG--MIIRGWAPQVVILGHPAIGAFLTHCGWNSTVEAVSAGVPMI 1092
Query: 387 TWPVAAEQFYNEKLVTEVLKTGVPVGVKKW----VMKVGDNVEWDAVEKAVKRVMEGE-E 441
TWPV EQFYNEKL+T+V GV VGV++W MK V D +EKA++R+M+G E
Sbjct: 1093 TWPVHDEQFYNEKLITQVRGIGVEVGVEEWSFIGFMKKKKIVGRDIIEKALRRLMDGGIE 1272
Query: 442 AYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSLSHHQHI 486
A E+R +A+ A AK+AV+E GSS+ L ALI++L+ H+ +
Sbjct: 1273 AVEIRKRAQEYAIKAKRAVQEGGSSHKNLMALIDDLKRQRGHKSL 1407
>TC79031 weakly similar to GP|13492674|gb|AAK28303.1
phenylpropanoid:glucosyltransferase 1 {Nicotiana
tabacum}, partial (32%)
Length = 1385
Score = 339 bits (869), Expect = 2e-93
Identities = 181/397 (45%), Positives = 260/397 (64%), Gaps = 10/397 (2%)
Frame = +1
Query: 94 VPAFFAAIRLLQQPFEELLLQQKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCA 153
+P + + +LQ E+L +P +V DMFFPW+ D A K GIPRI+FHG S+ + A
Sbjct: 22 IPKIYMGLYILQPDIEKLFETLQPDFIVTDMFFPWSADVAKKLGIPRIMFHGASYLARSA 201
Query: 154 SQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDS 213
+ ++ Y+P+ SDTD F I DLP ++MTRLQLP+ L + +A+L + IK+S
Sbjct: 202 AHSVEVYRPHLKAESDTDKFVIPDLPDELEMTRLQLPDWLRSPN----QYAELMKVIKES 369
Query: 214 EVRSYGVIVNSFYELENVYADYYREVLGIKEWHIGPFSI--HNRNKEEEIPSYRGKEASI 271
E +S+G + NSFY+LE+ Y D+Y++V+G K W +GP S+ + + ++ Y KE
Sbjct: 370 EKKSFGSVFNSFYKLESEYYDHYKKVMGTKSWGLGPVSLWANQDDSDKAARGYARKEEGA 549
Query: 272 DKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRT--QTEDG 329
+ LKWL++K SV+Y+ FGSM F SQL EIA LE SGHNFIWVVR + E+G
Sbjct: 550 KEEGWLKWLNSKPDGSVLYVSFGSMNKFPYSQLVEIAHALENSGHNFIWVVRKNEENEEG 729
Query: 330 DEWLPEGFEERTE--GKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMIT 387
+L E FE++ + GKG +I GW+PQ++ILE+ AIG V+HCGWN+V+E V G+P +T
Sbjct: 730 GVFLEE-FEKKMKESGKGYLIWGWAPQLLILENHAIGGLVSHCGWNTVVESVNVGLPTVT 906
Query: 388 WPVAAEQFYNEKLVTEVLKTGVPVGVKKW--VMKVGDN-VEWDAVEKAVKRVME-GEEAY 443
WP+ AE F+NEKLV +VLK GVPVG K+W + G V+ + + A++ +ME GEE
Sbjct: 907 WPLFAEHFFNEKLVVDVLKIGVPVGAKEWRNWNEFGSEVVKREDIGNAIRLMMEGGEEEV 1086
Query: 444 EMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSL 480
MR + K L+ AKKA++ GSSY+ + LI+ELRS+
Sbjct: 1087AMRKRVKELSVEAKKAIKVGGSSYNNMVELIQELRSI 1197
>TC87789 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (38%)
Length = 1081
Score = 249 bits (637), Expect = 1e-66
Identities = 128/257 (49%), Positives = 177/257 (68%), Gaps = 13/257 (5%)
Frame = +2
Query: 235 YYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFG 294
+Y + G K WH+GP S+ + +E+ S RG E +++ HE L WLD++ +NSV+Y+CFG
Sbjct: 5 HYEKATGHKVWHLGPTSLIRKTAQEK--SERGNEGAVNVHESLSWLDSERVNSVLYICFG 178
Query: 295 SMTHFLNSQLKEIAMGLEASGHNFIWVVRTQT-------EDGDEWLPEGFEERTEGK-GL 346
S+ +F + QL E+A +EASGH FIWVV + E+ ++WLP+GFEER GK GL
Sbjct: 179 SINYFSDKQLYEMACAIEASGHPFIWVVPEKKGKEDESEEEKEKWLPKGFEERNIGKKGL 358
Query: 347 IIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLK 406
IIRGW+PQV IL H A+G F+THCG NS +E V AGVPMITWPV +QFYNEKL+T+
Sbjct: 359 IIRGWAPQVKILSHPAVGGFMTHCGGNSTVEAVSAGVPMITWPVHGDQFYNEKLITQFRG 538
Query: 407 TGVPVGVKKW----VMKVGDNVEWDAVEKAVKRVME-GEEAYEMRNKAKMLAEMAKKAVE 461
GV VG +W V + V D++EKAV+R+M+ G+EA +R +A+ E A +A++
Sbjct: 539 IGVEVGATEWCTSGVAERKKLVSRDSIEKAVRRLMDGGDEAENIRLRAREFGEKAIQAIQ 718
Query: 462 EDGSSYSQLNALIEELR 478
