

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.6 + phase: 0
(160 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
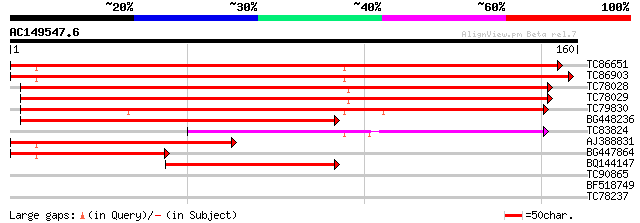
Score E
Sequences producing significant alignments: (bits) Value
TC86651 similar to PIR|F84555|F84555 similar to prolyl 4-hydroxy... 248 6e-67
TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylas... 174 1e-44
TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Ar... 162 7e-41
TC78029 similar to GP|21593296|gb|AAM65245.1 prolyl 4-hydroxylas... 152 4e-38
TC79830 similar to PIR|G84861|G84861 hypothetical protein At2g43... 150 2e-37
BG448236 similar to GP|21537370|gb putative prolyl 4-hydroxylase... 138 1e-33
TC83824 similar to GP|17381226|gb|AAL36425.1 unknown protein {Ar... 102 5e-23
AJ388831 weakly similar to GP|10177121|dbj prolyl 4-hydroxylase ... 80 5e-16
BG447864 weakly similar to GP|10177121|db prolyl 4-hydroxylase ... 61 2e-10
BQ144147 44 2e-05
TC90865 homologue to GP|9049326|dbj|BAA99336.1 orf129b {Beta vul... 29 0.72
BF518749 29 0.72
TC78237 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {... 26 7.9
>TC86651 similar to PIR|F84555|F84555 similar to prolyl 4-hydroxylase alpha
subunit [imported] - Arabidopsis thaliana, partial (85%)
Length = 1329
Score = 248 bits (634), Expect = 6e-67
Identities = 120/178 (67%), Positives = 138/178 (77%), Gaps = 22/178 (12%)
Frame = +2
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
MHKSTV D ETGKS D+ RTSSGTF+ RG DKI+RNIE++IADFTFIPVE+GE + +LH
Sbjct: 446 MHKSTVVDSETGKSKDSRVRTSSGTFLARGRDKIVRNIEKKIADFTFIPVEHGEGLQVLH 625
Query: 60 YEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL---------------------PWWNE 98
YEVGQKYEPH D+F DE NTKNGG+R+AT+LMYL PW+NE
Sbjct: 626 YEVGQKYEPHYDYFLDEFNTKNGGQRIATVLMYLTDVEEGGETVFPAAKGNFSNVPWYNE 805
Query: 99 LSDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRVGKW 156
LSDCGKKGLSIKPK GDALLFWSMKPD TLD S+HG CPVIKG+KWS TKW+RV ++
Sbjct: 806 LSDCGKKGLSIKPKRGDALLFWSMKPDATLDASSLHGGCPVIKGNKWSSTKWIRVNEY 979
>TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylase putative
{Arabidopsis thaliana}, partial (68%)
Length = 1310
Score = 174 bits (441), Expect = 1e-44
Identities = 88/180 (48%), Positives = 117/180 (64%), Gaps = 21/180 (11%)
Frame = +3
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
+ KS V D+E+GKS+ + RTSSG F+N+ D+I+ IE RIA +TF+PVENGES+ +LH
Sbjct: 402 LEKSMVADNESGKSIQSEVRTSSGMFLNKQQDEIVSGIEARIAAWTFLPVENGESMQVLH 581
Query: 60 YEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL--------------------PWWNEL 99
Y G+KYEPH DFF D+ N + GG RVAT+LMYL P
Sbjct: 582 YMNGEKYEPHFDFFHDKANQRLGGHRVATVLMYLSNVEKGGETIFPHAEGKLSQPKDESW 761
Query: 100 SDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRVGKWSIP 159
S+C KG ++KP+ GDALLF+S+ D T D S+HG+CPVI+G+KWS TKW+ V + P
Sbjct: 762 SECAHKGYAVKPRKGDALLFFSLHLDATTDSKSLHGSCPVIEGEKWSATKWIHVADFEKP 941
>TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Arabidopsis
thaliana}, partial (86%)
Length = 1217
Score = 162 bits (409), Expect = 7e-41
Identities = 78/175 (44%), Positives = 110/175 (62%), Gaps = 25/175 (14%)
Frame = +1
