

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.1 - phase: 0 /pseudo
(619 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
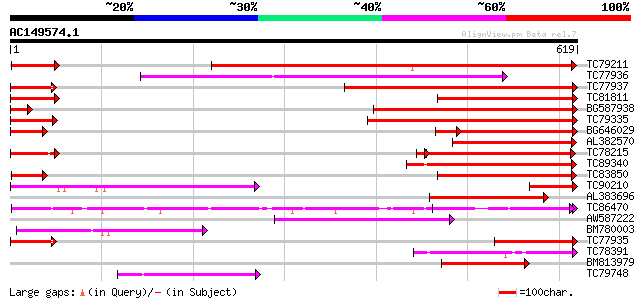
Score E
Sequences producing significant alignments: (bits) Value
TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Sol... 426 e-119
TC77936 weakly similar to PIR|T52319|T52319 P-glycoprotein-like ... 271 5e-73
TC77937 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 261 6e-70
TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gos... 256 1e-68
BG587938 homologue to GP|22655312|g putative ABC transporter {Ar... 256 2e-68
TC79335 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 249 3e-66
BG646029 similar to GP|14715462|d CjMDR1 {Coptis japonica}, part... 163 1e-43
AL382570 weakly similar to GP|10241621|em putative P-glycoprotei... 170 2e-42
TC78215 similar to GP|10177549|dbj|BAB10828. ABC transporter-lik... 167 1e-41
TC89340 weakly similar to GP|17427488|emb|CAD14006. PROBABLE COM... 161 8e-40
TC83850 weakly similar to GP|7300071|gb|AAF55241.1| CG4225 gene ... 159 4e-39
TC90210 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japo... 152 3e-37
AL383696 homologue to PIR|T47671|T4 P-glycoprotein-like - Arabid... 150 1e-36
TC86470 similar to PIR|F84919|F84919 glutathione-conjugate trans... 123 2e-28
AW587222 weakly similar to GP|5456699|gb| ATP-binding cassette m... 121 7e-28
BM780003 weakly similar to PIR|T06165|T0 multidrug resistance pr... 119 3e-27
TC77935 similar to PIR|E85023|E85023 probable P-glycoprotein-lik... 115 5e-26
TC78391 similar to PIR|F84919|F84919 glutathione-conjugate trans... 110 1e-24
BM813979 homologue to PIR|E85023|E85 probable P-glycoprotein-lik... 108 5e-24
TC79748 weakly similar to GP|9294577|dbj|BAB02858.1 multidrug re... 107 1e-23
>TC79211 similar to GP|4204793|gb|AAD10836.1| P-glycoprotein {Solanum
tuberosum}, partial (36%)
Length = 1667
Score = 426 bits (1094), Expect = e-119
Identities = 207/401 (51%), Positives = 299/401 (73%), Gaps = 3/401 (0%)
Frame = +1
Query: 221 SSMLSSRLETDATLLKTIVVDRSTILLQNVGLVVTALVIAFILNWRITLVVLATYPLIIS 280
S+ +S+RL DA +++ + DR ++++QN L++ A F+L WR+ LV++A +P++++
Sbjct: 7 SARISARLALDANNVRSAIGDRISVIVQNTALMLVACTAGFVLQWRLALVLIAVFPVVVA 186
Query: 281 GHIGEKLFMQGFGGNLSKAYLKANMLAGEAVSNIRTVAAFCAEEKVIDLYADELVEPSKR 340
+ +K+FM GF G+L A+ KA LAGEA++N+RTVAAF +E K++ L+A L P +R
Sbjct: 187 ATVLQKMFMTGFSGDLEAAHAKATQLAGEAIANVRTVAAFNSESKIVRLFASNLETPLQR 366
Query: 341 SFKRGQIAGIFYGISQFFIFSSYGLALWYGSVLLEKELASFKSIMKSFMVLIVTALAMGE 400
F +GQI+G YGI+QF +++SY L LWY S L++ ++ F ++ FMVL+V+A E
Sbjct: 367 CFWKGQISGSGYGIAQFALYASYALGLWYASWLVKHGISDFSKTIRVFMVLMVSANGAAE 546
Query: 401 TLALAPDLLKGNQMVSSIFDMIDRKSGIIHDVGEELMT---VEGMIELKRINFIYPSRPN 457
TL LAPD +KG + + S+FD++DR++ I D + + G +ELK ++F YP+RP+
Sbjct: 547 TLTLAPDFIKGGRAMRSVFDLLDRQTEIEPDDQDATPVPDRLRGEVELKHVDFSYPTRPD 726
Query: 458 VVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSL 517
+ +F+D NL + +GK+LALVG SG GKSS+I+LI RFYDPTSG++MIDGKDI+K NLKSL
Sbjct: 727 MPVFRDLNLRIRAGKTLALVGPSGCGKSSVIALIQRFYDPTSGRIMIDGKDIRKYNLKSL 906
Query: 518 RKQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGD 577
R+ I +V QEP LFAT+IY+NI YG + A+E+E+IEAA LA+AH FIS+LP+GY T G+
Sbjct: 907 RRHISVVPQEPCLFATTIYENIAYGHDSATEAEIIEAATLANAHKFISSLPDGYKTFVGE 1086
Query: 578 RGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESER 618
RGV LSGGQKQR+A+ARA LR +++LLDEATSALD ESER