E GSSY+ L ALI+EL+
Sbjct: 719 EGGSSYNNLLALIDELK 769
>TC78544 weakly similar to PIR|T03747|T03747 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(26%)
Length = 1248
Score = 242 bits (618), Expect(2) = 9e-66
Identities = 134/378 (35%), Positives = 207/378 (54%), Gaps = 13/378 (3%)
Frame = +3
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PL + P+ GH +P D+A LFAS+G VTI+TTP N ++K L + + +
Sbjct: 96 PLKLHFIPYPASGHMMPLCDIATLFASRGQHVTIITTPSNAQSLTKTLSSAALR-----L 260
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFF 126
T++FP + LP+G E++ S LL + + + + P C++AD F
Sbjct: 261 HTVEFPYQQVDLPKGVESMTSTTDPITTWKIHNGAMLLNEAVGDFVEKNPPDCIIADSAF 440
Query: 127 PWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTR 186
WA D A K IP + F+G+S F++ ++ T+ TD + +
Sbjct: 441 SWANDLAHKLQIPNLTFNGSSLFAVSIFHSLR-----------TNNLLHTDADADSDSSS 587
Query: 187 LQLPNTLTENDPISQSFAKLFEE----IKDSEVRSYGVIVNSFYELENVYA-DYYREVLG 241
+PN +N + K+ + D+ ++S G I+N+F EL+ +Y + G
Sbjct: 588 YVVPNLHHDNITLCSKPPKVLSMFIGMMLDTVLKSTGYIINNFVELDGEECVKHYEKTTG 767
Query: 242 IKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLN 301
K WH+GP S + +E+ + +G ++ + +HECL WL ++ +NSVVY+CFGS+ HF +
Sbjct: 768 HKAWHLGPTSFIRKTVQEK--AEKGNKSDVSEHECLNWLKSQRVNSVVYICFGSINHFSD 941
Query: 302 SQLKEIAMGLEASGHNFIWVVRTQ-------TEDGDEWLPEGFEERTEG-KGLIIRGWSP 353
QL EIA +EASGH FIWVV + E+ ++WLP+GFEER G KG IIRGW+P
Sbjct: 942 KQLYEIACAVEASGHPFIWVVPEKKGKEDEIEEEKEKWLPKGFEERNIGKKGFIIRGWAP 1121
Query: 354 QVMILEHEAIGAFVTHCG 371
QV+IL + A+G F+THCG
Sbjct: 1122QVLILSNPAVGGFLTHCG 1175
Score = 26.6 bits (57), Expect(2) = 9e-66
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = +2
Query: 379 VVAGVPMITWPVAAE 393
V AGVPMITWP A+
Sbjct: 1199 VGAGVPMITWPCHAD 1243
>TC92399 weakly similar to GP|5763524|dbj|BAA83484.1 UDP-glucose: flavonoid
7-O-glucosyltransferase {Scutellaria baicalensis},
partial (29%)
Length = 843
Score = 241 bits (616), Expect = 4e-64
Identities = 125/230 (54%), Positives = 163/230 (70%), Gaps = 14/230 (6%)
Frame = +1
Query: 265 RGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVV-- 322
RG+E+ + HECL WL++K NSVVY+CFGS HF + QL EIA G+EAS H FIWVV
Sbjct: 4 RGEESVVSVHECLSWLNSKQDNSVVYICFGSQCHFSDKQLYEIACGIEASSHEFIWVVPE 183
Query: 323 --RTQTEDGDE---WLPEGFEERTEGK--GLIIRGWSPQVMILEHEAIGAFVTHCGWNSV 375
RT+ ++ +E WLP+GFEER GK +II+GW+PQVMIL H A+GAF+THCGWNS
Sbjct: 184 KKRTENDNEEEKEKWLPKGFEERIIGKKKAMIIKGWAPQVMILSHTAVGAFMTHCGWNST 363
Query: 376 LEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKW-VMKVG--DNVEW-DAVEK 431
+E V AGVPMITWP+ EQFYNEKL+T+V GV VG +W +G + V W D +EK
Sbjct: 364 VEAVSAGVPMITWPMHGEQFYNEKLITQVHGIGVEVGATEWSTTGIGEREKVVWRDNIEK 543
Query: 432 AVKRVME-GEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSL 480
VKR+M+ G+EA ++R A+ E AK A++E GSS+S L A++ L+ L
Sbjct: 544 VVKRLMDSGDEAEKIRQHAREFGEKAKHAIKEGGSSHSNLTAVVNYLKRL 693
>TC80062 weakly similar to GP|4115534|dbj|BAA36410.1 UDP-glycose:flavonoid
glycosyltransferase {Vigna mungo}, partial (42%)
Length = 1071
Score = 230 bits (587), Expect = 8e-61
Identities = 115/222 (51%), Positives = 153/222 (68%), Gaps = 14/222 (6%)
Frame = +2
Query: 271 IDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGD 330
+ HECL WLD+K NSV+Y+CFGS++HF + QL EIA G+E SGH F+WVV + D
