

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149496.4 - phase: 0
(475 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
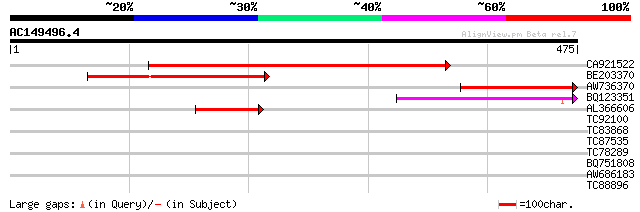
Score E
Sequences producing significant alignments: (bits) Value
CA921522 262 2e-70
BE203370 193 1e-49
AW736370 169 2e-42
BQ123351 110 9e-25
AL366606 75 5e-14
TC92100 similar to GP|15290106|dbj|BAB63798. contains EST D24276... 37 0.022
TC83868 31 0.91
TC87535 similar to GP|19310542|gb|AAL85004.1 unknown protein {Ar... 31 1.2
TC78289 similar to GP|15451044|gb|AAK96793.1 Unknown protein {Ar... 31 1.2
BQ751808 similar to GP|7769858|gb|A F12M16.20 {Arabidopsis thali... 30 2.6
AW686183 similar to GP|20197841|gb| hypothetical protein {Arabid... 29 4.5
TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown prot... 28 5.9
>CA921522
Length = 767
Score = 262 bits (669), Expect = 2e-70
Identities = 128/253 (50%), Positives = 165/253 (64%)
Frame = +3
Query: 117 SDLVHPHFTIKNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVD 176
+D V H + G + FLY++ + F K AF SI LL+YGL LFP++DNFVD
Sbjct: 6 TDEVKTHLITRGKFLGFSTDFLYERTTFFDKMGVAYAFNSILALLVYGLVLFPSLDNFVD 185
Query: 177 LNAIKIFLAKNPVPTLLANTYHSIHHRNIREGGLIVCCAPLLYRWYASHLTKSVFSKENS 236
+ AI+IFL++NPVPTLL +TY SIH R G I+CCA LLYRW SHL ++ N
Sbjct: 186 IKAIQIFLSRNPVPTLLGDTYLSIHRRTQAGRGTILCCAQLLYRWITSHLPRTPRFTTNP 365
Query: 237 GKASWSERIMPLTPADIVWVHAGTNTAGIIGSCGEFENVPLIGTCGGITYNPTLAMRQYG 296
WS+R+M LTPA++VW + II SCG+F NVPL+G GGI+YNPTLA Q+G
Sbjct: 366 ENLLWSKRLMSLTPAEVVWYDRVYDKGTIIDSCGKFANVPLLGMEGGISYNPTLARHQFG 545
Query: 297 YPMKGRPDSLSLSNEFYLNKNDHTNLRMRFVQAWHSIRKFDGIQLGRKQSFAHESYTQWV 356
YPM+ +P S+ L N +YLN +D T +R V+AWH+IR+ D QLG+K SYTQWV
Sbjct: 546 YPMERKPLSIYLENVYYLNADDSTGMREHVVRAWHTIRRRDKDQLGKKTGAISSSYTQWV 725
Query: 357 IDRATAFGMPYTL 369
IDRA GMPY +
Sbjct: 726 IDRAVQIGMPYKI 764
>BE203370
Length = 454
Score = 193 bits (491), Expect = 1e-49
Identities = 98/152 (64%), Positives = 113/152 (73%)
Frame = +1
Query: 66 HCFTFPDYQLVPTLEEYSFWVGLPVSEGEPFNGLEPSPKNATIAEALHLKPSDLVHPHFT 125
HCFTFPDYQL PTLEEYS+ VG PV + PF G EP PK ATIA+ALHL+ S L+ T
Sbjct: 1 HCFTFPDYQLAPTLEEYSYLVG*PVLDKVPFTGFEPIPKAATIADALHLETS-LIKAKLT 177
Query: 126 IKNNLQGLTAKFLYQKASDFVKAKKTNAFESIFTLLIYGLFLFPNMDNFVDLNAIKIFLA 185
IK NL L KFLYQ+ASDF K +AF SI LLIYGL LFPN+DN+VD++AI++FL
Sbjct: 178 IKGNLPSLPTKFLYQQASDFAKVDNVDAFYSILALLIYGLVLFPNVDNYVDIHAIQMFLT 357
Query: 186 KNPVPTLLANTYHSIHHRNIREGGLIVCCAPL 217
KNPVPTLLA+ YHSIH RN G I+ CAPL
Sbjct: 358 KNPVPTLLADMYHSIHDRNQVGRGAILGCAPL 453
>AW736370
Length = 488
Score = 169 bits (429), Expect = 2e-42
Identities = 83/98 (84%), Positives = 88/98 (89%)