Sbjct: 1087RGVQLSGGQKQRIAVARAFLRKAELMLLDEATSALDAESER 1209
Score = 59.7 bits (143), Expect = 3e-09
Identities = 27/53 (50%), Positives = 42/53 (78%), Gaps = 1/53 (1%)
Frame = +1
Query: 3 GRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISN-PNSLYSSLVQGQ 54
G+TT+IVAHRLSTI+NA++IAV++ G V E G+H +L+ N + +Y+ ++Q Q
Sbjct: 1243 GKTTIIVAHRLSTIRNANVIAVIDDGKVAEQGSHSQLMKNHQDGIYARMIQLQ 1401
>TC77936 weakly similar to PIR|T52319|T52319 P-glycoprotein-like protein
pgp3 [imported] - Arabidopsis thaliana, partial (32%)
Length = 1219
Score = 271 bits (693), Expect = 5e-73
Identities = 145/403 (35%), Positives = 237/403 (57%), Gaps = 3/403 (0%)
Frame = +1
Query: 144 ISHALVSYYMDWDSTCHEVKKIAFLFCGAAIVAITAYSIEHLSFGIMGERLTLRVRGIML 203
IS + +Y H+ K A +F A+ + FG+ G +L R+R +
Sbjct: 10 ISKMINIFYKPAHELRHDSKVWAIVFVAVAVATLLIIPCRFYFFGVAGGKLIQRIRNMCF 189
Query: 204 SAILKNEIGWFDDTRNTSSMLSSRLETDATLLKTIVVDRSTILLQNVGLVVTALVIAFIL 263
++ E+ WFD+ ++S L +RL TDA ++ +V D +L+QN+ + LVI+F
Sbjct: 190 EKVVHMEVSWFDEAEHSSGALGARLSTDAASVRALVGDALGLLVQNIATAIAGLVISFQA 369
Query: 264 NWRITLVVLATYPLI-ISGHIGEKLFMQGFGGNLSKAYLKANMLAGEAVSNIRTVAAFCA 322
+W++ +VLA PL+ ++G++ K+ ++GF + K Y +A+ +A +AV +IRTVA+FCA
Sbjct: 370 SWQLAFIVLALAPLLGLNGYVQVKV-LKGFSADAKKLYEEASQVANDAVGSIRTVASFCA 546
Query: 323 EEKVIDLYADELVEPSKRSFKRGQIAGIFYGISQFFIFSSYGLALWYGSVLLEKELASFK 382
E+KV++LY + P K+ +RG I+G +G+S F +++ + G+ L+E ++F
Sbjct: 547 EKKVMELYKQKCEGPIKKGVRRGIISGFGFGLSFFMLYAVDACVFYAGARLVEDGKSTFS 726
Query: 383 SIMKSFMVLIVTALAMGETLALAPDLLKGNQMVSSIFDMIDRKSGI--IHDVGEELMTVE 440
+ F L + A+ + ++ L PD +SIF ++D S I + G L V+
Sbjct: 727 DVFLVFFALTMAAMGVSQSGTLVPDSTNAKSAAASIFAILDHNSQIDSSDESGMTLEEVK 906
Query: 441 GMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSG 500
G IE ++F YP+ +V IF D L + SGK++ALVG SGSGKS++ISL+ RFYDP SG
Sbjct: 907 GDIEFNHVSFKYPTTLDVQIFNDLCLNIRSGKTVALVGESGSGKSTVISLLQRFYDPDSG 1086
Query: 501 KVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGK 543
+ +DG +I++M +K LR+Q+GLV QEP LF ++ NI Y K
Sbjct: 1087HITLDGIEIQRMQVKWLRQQMGLVSQEPILFNDTVXANIAYXK 1215
>TC77937 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (24%)
Length = 1164
Score = 261 bits (667), Expect = 6e-70
Identities = 140/257 (54%), Positives = 188/257 (72%), Gaps = 3/257 (1%)
Frame = +3
Query: 366 ALWYGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLLKGNQMVSSIFDMIDRK 425
+ + G+ L+E +SF + + F L + A+ + ++ +L PD K V+SIF ++DRK
Sbjct: 12 SFYAGARLVEDGKSSFSDVFRVFFALSMAAIGLSQSGSLLPDSTKAKSAVASIFAILDRK 191
Query: 426 SGI--IHDVGEELMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSG 483
S I + G L V+G IE K +NF YP+RP++ IF+D L + SGK++ALVG SGSG
Sbjct: 192 SLIDPTDESGITLEEVKGEIEFKHVNFKYPTRPDIQIFRDLCLNIHSGKTVALVGESGSG 371
Query: 484 KSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGK 543
KS++ISLI RFYDP SG + +DGK+I+ + +K LR+Q+GLV QEP LF +I NI YGK
Sbjct: 372 KSTVISLIQRFYDPDSGHITLDGKEIQSLQVKWLRQQMGLVSQEPVLFNDTIRANIAYGK 551
Query: 544 -EEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKI 602
+ASE+E+I AA+LA+AH FIS+L +GY T G+RGV LSGGQKQRVAIARAI++NPKI
Sbjct: 552 GGDASEAEIIAAAELANAHKFISSLQKGYDTVVGERGVQLSGGQKQRVAIARAIVKNPKI 731
Query: 603 LLLDEATSALDVESERV 619
LLLDEATSALD ESE+V
Sbjct: 732 LLLDEATSALDAESEKV 782
Score = 58.5 bits (140), Expect = 7e-09
Identities = 30/51 (58%), Positives = 37/51 (71%)
Frame = +3
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLV 51
M+ RTT+IVAHRLSTIK AD+IAVV+ G + E G HE L+ Y+SLV