Sbjct: 2 VSVHECLSWLDSKEDNSVLYICFGSISHFSDKQLYEIASGIENSGHKFVWVVPEKKGKED 181
Query: 331 E-------WLPEGFEERT--EGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVA 381
E WLP+GFEER KG II+GW+PQ MIL H +GAF+THCGWNS++E + A
Sbjct: 182 ESEEQKEKWLPKGFEERNILNKKGFIIKGWAPQAMILSHTVVGAFMTHCGWNSIVEAISA 361
Query: 382 GVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVM----KVGDNVEWDAVEKAVKRVM 437
G+PMITWPV EQFYNEKL+T V + GV VG +W + + V ++EKAV+R+M
Sbjct: 362 GIPMITWPVHGEQFYNEKLITVVQRIGVEVGATEWSLHGFQEKEKVVSRHSIEKAVRRLM 541
Query: 438 -EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELR 478
+G+EA E+R +A+ + A AVEE GSS++ L ALI +L+
Sbjct: 542 DDGDEAKEIRQRAQEFGKKATHAVEEGGSSHNNLLALINDLK 667
>TC80009 weakly similar to PIR|T03747|T03747 glucosyltransferase IS5a (EC
2.4.1.-) salicylate-induced - common tobacco, partial
(19%)
Length = 1118
Score = 213 bits (542), Expect = 1e-55
Identities = 120/354 (33%), Positives = 193/354 (53%), Gaps = 12/354 (3%)
Frame = +3
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PL + PF+ GH IP D+A +FAS G +VT++TTP N ++K+L + F + +
Sbjct: 102 PLKLHFIPFLASGHMIPLFDIATMFASHGHQVTVITTPSNAQSLTKSLSSAASFF--LRL 275
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFF 126
T+ FP + LP+G E++ S LL P E + + P C+++D +
Sbjct: 276 HTVDFPSEQVDLPKGIESMSSTTDSITSWKIHRGAMLLHGPIENFMEKDPPDCIISDSTY 455
Query: 127 PWATDSAAKFGIPRIVFHGTSFFSLCASQCMKK---YQPYKNVSSDTDLFEITDLPGNIK 183
PWA D A K IP + F+G S F++ + + + N SD+ F + + P I
Sbjct: 456 PWANDLAHKLQIPNLTFNGLSLFTISLVESLIRNNLLHSDTNSDSDSSSFLVPNFPHRI- 632
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYA-DYYREVLGI 242
TL+E P + +K + + ++ ++S +I+N+F EL+ +Y + G
Sbjct: 633 --------TLSEKPP--KVLSKFLKMMLETVLKSKALIINNFAELDGEECIQHYEKTTGR 782
Query: 243 KEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNS 302
K WH+GP S+ R +E+ + RG E ++ HECL WL+++ +N+V+Y+CFGS+ + +
Sbjct: 783 KVWHLGPTSLIRRTIQEK--AERGNEGEVNMHECLSWLNSQRVNAVLYICFGSINYLSDK 956
Query: 303 QLKEIAMGLEASGHNFIWVVRTQ-------TEDGDEWLPEGFEERTEGK-GLII 348
QL E+A +E SGH FIWVV + E+ ++WLP+GFEER K GLII
Sbjct: 957 QLYEMACAIETSGHPFIWVVPEKKGKEDESEEEKEKWLPKGFEERNISKMGLII 1118
>TC77222 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (76%)
Length = 1643
Score = 213 bits (542), Expect = 1e-55
Identities = 153/497 (30%), Positives = 238/497 (47%), Gaps = 18/497 (3%)
Frame = +3
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASK--GVRVTIVTTPLNKPPISKALEQSK 58
+ + +HI V P G H +P ++ +K + VT + L PP S K
Sbjct: 12 LTKMAKTIHIAVIPSPGFSHLVPIVEFSKRLVTNHPNFHVTCIIPSLGSPPDSSKSYLEK 191
Query: 59 IHFNNIDIQTIKFPCV-EAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKP 117
I N I +I P + + LP+G P++ ++ + Q + L +
Sbjct: 192 IPPN---INSIFLPPINKQDLPQGV-----YPAILIQQTVTLSLPSIHQALKSLSSKAPL 347
Query: 118 HCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSD-TDLFEIT 176
++AD F A D A +F + TS L + K + VS + DL E
Sbjct: 348 VAIIADSFAFEALDFAKEFNSLSYFYFPTSANVLSFILHLPKLD--EEVSCEFKDLQEPI 521
Query: 177 DLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYY 236
L G + + + LP + S ++F + S G+++NSFYELE+ +
Sbjct: 522 KLQGCVPINGIDLPTPTKDR---SSEAYRMFLQRAKSFYFVDGILINSFYELESSAVEAL 692
Query: 237 RE--VLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFG 294