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVG 63
+ D+ +G+S + RTSSG FI++ D I+ IE +I+ +TF+P ENGE + +L YE G
Sbjct: 235 AVADNLSGESKLSEVRTSSGMFISKNKDAIVSGIEDKISSWTFLPKENGEDIQVLRYEHG 414
Query: 64 QKYEPHPDFFTDEINTKNGGERVATMLMYLP-------------------------WWNE 98
QKY+PH D+F D++N GG RVAT+LMYL +
Sbjct: 415 QKYDPHYDYFADKVNIARGGHRVATVLMYLTNVTKGGETVFPNAELQESPRHKLSETDED 594
Query: 99 LSDCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRV 153
LS+CGKKG+++KP+ GDALLF+S+ P+ D LS+H CPVI+G+KWS TKW+ V
Sbjct: 595 LSECGKKGVAVKPRRGDALLFFSLHPNAIPDTLSLHAGCPVIEGEKWSATKWIHV 759
>TC78029 similar to GP|21593296|gb|AAM65245.1 prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(89%)
Length = 1126
Score = 152 bits (385), Expect = 4e-38
Identities = 74/173 (42%), Positives = 107/173 (61%), Gaps = 23/173 (13%)
Frame = +3
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVG 63
+ D+ +G S + RTSSG FI++ D I+ IE RI+ +TF+P ENGE + +L YE G
Sbjct: 327 AVADNLSGDSQLSDVRTSSGMFISKNKDPIVSGIEDRISAWTFLPKENGEDIQVLRYEHG 506
Query: 64 QKYEPHPDFFTDEINTKNGGERVATMLMYLP-----------------------WWNELS 100
QKY+PH D+F D++N GG R+AT+LMYL ++LS
Sbjct: 507 QKYDPHYDYFADKVNIVQGGHRLATVLMYLTNVTKGGETVFPEAEEPPRRRGSKKSSDLS 686
Query: 101 DCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMRV 153
+C KKG+++KP+ GDALLF+S+ + D S+H CPV++G+KWS TKW+ V
Sbjct: 687 ECAKKGIAVKPRRGDALLFFSLDTNAIPDTNSLHAGCPVLEGEKWSATKWIHV 845
>TC79830 similar to PIR|G84861|G84861 hypothetical protein At2g43080
[imported] - Arabidopsis thaliana, partial (87%)
Length = 1130
Score = 150 bits (380), Expect = 2e-37
Identities = 79/166 (47%), Positives = 104/166 (62%), Gaps = 17/166 (10%)
Frame = +1
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDK--ILRNIEQRIADFTFIPVENGESVNILHYE 61
+ VD TGK + + RTSSG F++ K ++ IE+RI+ ++ IP+ENGE + +L YE
Sbjct: 424 TVVDANTGKGIKSDVRTSSGMFLSHEERKYPMIHAIEKRISVYSQIPIENGELMQVLRYE 603
Query: 62 VGQKYEPHPDFFTDEINTKNGGERVATMLMYL-------------PWWNELSDCGK--KG 106
Q Y PH D+F+D N K GG+R+ATMLMYL +E S GK KG
Sbjct: 604 KNQYYRPHHDYFSDTFNLKRGGQRIATMLMYLGDNVEGGETHFPSAGSDECSCGGKLTKG 783
Query: 107 LSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWMR 152
L +KP G+A+LFWSM DG DP S+HG CPV+ G+KWS TKWMR
Sbjct: 784 LCVKPVKGNAVLFWSMGLDGQSDPDSVHGGCPVLAGEKWSATKWMR 921
>BG448236 similar to GP|21537370|gb putative prolyl 4-hydroxylase alpha
subunit {Arabidopsis thaliana}, partial (52%)
Length = 682
Score = 138 bits (347), Expect = 1e-33
Identities = 63/90 (70%), Positives = 76/90 (84%)
Frame = +3
Query: 4 STVDDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILHYEVG 63
S VD +TGKS ++ RTSSG F+ RG DKI++NIE+RIADFTFIPVENGE + +LHY VG
Sbjct: 309 SVVDSKTGKSTESRVRTSSGMFLKRGKDKIIQNIERRIADFTFIPVENGEGLQVLHYGVG 488
Query: 64 QKYEPHPDFFTDEINTKNGGERVATMLMYL 93
+KYEPH D+F DE NTKNGG+RVAT+LMYL
Sbjct: 489 EKYEPHYDYFLDEFNTKNGGQRVATVLMYL 578
>TC83824 similar to GP|17381226|gb|AAL36425.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 680
Score = 102 bits (255), Expect = 5e-23
Identities = 54/121 (44%), Positives = 73/121 (59%), Gaps = 19/121 (15%)
Frame = +3
Query: 51 NGESVNILHYEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL-----------PWWNEL 99