Sbjct: 807 MVERTTIIVAHRLSTIKGADLIAVVKNGVIAEKGKHEALLHKGGD-YASLV 956
>TC81811 similar to GP|6671365|gb|AAF23176.1| P-glycoprotein {Gossypium
hirsutum}, partial (17%)
Length = 850
Score = 256 bits (655), Expect = 1e-68
Identities = 131/152 (86%), Positives = 145/152 (95%)
Frame = +1
Query: 468 VPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQE 527
VPSGKS+ALVG SGSGKSS+ISLILRFYDPTSGKV+IDGKDI ++NLKSL K IGLVQQE
Sbjct: 1 VPSGKSVALVGQSGSGKSSVISLILRFYDPTSGKVLIDGKDITRINLKSLTKHIGLVQQE 180
Query: 528 PALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQK 587
PALFATSIY+NILYGKE AS+SEVIEAAKLA+AHNFISALPEGYSTK G+RGV LSGGQ+
Sbjct: 181 PALFATSIYENILYGKEGASDSEVIEAAKLANAHNFISALPEGYSTKVGERGVQLSGGQR 360
Query: 588 QRVAIARAILRNPKILLLDEATSALDVESERV 619
QRVAIARA+L+NP+ILLLDEATSALDVESER+
Sbjct: 361 QRVAIARAVLKNPEILLLDEATSALDVESERI 456
Score = 60.1 bits (144), Expect = 2e-09
Identities = 29/54 (53%), Positives = 37/54 (67%)
Frame = +1
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQGQ 54
M RTTV+VAHRLSTI+NAD I+V++ G ++E G H LI N + Y LV Q
Sbjct: 481 MQNRTTVMVAHRLSTIRNADQISVLQDGKIIEQGTHSSLIENKDGPYYKLVNLQ 642
>BG587938 homologue to GP|22655312|g putative ABC transporter {Arabidopsis
thaliana}, partial (19%)
Length = 780
Score = 256 bits (653), Expect = 2e-68
Identities = 131/224 (58%), Positives = 174/224 (77%), Gaps = 2/224 (0%)
Frame = +3
Query: 398 MGETLALAPDLLKGNQMVSSIFDMIDRKSGIIH--DVGEELMTVEGMIELKRINFIYPSR 455
+G++ K + IF +ID + GI + G EL TV G++ELK ++F YPSR
Sbjct: 3 LGQSAPSMAAFTKARVAAAKIFRIIDHQPGIDRNSESGLELETVTGLVELKNVDFSYPSR 182
Query: 456 PNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLK 515
P V+I DF+L VP+GK++ALVG SGSGKS+++SLI RFYDPTSG+VM+DG DIK + LK
Sbjct: 183 PEVLILNDFSLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPTSGQVMLDGHDIKTLKLK 362
Query: 516 SLRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKA 575
LR+QIGLV QEPALFAT+I +NIL G+ +A++ E+ EAA++A+AH+FI LPEG+ T+
Sbjct: 363 WLRQQIGLVSQEPALFATTIRENILLGRPDANQVEIEEAARVANAHSFIIKLPEGFETQV 542
Query: 576 GDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
G+RG+ LSGGQKQR+AIARA+L+NP ILLLDEATSALD ESE++
Sbjct: 543 GERGLQLSGGQKQRIAIARAMLKNPAILLLDEATSALDSESEKL 674
Score = 42.7 bits (99), Expect = 4e-04
Identities = 17/25 (68%), Positives = 24/25 (96%)
Frame = +3
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVV 25
MIGRTT+++AHRLSTI+ AD++AV+
Sbjct: 699 MIGRTTLVIAHRLSTIRKADLVAVI 773
>TC79335 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (23%)
Length = 1135
Score = 249 bits (635), Expect = 3e-66
Identities = 131/232 (56%), Positives = 175/232 (74%), Gaps = 3/232 (1%)
Frame = +2
Query: 391 LIVTALAMGETLALAPDLLKGNQMVSSIFDMIDRKSGI--IHDVGEELMTVEGMIELKRI 448
L + A+ + ++ + APD K +SIF MID+KS I + G L +++G IEL+ I
Sbjct: 8 LTMAAIGISQSSSFAPDSSKAKSATASIFGMIDKKSKIDPSEESGTTLDSIKGEIELRHI 187
Query: 449 NFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKD 508
+F YPSRP++ IF+D NL + SGK++ALVG SGSGKS++I+L+ RFYDP SG++ +DG +
Sbjct: 188 SFKYPSRPDIQIFRDLNLTIHSGKTVALVGESGSGKSTVIALLQRFYDPDSGEITLDGIE 367
Query: 509 IKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKEE-ASESEVIEAAKLADAHNFISAL 567
I+++ LK LR+Q+GLV QEP LF +I NI YGK A+E+E+I AA+LA+AH FIS L
Sbjct: 368 IRQLQLKWLRQQMGLVSQEPVLFNDTIRANIAYGKGGIATEAEIIAAAELANAHRFISGL 547
Query: 568 PEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
+GY T G+RG LSGGQKQRVAIARAI+++PKILLLDEATSALD ESERV