++ I + +GP + + + + D+HECLKWL + NSV+Y+ FG
Sbjct: 693 KQKGYGNISYFPVGPITQIGSSNNDVVG---------DEHECLKWLKNQPQNSVLYVSFG 845
Query: 295 SMTHFLNSQLKEIAMGLEASGHNFIWVVRTQT------------EDGDEWLPEGFEERTE 342
S Q+ EIA GLE SG FIW VR + ED ++LPEGF+ERT+
Sbjct: 846 SGGTLSQRQINEIAFGLELSGQRFIWGVRAPSDSVNAAYLESTNEDPLKFLPEGFQERTK 1025
Query: 343 GKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVT 402
KG I+ W+PQV IL+H ++G F++HCGWNSVLE + GVP++ WP+ AEQ N ++
Sbjct: 1026EKGFILPSWAPQVEILKHSSVGGFLSHCGWNSVLESMQEGVPIVAWPLFAEQAMNAVMLC 1205
Query: 403 EVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEE 462
+ LK + + + + VE D +K +MEGEE MR++ K L + A AV++
Sbjct: 1206DGLKVAI-----RLKFEDDEIVEKDKTANVIKCLMEGEEGKTMRDRMKSLKDYAVNAVKD 1370
Query: 463 DGSSYSQLNALIEELRS 479
+GSS L+ L + S
Sbjct: 1371EGSSIQNLSQLASQWES 1421
>BQ123034 weakly similar to GP|14334982|gb putative glucosyltransferase
{Arabidopsis thaliana}, partial (24%)
Length = 765
Score = 213 bits (541), Expect = 2e-55
Identities = 118/221 (53%), Positives = 153/221 (68%), Gaps = 10/221 (4%)
Frame = +2
Query: 270 SIDKH-ECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQT-- 326
S+ KH E L WL++K SV+Y+ FGS T +QL EI GLE SGHNFIWV++
Sbjct: 56 SLGKHTELLNWLNSKENESVLYVSFGSFTRLPYAQLVEIVHGLENSGHNFIWVIKRDDTD 235
Query: 327 EDGDEWLPEGFEERTE--GKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVP 384
EDG+ +L E FEER + KG II W+PQ++IL+H A G VTHCGWNS LE + AG+P
Sbjct: 236 EDGEGFLQE-FEERIKESSKGYIIWDWAPQLLILDHPATGGIVTHCGWNSTLESLNAGLP 412
Query: 385 MITWPVAAEQFYNEKLVTEVLKTGVPVGVKK---WV-MKVGDNVEWDAVEKAVKRVM-EG 439
MITWP+ AEQFYNEKL+ +VLK GVPVG K+ W+ + V V + +EK VK +M G
Sbjct: 413 MITWPIFAEQFYNEKLLVDVLKIGVPVGAKENKLWLDISVEKVVRREEIEKTVKILMGSG 592
Query: 440 EEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSL 480
+E+ EMR +AK L+E AK+ +EE G SY+ L LI+EL+SL
Sbjct: 593 QESKEMRMRAKKLSEAAKRTIEEGGDSYNNLIQLIDELKSL 715
>TC81362 similar to GP|19911201|dbj|BAB86927. glucosyltransferase-9 {Vigna
angularis}, partial (70%)
Length = 1347
Score = 201 bits (510), Expect = 7e-52
Identities = 120/329 (36%), Positives = 183/329 (55%), Gaps = 8/329 (2%)
Frame = +1
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
+ + N H ++FP + GH IP ID+AKL A +GV VTI TTP N + L ++
Sbjct: 82 LQANINVPHFVLFPLIAQGHIIPMIDIAKLLAQRGVIVTIFTTPKNASRFTSVLSRAVSS 261
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSI---PSVSFVPAFFAAIRLLQQPFEELL--LQQ 115
I I T+ FP + GLP+GCEN D + ++ F A+ LLQ+ E+L L
Sbjct: 262 GLQIKIVTLNFPSKQVGLPDGCENFDMVNISKDMNMKYNLFHAVSLLQKEGEDLFDKLSP 441
Query: 116 KPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQP---YKNVSSDTDL 172
KP C+++D W + A K IPRI FHG F+L CM K ++++S+T+
Sbjct: 442 KPSCIISDFCITWTSQIAEKHHIPRISFHGFCCFTL---HCMFKVHTSNILESINSETEF 612
Query: 173 FEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVY 232
F I +P I++T+ Q+P T+ E + FA E+++++E++SYGVI+NSF ELE Y
Sbjct: 613 FSIPGIPDKIQVTKEQIPGTVKEEK--MKGFA---EKMQEAEMKSYGVIINSFEELEKEY 777
Query: 233 ADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMC 292
+ Y++V K W +GP ++ N++ ++ + RG ASI +H CL +LD SVVY+C
Sbjct: 778 VNDYKKVRNDKVWCVGPVALCNKDGLDK--AQRGNIASISEHNCLNFLDLHKPKSVVYVC 951
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWV 321
G F + I G+ S N I++