+GE+ NIL YEVGQ+Y H D F + +RVA+ L+YL P+ N L
Sbjct: 3 HGEAFNILRYEVGQRYNSHYDAFNPDEYGPQKSQRVASFLLYLTDVEEGGETMFPFENGL 182
Query: 100 S--------DCGKKGLSIKPKMGDALLFWSMKPDGTLDPLSMHGACPVIKGDKWSCTKWM 151
+ DC GL +KP+ GD LLF+S+ P+GT+D S+HG+CPVIKG+KW TKW+
Sbjct: 183 NMDGTYGYEDC--VGLRVKPRQGDGLLFYSLLPNGTIDQTSLHGSCPVIKGEKWVATKWI 356
Query: 152 R 152
R
Sbjct: 357 R 359
>AJ388831 weakly similar to GP|10177121|dbj prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(33%)
Length = 505
Score = 79.7 bits (195), Expect = 5e-16
Identities = 39/65 (60%), Positives = 51/65 (78%), Gaps = 1/65 (1%)
Frame = +3
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFTFIPVENGESVNILH 59
MHKS V D+ETG VD+S RTSSG F+ RG D+I++NIE+RIADFTFIP E+GE+ N L
Sbjct: 291 MHKSAVIDEETGNGVDSSERTSSGAFLKRGSDRIVKNIERRIADFTFIPXEHGENFNGLX 470
Query: 60 YEVGQ 64
++ +
Sbjct: 471 MKLAK 485
>BG447864 weakly similar to GP|10177121|db prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(50%)
Length = 639
Score = 61.2 bits (147), Expect = 2e-10
Identities = 31/46 (67%), Positives = 35/46 (75%), Gaps = 1/46 (2%)
Frame = +3
Query: 1 MHKSTV-DDETGKSVDNSARTSSGTFINRGHDKILRNIEQRIADFT 45
MHKSTV D ETGKS D+ RT SGTF+ RG D I RNI ++IADFT
Sbjct: 501 MHKSTVVDSETGKSKDSXVRTXSGTFLARGRDXIXRNIXKKIADFT 638
>BQ144147
Length = 729
Score = 44.3 bits (103), Expect = 2e-05
Identities = 18/49 (36%), Positives = 32/49 (64%)
Frame = +2
Query: 45 TFIPVENGESVNILHYEVGQKYEPHPDFFTDEINTKNGGERVATMLMYL 93
T +P+ENG++++I Y+ G + P+ ++ T +IN GG R T+LM +
Sbjct: 92 TSLPIENGDNLHIWRYKHGHNHNPNDNYSTHKINIVQGGHRPPTVLMLI 238
>TC90865 homologue to GP|9049326|dbj|BAA99336.1 orf129b {Beta vulgaris},
partial (16%)
Length = 940
Score = 29.3 bits (64), Expect = 0.72
Identities = 13/32 (40%), Positives = 20/32 (61%)
Frame = +3
Query: 88 TMLMYLPWWNELSDCGKKGLSIKPKMGDALLF 119
T+ +Y+ W+ S C K L I+P +GD +LF
Sbjct: 768 TLQLYISSWS--SSCPIKSLQIEPNVGDEILF 857
>BF518749
Length = 416
Score = 29.3 bits (64), Expect = 0.72
Identities = 10/29 (34%), Positives = 21/29 (71%)
Frame = +1
Query: 31 DKILRNIEQRIADFTFIPVENGESVNILH 59
D I++ IE+R++ +TF+ EN + + ++H
Sbjct: 328 DDIVKRIEERLSVWTFLSKENSKPLQVMH 414
>TC78237 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {Arabidopsis
thaliana}, partial (59%)
Length = 1166
Score = 25.8 bits (55), Expect = 7.9
Identities = 11/34 (32%), Positives = 16/34 (46%)
Frame = +2
Query: 72 FFTDEINTKNGGERVATMLMYLPWWNELSDCGKK 105
FF ++T R + +PWW S CG+K
Sbjct: 530 FFVRIVSTTTSSSR-SYKFECVPWWRPRSSCGRK 628
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.317 0.136 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,723,904
Number of Sequences: 36976
Number of extensions: 80804
Number of successful extensions: 360
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 351
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 355
length of query: 160
length of database: 9,014,727
effective HSP length: 88
effective length of query: 72
effective length of database: 5,760,839
effective search space: 414780408
effective search space used: 414780408
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149547.6