Sbjct: 548 QQGYDTIVGERGTQLSGGQKQRVAIARAIIKSPKILLLDEATSALDAESERV 703
Score = 70.5 bits (171), Expect = 2e-12
Identities = 34/52 (65%), Positives = 42/52 (80%)
Frame = +2
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQ 52
M+ RTTV+VAHRLSTIKNAD+IAVV+ G +VE G HE LI+ + Y+SLVQ
Sbjct: 728 MVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGRHETLINVKDGFYASLVQ 883
>BG646029 similar to GP|14715462|d CjMDR1 {Coptis japonica}, partial (16%)
Length = 680
Score = 163 bits (413), Expect(2) = 1e-43
Identities = 85/133 (63%), Positives = 106/133 (78%), Gaps = 1/133 (0%)
Frame = -3
Query: 488 ISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKE-EA 546
ISL+ RFYDP SG++ +DG +I+K+ LK R+Q+GLV QEP LF +I NI YGK A
Sbjct: 606 ISLLQRFYDPDSGQIKLDGTEIQKLQLKWFRQQMGLVSQEPVLFNDTIRANIAYGKGGNA 427
Query: 547 SESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLD 606
+E+EVI AA+LA+AHNFIS+L +GY T G+RG+ LSGGQKQ VAIARAI+ P+ILLLD
Sbjct: 426 TEAEVIAAAELANAHNFISSLQQGYDTIVGERGIQLSGGQKQPVAIARAIVNRPRILLLD 247
Query: 607 EATSALDVESERV 619
EATSALD ESE+V
Sbjct: 246 EATSALDAESEKV 208
Score = 50.1 bits (118), Expect = 3e-06
Identities = 23/40 (57%), Positives = 31/40 (77%)
Frame = -3
Query: 2 IGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELIS 41
+ RTT++VAHRLSTIK A+ IAVV+ G + E G H+ LI+
Sbjct: 180 VDRTTIVVAHRLSTIKGANSIAVVKNGVIEEKGKHDILIN 61
Score = 31.6 bits (70), Expect(2) = 1e-43
Identities = 13/29 (44%), Positives = 23/29 (78%)
Frame = -2
Query: 465 NLIVPSGKSLALVGHSGSGKSSIISLILR 493
+L + SG+++ALVG SGSGKS++ ++ +
Sbjct: 676 SLTIHSGQTVALVGESGSGKSTVDLIVAK 590
>AL382570 weakly similar to GP|10241621|em putative P-glycoprotein {Oryza
sativa}, partial (17%)
Length = 481
Score = 170 bits (430), Expect = 2e-42
Identities = 84/135 (62%), Positives = 106/135 (78%)
Frame = +3
Query: 484 KSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGK 543
KSS+++L+L+FYDP GKV+IDGKD+++ NL+ LR QIGLVQQEP LF SI +NI YG
Sbjct: 3 KSSVLALLLKFYDPVVGKVLIDGKDLREYNLRWLRTQIGLVQQEPLLFNCSIRENICYGN 182
Query: 544 EEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKIL 603
A ESE++E A+ A+ H F+S LP GY+T G++G LSGGQKQR+AIAR +L+ P IL
Sbjct: 183 NGAFESEIVEVAREANIHEFVSNLPNGYNTVVGEKGCQLSGGQKQRIAIARTLLKKPAIL 362
Query: 604 LLDEATSALDVESER 618
LLDEATSALD SER
Sbjct: 363 LLDEATSALDA*SER 407
>TC78215 similar to GP|10177549|dbj|BAB10828. ABC transporter-like protein
{Arabidopsis thaliana}, partial (36%)
Length = 1176
Score = 167 bits (422), Expect(2) = 1e-41
Identities = 88/163 (53%), Positives = 116/163 (70%), Gaps = 1/163 (0%)
Frame = +1
Query: 456 PNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLK 515
P ++ K N+ + G +ALVG SG GK++I +LI RFYDPT G ++++G + +++ K
Sbjct: 34 P*YMVLKGINIKLQPGSKVALVGPSGGGKTTIANLIERFYDPTKGNILVNGVPLVEISHK 213
Query: 516 SLRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIE-AAKLADAHNFISALPEGYSTK 574
L ++I +V QEP LF SI +NI YG + E IE AAK+A+AH FIS PE Y T
Sbjct: 214 HLHRKISIVSQEPTLFNCSIEENIAYGFDGKIEDADIENAAKMANAHEFISNFPEKYKTF 393
Query: 575 AGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESE 617
G+RG+ LSGGQKQR+AIARA+L +PKILLLDEATSALD ESE
Sbjct: 394 VGERGIRLSGGQKQRIAIARALLMDPKILLLDEATSALDAESE 522
Score = 59.7 bits (143), Expect = 3e-09
Identities = 27/54 (50%), Positives = 41/54 (75%)
Frame = +1
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQGQ 54
M GRT +++AHRLST+K A+ +AVV G +VE+G H+EL+ N +Y++LV+ Q