Sbjct: 952 LGKFMQFNSFTTY*IGFGI-TSNKNSIYL 1035
Score = 149 bits (377), Expect = 2e-36
Identities = 71/135 (52%), Positives = 101/135 (74%), Gaps = 4/135 (2%)
Frame = +2
Query: 284 NINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRT---QTEDGDEWLP-EGFEE 339
N N + S+ + + SQL E+A+GL+A+ FIWV+R ++E+ ++W+ E FEE
Sbjct: 926 NQNLLFMFVLESLCNLIPSQLIELALGLQATKIPFIWVIREGIYKSEELEKWISDEKFEE 1105
Query: 340 RTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEK 399
R +G+GLIIRGW+PQ++IL H +IG F+THCGWNS LEG+ GVPM+TWP+ A+QF NEK
Sbjct: 1106 RNKGRGLIIRGWAPQMVILSHSSIGGFLTHCGWNSTLEGISFGVPMVTWPLFADQFLNEK 1285
Query: 400 LVTEVLKTGVPVGVK 414
LVT+VL+ GV +GV+
Sbjct: 1286 LVTQVLRIGVSLGVE 1330
>CA918173 weakly similar to GP|11034537|dbj putative glucosyl transferase
{Oryza sativa (japonica cultivar-group)}, partial (19%)
Length = 769
Score = 200 bits (508), Expect = 1e-51
Identities = 113/227 (49%), Positives = 150/227 (65%), Gaps = 5/227 (2%)
Frame = +2
Query: 178 LPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYR 237
LP I+MTRL L N EN+ ++ +FE + +S RSYG + NSF+ELE+ Y ++
Sbjct: 17 LPHTIEMTRLHLHNWERENNAMTA----IFEPMYESAERSYGSLYNSFHELESDYEKLFK 184
Query: 238 EVLGIKEWHIGPFSIHNRNKEEEIPSYRGK-EASIDKH-ECLKWLDTKNINSVVYMCFGS 295
+GIK W +GP S NK++E + RG E S+ KH E L WL++K SV+Y+ FGS
Sbjct: 185 TTIGIKSWSVGPVSAW-ANKDDERKANRGHIEKSLGKHTELLNWLNSKENESVLYVSFGS 361
Query: 296 MTHFLNSQLKEIAMGLEASGHNFIWVVRT-QTEDGDEWLPEGFEERTE--GKGLIIRGWS 352
+ ++QL EIA GLE SGHNFIWV++ + EDG+ +L E FE+R + KG II W+
Sbjct: 362 LIRLPHAQLVEIAHGLENSGHNFIWVIKNNKDEDGEGFLQE-FEKRMKESNKGYIIWDWA 538
Query: 353 PQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEK 399
PQ++ILE+ AIG VTHCGWNS LE V AG+PMITWPV AE F K
Sbjct: 539 PQLLILEYPAIGGIVTHCGWNSTLESVNAGLPMITWPVFAEDFTMRK 679
>TC77223 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (50%)
Length = 1628
Score = 197 bits (502), Expect = 6e-51
Identities = 150/496 (30%), Positives = 237/496 (47%), Gaps = 23/496 (4%)
Frame = +3
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASK--GVRVTIVTTPLNKPPISKALEQSKIHFNNI- 64
+HI V P G H +P ++ K + VT + L PP + SK + I
Sbjct: 24 IHIAVIPSPGFSHLVPIVEFTKRLVTNHPNFHVTCIIPSLGSPP-----DSSKSYLETIP 188
Query: 65 -DIQTIKFPCV-EAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVA 122
+I +I P + + LP+G P V ++ + Q E L + ++A
Sbjct: 189 PNINSIFLPPINKQDLPQGVH-----PGVLIQLTVTHSLPSIHQALESLTSKTPLVAIIA 353
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNI 182
D F A D A +F ++ S F L + K + DL E L G +
Sbjct: 354 DTFAFEALDFAKEFNSLSYLYFPCSSFVLSLLLHLPKLDEEFSCEYK-DLQEPIKLQGCV 530
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELEN--VYADYYREVL 240
+ + LP + S K++ + S G+++NSF ELE+ + A +
Sbjct: 531 PINGIDLPAATKDR---SNEGYKMYIQRAKSMYFVDGILINSFIELESSAIKALELKGYG 701
Query: 241 GIKEWHIGPFS---IHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMT 297
I + +GP + + N + +E+ ECLKWL + NSV+Y+ FGS
Sbjct: 702 KIDFFPVGPITQTGLSNNDVGDEL-------------ECLKWLKNQPQNSVLYVSFGSGG 842
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQT------------EDGDEWLPEGFEERTEGKG 345
+Q+ E+A GLE SG FIWV+R + ED ++LP+GF ERT+ KG
Sbjct: 843 TLSQTQINELAFGLELSGQRFIWVLRAPSDSVSAAYLEATNEDPLKFLPKGFLERTKEKG 1022
Query: 346 LIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVL 405
