

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149546.1 - phase: 0
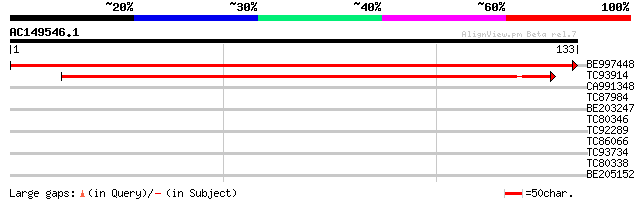
(133 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE997448 similar to GP|15208467|gb| kinesin heavy chain {Zea may... 266 1e-72
TC93914 weakly similar to GP|7270878|emb|CAB80558.1 kinesin like... 119 5e-28
CA991348 similar to PIR|D84685|D84 hypothetical protein At2g2848... 28 1.1
TC87984 weakly similar to GP|10177368|dbj|BAB10659. contains sim... 27 3.1
BE203247 26 5.2
TC80346 weakly similar to GP|20453181|gb|AAM19831.1 AT4g10060/T5... 26 5.2
TC92289 weakly similar to GP|20197927|gb|AAD24632.2 hypothetical... 25 6.8
TC86066 PIR|S46316|S46316 aspartate transaminase (EC 2.6.1.1) - ... 25 6.8
TC93734 similar to PIR|T46027|T46027 hypothetical protein T10K17... 25 8.9
TC80338 weakly similar to GP|20127018|gb|AAM10936.1 putative bHL... 25 8.9
BE205152 similar to GP|19347965|gb putative helicase {Arabidopsi... 25 8.9
>BE997448 similar to GP|15208467|gb| kinesin heavy chain {Zea mays}, partial
(9%)
Length = 818
Score = 266 bits (681), Expect = 1e-72
Identities = 133/133 (100%), Positives = 133/133 (100%)
Frame = +1
Query: 1 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 60
MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG
Sbjct: 253 MEVELRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWG 432
Query: 61 ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRK 120
ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRK
Sbjct: 433 ISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRK 612
Query: 121 KSFGWTSSMKHIL 133
KSFGWTSSMKHIL
Sbjct: 613 KSFGWTSSMKHIL 651
>TC93914 weakly similar to GP|7270878|emb|CAB80558.1 kinesin like protein
{Arabidopsis thaliana}, partial (11%)
Length = 728
Score = 119 bits (297), Expect = 5e-28
Identities = 59/116 (50%), Positives = 84/116 (71%)
Frame = +1
Query: 13 DNQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQL 72
+NQ +D +T+T SS R LRRE++ML + M+++LS+ ER ++ +WGI ++SK RR+QL
Sbjct: 40 ENQSEKDSQTITLTSSVRALRREKEMLMKLMRKRLSEEERKRLFNEWGIGLNSKRRRMQL 219
Query: 73 AHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRKKSFGWTSS 128
A RLW TD+NHV +SA IVA+LV E +A KEMFGL+F P+ R +S+ W SS
Sbjct: 220 ADRLWCNTDMNHVMQSAAIVARLVRFSEQGRALKEMFGLSFTPQLTR-RSYSWKSS 384
>CA991348 similar to PIR|D84685|D84 hypothetical protein At2g28480
[imported] - Arabidopsis thaliana, partial (52%)
Length = 658
Score = 28.1 bits (61), Expect = 1.1
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Frame = +1
Query: 5 LRRLSYLKDNQILEDGRTLTPESSKRYLRRERQMLSR--QMQRKLSKSER 52
LR +SY K N +++DG+T R+++ + +MQRK K +R
Sbjct: 13 LREMSYGKVNLVIKDGKTKFETHEVEAPRKDKWKTKKRLKMQRKREKEKR 162
>TC87984 weakly similar to GP|10177368|dbj|BAB10659. contains similarity to
heat shock transcription factor HSF30~gene_id:K16L22.13,
partial (5%)
Length = 696
Score = 26.6 bits (57), Expect = 3.1
Identities = 19/63 (30%), Positives = 32/63 (50%)
Frame = -3
Query: 30 RYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINHVRESA 89
R LRR R +L++ + K S ++ WG+ +S K + + HRL H+RE+
Sbjct: 343 RDLRRVRTVLTKTEKIKNCSSLSNS*I--WGLELSQKSKASRFFHRL------RHLRETR 188
Query: 90 TIV 92
+V
Sbjct: 187 GVV 179
>BE203247
Length = 475