Sbjct: 553 MKGRTVLVIAHRLSTVKTANTVAVVSDGQIVESGTHDELLEK-NGVYTALVKRQ 711
Score = 21.6 bits (44), Expect(2) = 1e-41
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = +2
Query: 445 LKRINFIYPSRPNV 458
L + F YPSRPN+
Sbjct: 2 LNDVWFSYPSRPNI 43
>TC89340 weakly similar to GP|17427488|emb|CAD14006. PROBABLE COMPOSITE
ATP-BINDING TRANSMEMBRANE ABC TRANSPORTER PROTEIN
{Ralstonia solanacearum}, partial (32%)
Length = 795
Score = 161 bits (407), Expect = 8e-40
Identities = 80/185 (43%), Positives = 118/185 (63%)
Frame = +1
Query: 434 EELMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILR 493
E L G I K + F Y P KD + G + A VG SG GKS++ L+ R
Sbjct: 79 EPLRDCNGNIRWKNVGFNYD--PKNKALKDLDFECKPGSTTAFVGESGGGKSTVFRLMFR 252
Query: 494 FYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIE 553
+Y+ TSG + IDG+D+K + + S+R IG+V Q+ LF S+ N+ Y + EA++ E+ +
Sbjct: 253 YYNTTSGSIEIDGRDVKDLTIDSVRSYIGVVPQDTTLFNESLMYNLRYARPEATDEEIYD 432
Query: 554 AAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALD 613
A + A H+ I + P+ Y TK G+RG+ LSGG++QRVAIAR I+++PKI++LDEATSALD
Sbjct: 433 ACRAASIHDRIMSFPDRYDTKVGERGLKLSGGERQRVAIARTIIKSPKIIMLDEATSALD 612
Query: 614 VESER 618
+E+
Sbjct: 613 AHTEQ 627
Score = 38.5 bits (88), Expect = 0.008
Identities = 19/38 (50%), Positives = 26/38 (68%)
Frame = +1
Query: 3 GRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELI 40
GRT +I+AHRLSTI +AD I V + G+H+EL+
Sbjct: 661 GRTLLIIAHRLSTITHADQIIVPQQWDY**KGHHKELV 774
>TC83850 weakly similar to GP|7300071|gb|AAF55241.1| CG4225 gene product
{Drosophila melanogaster}, partial (23%)
Length = 816
Score = 159 bits (401), Expect = 4e-39
Identities = 79/151 (52%), Positives = 107/151 (70%)
Frame = +2
Query: 468 VPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQE 527
V G A+VG SGSGKS+ + L+ RFYD G V +DG DIK + L SLR+ IG+V Q+
Sbjct: 47 VAPGTKTAIVGESGSGKSTCLKLLFRFYDVQEGAVTVDGHDIKHLKLDSLRRNIGVVPQD 226
Query: 528 PALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQK 587
LF +I N+LY +AS+++V EA K A+ H I P+GY TK G+RG+ LSGG++
Sbjct: 227 TVLFNATIMYNLLYANPKASQADVFEACKAANIHERILNFPDGYETKVGERGLKLSGGER 406
Query: 588 QRVAIARAILRNPKILLLDEATSALDVESER 618
QR+AIARAIL++ +ILLLDEAT++LD +ER
Sbjct: 407 QRIAIARAILKDARILLLDEATASLDSHTER 499
Score = 47.0 bits (110), Expect = 2e-05
Identities = 20/39 (51%), Positives = 27/39 (68%)
Frame = +2
Query: 3 GRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELIS 41
GRTT+ +AHRLSTI +D I V+ +VE G H EL++
Sbjct: 533 GRTTITIAHRLSTITTSDQIVVLHKSKIVERGTHSELLA 649
>TC90210 similar to GP|14715462|dbj|BAB62040. CjMDR1 {Coptis japonica},
partial (26%)
Length = 1158
Score = 152 bits (385), Expect = 3e-37
Identities = 97/298 (32%), Positives = 155/298 (51%), Gaps = 26/298 (8%)
Frame = +2
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLV-------QG 53
M+ RTTV+VAHRLST++NADMIAV+ G +VE G H EL+ +P YS L+ +
Sbjct: 182 MVNRTTVVVAHRLSTVRNADMIAVIHRGKMVEKGTHSELLKDPEGAYSQLIRLQEVNKES 361
Query: 54 QPSPD---------PSLGQSSSLKN-SAEISHAATIGGSF-HSDRSSIGHA----LADEP 98
+ + D S QSS K+ IS ++IG S HS S G +AD
Sbjct: 362 EETTDHHGKRELSAESFRQSSQRKSLQRSISRGSSIGNSSRHSFSVSFGLPTGVNVADPD 541
Query: 99 RSVV----KPRHVSLIRLYSMIGPYWSYGVFGTLAAFTTGALMPLFALGISHALVSYYMD 154
V K + V L RL S+ P + G+LAA G ++P+F + IS + ++Y
Sbjct: 542 LEKVPTKEKEQEVPLRRLASLNKPEIPVLLIGSLAAIANGVILPIFGVLISSVIKTFYEP 721
Query: 155 WDSTCHEVKKIAFLFCGAAIVAITAYSIEHLSFGIMGERLTLRVRGIMLSAILKNEIGWF 214
+D + K A +F + ++ F + G +L R+R + ++ E+GW+