LI+ W+PQV IL+ +++G F++HCGWNSVLE + GVP++ WP+ AEQ N +++ L
Sbjct: 1023LILPSWAPQVQILKEKSVGGFLSHCGWNSVLESMQEGVPIVAWPLFAEQAMNAVMLSNDL 1202
Query: 406 KTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVE-EDG 464
K + + + + VE D + +K +MEGEE MR++ K L + A KA+ +DG
Sbjct: 1203KVAI-----RLKFEDDEIVEKDKIANVIKCLMEGEEGKAMRDRMKSLRDYATKALNVKDG 1367
Query: 465 SSYSQLNALIEELRSL 480
SS L+ L ++ +
Sbjct: 1368SSIQTLSHLASQMEMI 1415
>TC85817 weakly similar to PIR|B85014|B85014 probable flavonol
glucosyltransferase [imported] - Arabidopsis thaliana,
partial (83%)
Length = 1736
Score = 195 bits (496), Expect = 3e-50
Identities = 155/492 (31%), Positives = 243/492 (48%), Gaps = 25/492 (5%)
Frame = +3
Query: 7 PLHILVFPFMGHGHTIPTIDMAK--LFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNI 64
P +++ P G GH IP I+ AK + ++ +++T P PP SKA +
Sbjct: 153 PPMVVMLPSPGMGHLIPMIEFAKRIIILNQNLQITFFI-PTEGPP-SKAQKTVLQSLPKF 326
Query: 65 DIQTIKFPCVEAGLP--EGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVA 122
T P + LP G E + S+ + +P+ L+Q F L VV
Sbjct: 327 ISHTFLPPVSFSDLPPNSGIETIISLTVLRSLPS-------LRQNFNTLSETHTITAVVV 485
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITD---LP 179
D+F A D A +F +P+ VF+ ++ +L + Y P + + E+T+ +P
Sbjct: 486 DLFGTDAFDVAREFNVPKYVFYPSTAMALS----LFLYLPRLDEEVHCEFRELTEPVKIP 653
Query: 180 GNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE-NVYADYYRE 238
G I + L + L D + ++ +F K + G+I NSF ELE + +E
Sbjct: 654 GCIPIHGKYLLDPL--QDRKNDAYQSVFRNAKRYR-EADGLIENSFLELEPGPIKELLKE 824
Query: 239 VLGIKEWH-IGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMT 297
G +++ +GP +E E+ E LKWLD + SV+++ FGS
Sbjct: 825 EPGKPKFYPVGPLV----KREVEVGQIGPNS------ESLKWLDNQPHGSVLFVSFGSGG 974
Query: 298 HFLNSQLKEIAMGLEASGHNFIWVVRTQTE--------------DGDEWLPEGFEERTEG 343
+ Q+ E+A+GLE S F+WVVR+ + D ++LP GF ERT+G
Sbjct: 975 TLSSKQIVELALGLEMSEQRFLWVVRSPNDKVANASYFSAETDSDPFDFLPNGFLERTKG 1154
Query: 344 KGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTE 403
+GL++ W+PQ +L H + G F+THCGWNSVLE VV GVP++ WP+ AEQ N ++TE
Sbjct: 1155RGLVVSSWAPQPQVLAHGSTGGFLTHCGWNSVLESVVNGVPLVVWPLYAEQKMNAVMLTE 1334
Query: 404 VLKTGVPVGVKKWVMKVGDN--VEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVE 461
+K G+ VG+N VE + VK +MEGEE ++R + K L E A K +
Sbjct: 1335DVKVGLR-------PNVGENGLVERLEIASVVKCLMEGEEGKKLRYQMKDLKEAASKTLG 1493
Query: 462 EDGSSYSQLNAL 473
E+G+S + ++ L
Sbjct: 1494ENGTSTNHISNL 1529
>TC85899 weakly similar to GP|7385017|gb|AAF61647.1| UDP-glucose:salicylic
acid glucosyltransferase {Nicotiana tabacum}, partial
(47%)
Length = 1617
Score = 194 bits (493), Expect = 6e-50
Identities = 143/485 (29%), Positives = 230/485 (46%), Gaps = 9/485 (1%)
Frame = +2
Query: 2 DSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHF 61
+ +++ H L+ P+ GH P I +K KGV++T++T IS +
Sbjct: 23 EKKNHAPHCLILPYPAQGHMNPMIQFSKRLIEKGVKITLITVTSFWKVISNK------NL 184
Query: 62 NNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL-----LQQK 116
+ID+++I E GL E+++ + ++ Q ELL +
Sbjct: 185 TSIDVESISDGYDEGGLL-AAESLEDYKETFW--------KVGSQTLSELLHKLSSSENP 337
Query: 117 PHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEIT 176
P+CV+ D F PW D FG+ + F F C+ + + K + E