Score = 25.8 bits (55), Expect = 5.2
Identities = 17/67 (25%), Positives = 31/67 (45%), Gaps = 1/67 (1%)
Frame = -3
Query: 23 LTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRR-LQLAHRLWSETD 81
+ P S + ++ R + +++MQ RDN+ + W + RR +L H +
Sbjct: 200 IMPPSLLQQIKLARVLATKRMQNVTPTKIRDNLVIVWNLMHGYTMRRCTKLCH--GCPVE 27
Query: 82 INHVRES 88
I HV +S
Sbjct: 26 IQHVIQS 6
>TC80346 weakly similar to GP|20453181|gb|AAM19831.1 AT4g10060/T5L19_190
{Arabidopsis thaliana}, partial (16%)
Length = 801
Score = 25.8 bits (55), Expect = 5.2
Identities = 16/58 (27%), Positives = 27/58 (45%)
Frame = +3
Query: 44 QRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEP 101
QRKL+ N+ ++ +S + +RLW HVRE A+ +G ++P
Sbjct: 318 QRKLNNHANSNVPSEFTLSFKEMIHLAPIGYRLW-----RHVREEAS--KGRIGMIDP 470
>TC92289 weakly similar to GP|20197927|gb|AAD24632.2 hypothetical protein
{Arabidopsis thaliana}, partial (7%)
Length = 1055
Score = 25.4 bits (54), Expect = 6.8
Identities = 13/58 (22%), Positives = 27/58 (46%)
Frame = +3
Query: 46 KLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQ 103
+L + E+ N +L+W + + L +A R W T + + A +++ T D+
Sbjct: 693 RLKQQEQFNRHLEWHATKEREQNGLTVASRRWYVTSDDWIASKAECLSESEFTDSVDE 866
>TC86066 PIR|S46316|S46316 aspartate transaminase (EC 2.6.1.1) - alfalfa,
partial (76%)
Length = 1359
Score = 25.4 bits (54), Expect = 6.8
Identities = 14/27 (51%), Positives = 18/27 (65%)
Frame = +3
Query: 47 LSKSERDNMYLKWGISMSSKHRRLQLA 73
L+KS+ DNM KW I M +K R+ LA
Sbjct: 900 LNKSQSDNMTNKWHIYM-TKDGRISLA 977
>TC93734 similar to PIR|T46027|T46027 hypothetical protein T10K17.260 -
Arabidopsis thaliana, partial (10%)
Length = 686
Score = 25.0 bits (53), Expect = 8.9
Identities = 14/58 (24%), Positives = 31/58 (53%)
Frame = +1
Query: 14 NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQ 71
NQ+ + L+P++ + LR ++QML Q ++ + S + + L+ + + + LQ
Sbjct: 268 NQLHKFWSELSPQARQELLRIDKQMLFEQARKNMYCSRCNGLLLEGFLQIVMYGKSLQ 441
>TC80338 weakly similar to GP|20127018|gb|AAM10936.1 putative bHLH
transcription factor {Arabidopsis thaliana}, partial
(15%)
Length = 1128
Score = 25.0 bits (53), Expect = 8.9
Identities = 8/23 (34%), Positives = 14/23 (60%)
Frame = -2
Query: 74 HRLWSETDINHVRESATIVAKLV 96
H LW+ D +H+R S I+ ++
Sbjct: 1067 HTLWTRIDNSHLRSSILIIRSII 999
>BE205152 similar to GP|19347965|gb putative helicase {Arabidopsis thaliana},
partial (27%)
Length = 626
Score = 25.0 bits (53), Expect = 8.9
Identities = 15/52 (28%), Positives = 23/52 (43%)
Frame = +3
Query: 59 WGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFG 110
W S +K R ++AH LW ++ ++ E+ T V D E FG
Sbjct: 246 WVFSTYAKFRPSKIAH*LWEASNT*YIIETLTSRKSSCFVVCSDDQDAEYFG 401
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,541,687
Number of Sequences: 36976
Number of extensions: 35621
Number of successful extensions: 211
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 209
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 211
length of query: 133
length of database: 9,014,727
effective HSP length: 86
effective length of query: 47
effective length of database: 5,834,791
effective search space: 274235177
effective search space used: 274235177
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC149546.1