Sbjct: 722 FDEMKKDSKFWAIMFMLLGLASLVVIPARGYFFSVAGCKLIQRIRLLCFEKVVNMEVGWY 901
Query: 215 DDTRNTSSMLSSRLETDATLLKTIVVDRSTILLQNVGLVVTALVIAFILNWRITLVVL 272
D+ N+S + +RL DA ++ +V D +L+QN+ + L+IAFI +W++ L++L
Sbjct: 902 DEPENSSGAVGARLSADAASVRALVGDALGLLVQNLASALAGLIIAFIASWQLALIIL 1075
Score = 81.6 bits (200), Expect = 8e-16
Identities = 40/52 (76%), Positives = 44/52 (83%)
Frame = +2
Query: 568 PEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
P+G T GD G LSGGQKQR+AIARAIL+NP+ILLLDEATSALD ESERV
Sbjct: 2 PQGLDTMVGDHGTQLSGGQKQRIAIARAILKNPRILLLDEATSALDAESERV 157
>AL383696 homologue to PIR|T47671|T4 P-glycoprotein-like - Arabidopsis
thaliana, partial (9%)
Length = 391
Score = 150 bits (380), Expect = 1e-36
Identities = 72/130 (55%), Positives = 104/130 (79%)
Frame = +1
Query: 459 VIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLR 518
++ +F+L V G+++A+VG SGSGKS+IISLI RFYDP +G+V++DG+D+K NL+ LR
Sbjct: 1 LVLSNFSLKVNGGQTVAVVGVSGSGKSTIISLIERFYDPVAGQVLLDGRDLKLYNLRWLR 180
Query: 519 KQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDR 578
+GL+QQEP +F+T+I +NI+Y + ASE+E+ EAA++A+AH+FIS+LP GY T G R
Sbjct: 181 SHLGLIQQEPIIFSTTIRENIIYARHNASEAEMKEAARIANAHHFISSLPHGYDTHVGMR 360
Query: 579 GVLLSGGQKQ 588
GV L+ GQKQ
Sbjct: 361 GVDLTPGQKQ 390
>TC86470 similar to PIR|F84919|F84919 glutathione-conjugate transporter AtMRP4
[imported] - Arabidopsis thaliana, partial (57%)
Length = 3175
Score = 123 bits (308), Expect = 2e-28
Identities = 142/656 (21%), Positives = 292/656 (43%), Gaps = 39/656 (5%)
Frame = +1
Query: 3 GRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQGQPSPDPSLG 62
G+T V+V H++ + N D I V+ G +V++G + +L+ + + LV + +
Sbjct: 676 GKTIVLVTHQVDFLHNVDRIVVMRDGMIVQSGRYNDLLDSGLD-FGVLVAAHETSMELVE 852
Query: 63 QSSSL----KNSAEISHAATIGGSFHSDRSSIGHALA-DEPRS------VVKPRHVSLIR 111
Q +++ N IS +A+I ++R + G + + D+P S +VK +
Sbjct: 853 QGAAVPGENSNKLMISKSASI-----NNRETNGESNSLDQPNSAKGSSKLVKEEERETGK 1017
Query: 112 LYSMIGPYWSYGVFGTLAAFTTGALMPLFALGISHALVSYYMDWDSTCHEVK---KIAFL 168
+ I + FG L L+ + + Y++ ++++ + + F+
Sbjct: 1018 VSFNIYKRYCTEAFGWAGILAVLFLSVLWQASMMAS--DYWLAFETSVERAEVFNPVVFI 1191
Query: 169 FCGAAI--VAITAYSIEHLSFGIMGERLTLRVRGIMLSAILKNEIGWFDDTRNTSSMLSS 226
AAI V++ + S I G + +L++IL + ++D T S + S
Sbjct: 1192 SIYAAITIVSVILIVVRSYSVTIFGLKTAQIFFNQILTSILHAPMSFYDTT--PSGRILS 1365
Query: 227 RLETDATLLKTIVVDRSTILLQNVGLVVTALVIAFILNWRITLVVLATYPLIISGHIGEK 286
R TD T + + ++ V++ ++I +W +++ PL+
Sbjct: 1366 RASTDQTNVDIFIPLFINFVVAMYITVISIVIITCQNSWPTAFLLI---PLVWLN----- 1521
Query: 287 LFMQGFGGNLSKAYLKANMLA--------GEAVSNIRTVAAFCAEEKVIDLYADELVEPS 338
++ +G+ + S+ + + + E++S + TV AF ++K L + V +
Sbjct: 1522 IWYRGYFLSTSRELTRLDSITKAPVIVHFSESISGVMTVRAF-RKQKEFRLENFKRVNSN 1698
Query: 339 KRSFKRGQIAGIFYGI------SQFFIFSSYGLALWYGSVLLEKELASFKSIMKSFMVLI 392
R + + G S F S+ + L +++ + + S S ++
Sbjct: 1699 LRMDFHNYSSNAWLGFRLELLGSLVFCLSALFMILLPSNIIKPENVGLSLSYGLSLNSVL 1878
Query: 393 VTALAMGETLALAPDLLKGNQMVSSIFDMIDRKSGIIHDVGEELMTV--------EGMIE 444
A+ M + N+MVS + I + S I + + +G ++
Sbjct: 1879 FWAIYMSCFIE--------NKMVS--VERIKQFSNIPSEAAWNIKDRSPPPNWPGQGHVD 2028
Query: 445 LKRINFIYPSRPNV-VIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVM 503
+K + Y RPN ++ K L + G+ + +VG +GSGKS++I + R +PT GK++
Sbjct: 2029 IKDLQVRY--RPNTPLVLKGITLSISGGEKVGVVGRTGSGKSTLIQVFFRLVEPTGGKII 2202
Query: 504 IDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNF 563