Sbjct: 338 PNCVIFDAFLPWVLDVGKSFGLVGVAF----FTQSCSVNSVYYHTHEKLIELPLTQSEYL 505
Query: 177 DLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYY 236
LPG K+ LP+ L + F + + + + ++ NS YELE D+
Sbjct: 506 -LPGLPKLAPGDLPSFLYKYGSYPGYFDIVVNQFANIGKADW-ILANSIYELEPEVVDWL 679
Query: 237 REVLGIKEWHIGP----FSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMC 292
++ +K IGP + R K+++ Y + + C+KWL+ K SVVY
Sbjct: 680 VKIWPLKT--IGPSVPSMLLDKRLKDDK--EYGVSLSDPNTEFCIKWLNDKPKGSVVYAS 847
Query: 293 FGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWS 352
FGSM Q +E+A+GL+ S F+WVVR E LP+GF E ++ KGLI+ W
Sbjct: 848 FGSMAGLSEEQTQELALGLKDSESYFLWVVR---ECDQSKLPKGFVESSK-KGLIVT-WC 1012
Query: 353 PQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVG 412
PQ+++L HEA+G FVTHCGWNS LE + GVP+I P+ +Q N KL+ +V K GV
Sbjct: 1013PQLLVLTHEALGCFVTHCGWNSTLEALSIGVPLIAMPLWTDQVTNAKLIADVWKMGV--- 1183
Query: 413 VKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNA 472
+ V + V + ++ +K ++E E+ E++ A +AK +V+E G S +
Sbjct: 1184--RAVADEKEIVRSETIKNCIKEIIETEKGNEIKKNALKWKNLAKSSVDEGGRSDKNIEE 1357
Query: 473 LIEEL 477
+ L
Sbjct: 1358FVAAL 1372
>TC85228 weakly similar to GP|19911209|dbj|BAB86931. glucosyltransferase-13
{Vigna angularis}, partial (31%)
Length = 3139
Score = 194 bits (492), Expect = 8e-50
Identities = 151/484 (31%), Positives = 229/484 (47%), Gaps = 16/484 (3%)
Frame = -1
Query: 6 NPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNID 65
N I + P G H IP ++ AKL I P++ ++ QS ++ +
Sbjct: 2782 NKTCIAMVPSPGLSHLIPQVEFAKLLLQHHNESHITFLIPTLGPLTPSM-QSILNTLPPN 2606
Query: 66 IQTIKFPCVEA-GLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADM 124
+ P V LP E PS +I L + + LL + +V M
Sbjct: 2605 MNFTVLPQVNIEDLPHNLE-----PSTQMKLIVKHSIPFLHEEVKSLLSKTNLVALVCSM 2441
Query: 125 FFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPG-NIK 183
F A D A F + +F + + + +E+ ++PG +I
Sbjct: 2440 FSTDAHDVAKHFNLLSYLFFSSGAVLFSFFLTLPNLDDAASTQFLGSSYEMVNVPGFSIP 2261
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSF--YELENVYADYYREVLG 241
+LP+ + S ++ + + + S + GVI+N+F ELE V RE
Sbjct: 2260 FHVKELPDPFN-CERSSDTYKSILDVCQKSSLFD-GVIINTFSNLELEAVRVLQDREKPS 2087
Query: 242 IKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLN 301
+ + +GP + N E + CL+WL+ + +SV+++ FGS
Sbjct: 2086 V--FPVGPIIRNESNNEANMSV------------CLRWLENQPPSSVIFVSFGSGGTLSQ 1949
Query: 302 SQLKEIAMGLEASGHNFIWVVRT------------QTEDGDEWLPEGFEERTEGKGLIIR 349
QL E+A GLE SGH F+WVVR Q + E+LP GF ERT+ KGL++
Sbjct: 1948 DQLNELAFGLELSGHKFLWVVRAPSKHSSSAYFNGQNNEPLEYLPNGFVERTKEKGLVVT 1769
Query: 350 GWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGV 409
W+PQV IL H +IG F++HCGW+S LE VV GVP+I WP+ AEQ N KL+T+VLK
Sbjct: 1768 SWAPQVEILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLK--- 1598
Query: 410 PVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQ 469
V V+ V ++ + V KA+KR+MEG+E++E+R K K L+ A + E GSS
Sbjct: 1597 -VAVRPKVDGETGIIKREEVSKALKRIMEGDESFEIRKKIKELSVSAATVLSEHGSSRKA 1421
Query: 470 LNAL 473
L+ L
Sbjct: 1420 LSTL 1409
>TC85231 similar to GP|13508844|emb|CAC35167. arbutin synthase {Rauvolfia
serpentina}, partial (34%)
Length = 1602
Score = 193 bits (490), Expect = 1e-49
Identities = 148/483 (30%), Positives = 236/483 (48%), Gaps = 19/483 (3%)