Frame = -2
Query: 378 PEIPLPLLPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGLLEQEAYDSRQKDVI 437
P PLP+ T++EYQERL EAEREIH WKRE QKKD+DYETVMGLLEQEAYDSR KDVI
Sbjct: 478 PAPPLPMTFDTEKEYQERLTEAEREIHRWKREYQKKDQDYETVMGLLEQEAYDSR*KDVI 299
Query: 438 IAKLNERIKEKDAALDRIPGRKKKRMDLFDGPHSDFED 475
IAKLNERIKE DAALDRIPGRKK+RMDLFDGPHSDFED
Sbjct: 298 IAKLNERIKENDAALDRIPGRKKRRMDLFDGPHSDFED 185
>BQ123351
Length = 725
Score = 110 bits (276), Expect = 9e-25
Identities = 60/153 (39%), Positives = 92/153 (59%), Gaps = 2/153 (1%)
Frame = +2
Query: 325 RFVQAWHSIRKFDGIQLGRKQSFAHESYTQWVIDRATAFGMPYTLPRFLSSTVPEIPLPL 384
+F+QAW ++ K + QLGR+ +ESYT+WVIDRA G+P+ L R SST LP+
Sbjct: 5 KFIQAWRAVHKREKGQLGRRS*LVYESYTKWVIDRAAKNGIPFPLQRLPSSTTLSPSLPM 184
Query: 385 LPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGLLEQEAYDSRQKDVIIAKLNER 444
+TKEE Q LAE RE W+ + + + T+ G +EQ+ ++ + + + ++
Sbjct: 185 TSKTKEEAQYLLAEMTREKDTWRIRYMEAESEIGTLRGQVEQKDHELLKMRQQMIERDDL 364
Query: 445 IKEKDAALDRIPGRKKK--RMDLFDGPHSDFED 475
++EKD L + +K++ MDLFDGP SDFED
Sbjct: 365 LQEKDKLLKKHITKKQRMDSMDLFDGPDSDFED 463
>AL366606
Length = 393
Score = 75.1 bits (183), Expect = 5e-14
Identities = 34/57 (59%), Positives = 45/57 (78%)
Frame = -1
Query: 156 SIFTLLIYGLFLFPNMDNFVDLNAIKIFLAKNPVPTLLANTYHSIHHRNIREGGLIV 212
+I L GL LFPN+DNFVD+NAI++F +KNPVPTLLA+TYH+IH R ++ G I+
Sbjct: 381 AIXLCLSTGLILFPNLDNFVDMNAIEVFHSKNPVPTLLADTYHAIHDRTLKGRGYIL 211
>TC92100 similar to GP|15290106|dbj|BAB63798. contains EST
D24276(R1622)~similar to Arabidopsis thaliana chromosome
5 K16F4.11~unknown protein, partial (71%)
Length = 1083
Score = 36.6 bits (83), Expect = 0.022
Identities = 26/80 (32%), Positives = 41/80 (50%)
Frame = +2
Query: 348 AHESYTQWVIDRATAFGMPYTLPRFLSSTVPEIPLPLLPQTKEEYQERLAEAEREIHMWK 407
A +S + +RA AF L + + EI L L + E ++AEAERE H K
Sbjct: 767 AVQSIKATIAERANAFRRQCELAETMK--LKEINLDKLMLIRSE---KVAEAEREYHELK 931
Query: 408 RECQKKDKDYETVMGLLEQE 427
E ++ K +ET++ L+ +E
Sbjct: 932 AESEQATKTFETIVKLMSEE 991
>TC83868
Length = 828
Score = 31.2 bits (69), Expect = 0.91
Identities = 20/85 (23%), Positives = 38/85 (44%)
Frame = +3
Query: 378 PEIPLPLLPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGLLEQEAYDSRQKDVI 437
PE+ + E +RL E+E EI + + +KKD + + +
Sbjct: 177 PEVEKDAYLKQIEALNQRLKESELEIESLREQLRKKDLQVDDSKEVKVDHEIEKDAHLKE 356
Query: 438 IAKLNERIKEKDAALDRIPGRKKKR 462
I LN+R+KE +A ++ + +K+
Sbjct: 357 IEALNQRLKESEAEIESSREQLRKK 431
>TC87535 similar to GP|19310542|gb|AAL85004.1 unknown protein {Arabidopsis
thaliana}, partial (86%)
Length = 893
Score = 30.8 bits (68), Expect = 1.2
Identities = 15/57 (26%), Positives = 31/57 (54%)