IDG DI + L LR + G++ QEP LF ++ NI + ++ E+ ++ +
Sbjct: 2203 IDGIDICALGLHDLRSRFGIIPQEPVLFEGTVRSNI-DPTGQYTDDEIWKSLDRCQLKDT 2379
Query: 564 ISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
+++ PE + D G S GQ+Q + + R +L+ ++L +DEAT+++D +++ V
Sbjct: 2380 VASKPEKLDSLVVDNGDNWSVGQRQLLCLGRVMLKQSRLLFMDEATASVDSQTDAV 2547
Score = 75.5 bits (184), Expect = 6e-14
Identities = 52/155 (33%), Positives = 80/155 (51%)
Frame = +1
Query: 462 KDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQI 521
K+ NL V G+ A+VG GSGKSS+++ IL SGKV + G
Sbjct: 211 KNINLKVNKGELTAIVGTVGSGKSSLLASILGEMHRNSGKVQVCGST------------- 351
Query: 522 GLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVL 581
V Q + +I +NIL+G + + E ++ + + G T+ G+RG+
Sbjct: 352 AYVAQTSWIQNGTIEENILFGLP-MNRQKYNEIIRVCCLEKDLEMMEYGDQTEIGERGIN 528
Query: 582 LSGGQKQRVAIARAILRNPKILLLDEATSALDVES 616
LSGGQKQR+ +ARA+ ++ I LLD+ SA+D +
Sbjct: 529 LSGGQKQRIQLARAVYQDCDIYLLDDVFSAVDAHT 633
>AW587222 weakly similar to GP|5456699|gb| ATP-binding cassette multidrug
transport protein ATRC {Emericella nidulans}, partial
(3%)
Length = 634
Score = 121 bits (304), Expect = 7e-28
Identities = 64/198 (32%), Positives = 114/198 (57%), Gaps = 2/198 (1%)
Frame = +2
Query: 290 QGFGGNLSKAYLKANMLAGEAVSNIRTVAAFCAEEKVIDLYADELVEPSKRSFKRGQIAG 349
+GF G+ S + LA E+ +NIRT+A+FC EE+V++ L P K+ K G
Sbjct: 41 KGFSGDYSATHSDLVALASESTTNIRTIASFCHEEQVLEKAKTYLDIPKKKYRKESIKYG 220
Query: 350 IFYGISQFFIFSSYGLALWYGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLL 409
I G S ++ +ALWY ++L+++ ASF++ ++++ + +T ++ E L P ++
Sbjct: 221 IIQGFSLCLWNIAHAVALWYTTILVDRRQASFENGIRAYQIFSLTVPSITELYTLIPTVI 400
Query: 410 KGNQMVSSIFDMIDRKSGIIHDVGEELM--TVEGMIELKRINFIYPSRPNVVIFKDFNLI 467
M++ F +DRK+ I D+ ++ ++G +E + +NF YP RP V + +F+L
Sbjct: 401 TAINMLTPAFKTLDRKTEIEPDIPDDSQPDRIQGNVEFENVNFKYPLRPTVTVLDNFSLQ 580
Query: 468 VPSGKSLALVGHSGSGKS 485
+ +G +A VG SG+GKS
Sbjct: 581 IEAGSKVAFVGPSGAGKS 634
>BM780003 weakly similar to PIR|T06165|T0 multidrug resistance protein 1
homolog - barley, partial (12%)
Length = 665
Score = 119 bits (299), Expect = 3e-27
Identities = 77/222 (34%), Positives = 115/222 (51%), Gaps = 13/222 (5%)
Frame = +2
Query: 8 IVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSL--VQGQPSPDPSLGQSS 65
+VAH+LSTI+NAD+IAVV G ++E+G H ELI+ PN Y+ L +Q Q S + Q+
Sbjct: 2 VVAHKLSTIRNADLIAVVSNGCIIESGTHNELINTPNGHYAKLAKLQTQLSINDDQDQNQ 181
Query: 66 SLKNSAEISHAATIGGSFHSDRSSIGHALADEPR------SVVKPRH----VSLIRLYSM 115
+ + S +S A + G + RSS L P S V H S RL +
Sbjct: 182 NQEQSILLSAARSSAGRTSTARSS-PLILPKSPLPNDIIISQVSHSHSHPSPSFSRLLFL 358
Query: 116 IGPYWSYGVFGTLAAFTTGALMPLFALGISHALVSYY-MDWDSTCHEVKKIAFLFCGAAI 174
P W G+ GTL+A G++ PL+AL I + +++ H + + +F ++
Sbjct: 359 NSPEWKQGLIGTLSAIANGSIQPLYALTIGGMISAFFAKSHQEMKHRIMNYSLIFTALSV 538
Query: 175 VAITAYSIEHLSFGIMGERLTLRVRGIMLSAILKNEIGWFDD 216
+IT +H +F MG +LT R+R ML IL E WFD+
Sbjct: 539 ASITLNLFQHYNFAYMGAKLTKRIRLCMLEKILTFETAWFDE 664
>TC77935 similar to PIR|E85023|E85023 probable P-glycoprotein-like protein
[imported] - Arabidopsis thaliana, partial (12%)
Length = 869
Score = 115 bits (288), Expect = 5e-26
Identities = 59/91 (64%), Positives = 75/91 (81%), Gaps = 1/91 (1%)
Frame = +2
Query: 530 LFATSIYKNILYGKE-EASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQ 588
LF ++ NI YGK +A+E+E++ AA+LA+AH FI +L +GY T G+RG+ LSGGQKQ
Sbjct: 65 LFNDTVRANIAYGKGGDATEAEIVAAAELANAHQFIGSLQKGYDTIVGERGIQLSGGQKQ 244