Frame = +3
Query: 10 ILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQTI 69
I + P G H IP ++ AKL I P++ ++ QS ++ ++ I
Sbjct: 66 IAMVPSPGLSHLIPLVEFAKLLLQNHNEYHITFLIPTLGPLTPSM-QSILNTLPPNMNFI 242
Query: 70 KFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFFPWA 129
P + + + N+D + + +I L + + LL + + +V MF A
Sbjct: 243 VLP--QVNIEDLPHNLDPATQMKLIVKH--SIPFLYEEVKSLLSKTRLVALVFSMFSTDA 410
Query: 130 TDSAAKFGIPRIVFH--GTSFFSLCASQCMKKYQPYKNVSSDTDL----FEITDLPG-NI 182
D A F + +F G FSL + P + ++ T +E ++PG +I
Sbjct: 411 HDVAKHFNLLSYLFFSSGAVLFSLFLTI------PNLDEAASTQFLGSSYETVNIPGFSI 572
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGI 242
+ +LP+ + S ++ + + + + GVI+N+F +LE ++
Sbjct: 573 PLHIKELPDPFI-CERSSDAYKSILDVCQKLSLFD-GVIMNTFTDLEPEVIRVLQDREKP 746
Query: 243 KEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNS 302
+ +GP + N E + CL+WL+ + +SV+++ FGS
Sbjct: 747 SVYPVGPMIRNESNNEANMSM------------CLRWLENQQPSSVLFVSFGSGGTLSQD 890
Query: 303 QLKEIAMGLEASGHNFIWVVRT------------QTEDGDEWLPEGFEERTEGKGLIIRG 350
QL EIA GLE SGH F+WVVR Q D E+LP GF ERT+ GL++
Sbjct: 891 QLNEIAFGLELSGHKFLWVVRAPSKNSSSAYFSGQNNDPLEYLPNGFLERTKENGLVVAS 1070
Query: 351 WSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVP 410
W+PQV IL H +IG F++HCGW+S LE VV GVP+I WP+ AEQ N KL+T+VLK
Sbjct: 1071WAPQVEILGHGSIGGFLSHCGWSSTLESVVNGVPLIAWPLFAEQRMNAKLLTDVLK---- 1238
Query: 411 VGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQL 470
V V+ V ++ + V KA+KR+M+G+E++E+R K K L+ A + E GSS L
Sbjct: 1239VAVRPKVDDETGIIKQEEVAKAIKRIMKGDESFEIRKKIKELSVGAATVLSEHGSSRKAL 1418
Query: 471 NAL 473
++L
Sbjct: 1419SSL 1427
>BG450101 similar to GP|4115534|dbj UDP-glycose:flavonoid glycosyltransferase
{Vigna mungo}, partial (21%)
Length = 646
Score = 189 bits (479), Expect(2) = 2e-48
Identities = 92/176 (52%), Positives = 119/176 (67%), Gaps = 8/176 (4%)
Frame = +1
Query: 232 YADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYM 291
Y YY G K WH+GP S+ + +E+ + RG+E+++ ECL WL++K NSV+Y+
Sbjct: 97 YIKYYVSSTGHKAWHLGPASLIRKTVQEK--AERGQESAVSVQECLSWLNSKRHNSVLYI 270
Query: 292 CFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDE-------WLPEGFEERTEGK 344
FGS+ + + QL EIA +EASGH+FIWV+ DE WLP+GFEER GK
Sbjct: 271 SFGSLCRYQDKQLYEIACAIEASGHDFIWVIPLNNGKEDESEEEKQKWLPKGFEERNIGK 450
Query: 345 -GLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEK 399
GLIIRGW+PQV+IL H A+GAF+THCGWNS +E V AGVPMITWP EQF + K
Sbjct: 451 KGLIIRGWAPQVLILSHPAVGAFMTHCGWNSTVEAVGAGVPMITWPSHGEQFLHRK 618
Score = 22.3 bits (46), Expect(2) = 2e-48
Identities = 9/14 (64%), Positives = 10/14 (71%)
Frame = +2
Query: 395 FYNEKLVTEVLKTG 408
FY EKL+TEV G
Sbjct: 605 FYTEKLITEVRXIG 646
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,845,644
Number of Sequences: 36976
Number of extensions: 227870
Number of successful extensions: 1428
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 1222
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1237
length of query: 489
length of database: 9,014,727
effective HSP length: 100
effective length of query: 389
effective length of database: 5,317,127
effective search space: 2068362403
effective search space used: 2068362403
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149574.3