Frame = +1
Query: 393 QERLAEAEREIHMWKRECQKKDKDYETVMGLLEQEAYDSRQKDVIIAKLNERIKEKD 449
++R+ +E+ CQKK+K+Y+ + ++ +R+K +I KL E + E +
Sbjct: 445 RKRIDSVNKELKPLGHTCQKKEKEYKDALEAFNEK---NREKVQLITKLMELVGESE 606
>TC78289 similar to GP|15451044|gb|AAK96793.1 Unknown protein {Arabidopsis
thaliana}, partial (15%)
Length = 1020
Score = 30.8 bits (68), Expect = 1.2
Identities = 15/43 (34%), Positives = 28/43 (64%)
Frame = +1
Query: 391 EYQERLAEAEREIHMWKRECQKKDKDYETVMGLLEQEAYDSRQ 433
++QE+L E ER IH +R+ ++KD++ + L+ EA ++Q
Sbjct: 613 KFQEQLCERERAIHELERKMEEKDRELHKIK--LDNEAAWAKQ 735
>BQ751808 similar to GP|7769858|gb|A F12M16.20 {Arabidopsis thaliana},
partial (2%)
Length = 661
Score = 29.6 bits (65), Expect = 2.6
Identities = 19/39 (48%), Positives = 22/39 (55%)
Frame = +1
Query: 361 TAFGMPYTLPRFLSSTVPEIPLPLLPQTKEEYQERLAEA 399
T G P T PR SSTVP P P Q + YQ+RL+ A
Sbjct: 229 TPLGPPQTGPR-QSSTVPA-PAPSFAQPQSPYQQRLSAA 339
>AW686183 similar to GP|20197841|gb| hypothetical protein {Arabidopsis
thaliana}, partial (31%)
Length = 655
Score = 28.9 bits (63), Expect = 4.5
Identities = 23/95 (24%), Positives = 40/95 (41%), Gaps = 4/95 (4%)
Frame = +3
Query: 137 FLYQKASDFVKAKKTNAFESIFT-LLIYGLFLFPNMDNFVDLN---AIKIFLAKNPVPTL 192
FLY K F+ + + I LL G+F FP+ F+ ++ + IF+ + +
Sbjct: 150 FLYPKYPYFINRNNFHIYGKIQQ*LLFLGIFFFPSSSLFIHVHPFLCVSIFMV-HKL*VR 326
Query: 193 LANTYHSIHHRNIREGGLIVCCAPLLYRWYASHLT 227
+ +S + +IV C L +W H T
Sbjct: 327 VRGQVYSGEDFVVPCATIIVACCSLSLKWRIVHFT 431
>TC88896 weakly similar to GP|6728968|gb|AAF26966.1| unknown protein
{Arabidopsis thaliana}, partial (27%)
Length = 1097
Score = 28.5 bits (62), Expect = 5.9
Identities = 17/71 (23%), Positives = 34/71 (46%), Gaps = 6/71 (8%)
Frame = +2
Query: 385 LPQTKEEYQERLAEAEREIHMWKRECQKKDKDYETVMGLLEQE------AYDSRQKDVII 438
L +KE+Y+ L +A EI K + + + YE+++ E + ++ + D++
Sbjct: 722 LEASKEKYESMLNDAHHEIEDLKSDLEASKEKYESMLNDAHHEIDVLTSSIENSKMDILN 901
Query: 439 AKLNERIKEKD 449
+K KE D
Sbjct: 902 SKAEWEQKEHD 934
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.138 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,767,578
Number of Sequences: 36976
Number of extensions: 240811
Number of successful extensions: 1230
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1208
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1226
length of query: 475
length of database: 9,014,727
effective HSP length: 100
effective length of query: 375
effective length of database: 5,317,127
effective search space: 1993922625
effective search space used: 1993922625
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149496.4