Query: 589 RVAIARAILRNPKILLLDEATSALDVESERV 619
RVAIARAI++NPKILLLDEATSALD ESE+V
Sbjct: 245 RVAIARAIVKNPKILLLDEATSALDAESEKV 337
Score = 58.5 bits (140), Expect = 7e-09
Identities = 30/51 (58%), Positives = 37/51 (71%)
Frame = +2
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLV 51
M+ RTT+IVAHRLSTIK AD+IAVV+ G + E G HE L+ Y+SLV
Sbjct: 362 MVERTTIIVAHRLSTIKGADLIAVVKNGVIAEKGKHEALLHKGGD-YASLV 511
>TC78391 similar to PIR|F84919|F84919 glutathione-conjugate transporter
AtMRP4 [imported] - Arabidopsis thaliana, partial (17%)
Length = 1265
Score = 110 bits (276), Expect = 1e-24
Identities = 65/184 (35%), Positives = 110/184 (59%), Gaps = 5/184 (2%)
Frame = +2
Query: 441 GMIELKRINFIY-PSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTS 499
G IEL + Y P+ P ++ K +L + G+ + +VG +GSGKS++I ++ R +P++
Sbjct: 86 GTIELNNLQVRYRPTTP--LVLKGVSLTIEGGEKVGVVGRTGSGKSTLIQVLFRLIEPSA 259
Query: 500 GKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNI----LYGKEEASESEVIEAA 555
GK++IDG +I + L LR + G++ QEP LF ++ NI LY +EE +S +E
Sbjct: 260 GKIIIDGINISNVGLHDLRSRFGIIPQEPVLFQGTVRTNIDPLGLYSEEEIWKS--LERC 433
Query: 556 KLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVE 615
+L +A ++A PE D G S GQ+Q + + R +L+ KIL +DEAT+++D +
Sbjct: 434 QLKEA---VAAKPEKLDALVVDGGDNWSVGQRQLLCLGRIMLKRSKILFMDEATASVDSQ 604
Query: 616 SERV 619
++ V
Sbjct: 605 TDAV 616
>BM813979 homologue to PIR|E85023|E85 probable P-glycoprotein-like protein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 434
Score = 108 bits (271), Expect = 5e-24
Identities = 55/97 (56%), Positives = 77/97 (78%), Gaps = 1/97 (1%)
Frame = +3
Query: 472 KSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALF 531
+++ALVG SGSGKS++I+L+ RFYDP SG++ +DG +I+++ LK LR+Q+GLV QEP LF
Sbjct: 135 QTVALVGESGSGKSTVIALLQRFYDPDSGEITLDGIEIRQLQLKWLRQQMGLVSQEPVLF 314
Query: 532 ATSIYKNILYGKEE-ASESEVIEAAKLADAHNFISAL 567
+I NI YGK A+E+E+I AA+LA+AH FIS L
Sbjct: 315 NDTIRANIAYGKGGIATEAEIIAAAELANAHRFISGL 425
>TC79748 weakly similar to GP|9294577|dbj|BAB02858.1 multidrug resistance
p-glycoprotein; ABC transporter-like protein
{Arabidopsis thaliana}, partial (12%)
Length = 483
Score = 107 bits (267), Expect = 1e-23
Identities = 56/158 (35%), Positives = 94/158 (59%), Gaps = 2/158 (1%)
Frame = +2
Query: 118 PYWSYGVFGTLAAFTTGALMPLFALGISHALVSYYMDWDSTCHEVKK--IAFLFCGAAIV 175
P W GV G L A +GA+ P+ A + L+S Y + D++ + K +A +F G +
Sbjct: 8 PEWGRGVLGVLGAIGSGAVQPINAYCVG-LLISVYFEPDTSKMKSKARALALVFLGIGVF 184
Query: 176 AITAYSIEHLSFGIMGERLTLRVRGIMLSAILKNEIGWFDDTRNTSSMLSSRLETDATLL 235
++H +F +MGERLT R+R ML ++ EIGWFD NTS+ + +RL ++A L+
Sbjct: 185 NFFTSILQHYNFAVMGERLTKRIREKMLEKLMSFEIGWFDHEDNTSAAICARLASEANLV 364
Query: 236 KTIVVDRSTILLQNVGLVVTALVIAFILNWRITLVVLA 273
+++V DR ++L Q + + A + +L WR++LV++A
Sbjct: 365 RSLVGDRMSLLAQAIFGSIFAYTVGLVLTWRLSLVMIA 478
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,600,713
Number of Sequences: 36976
Number of extensions: 189596
Number of successful extensions: 1222
Number of sequences better than 10.0: 125
Number of HSP's better than 10.0 without gapping: 1163
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1191
length of query: 619
length of database: 9,014,727
effective HSP length: 102
effective length of query: 517
effective length of database: 5,243,175
effective search space: 2710721475
effective search space used: 2710721475
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149574.1


