

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149496.11 - phase: 0 /pseudo
(1173 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
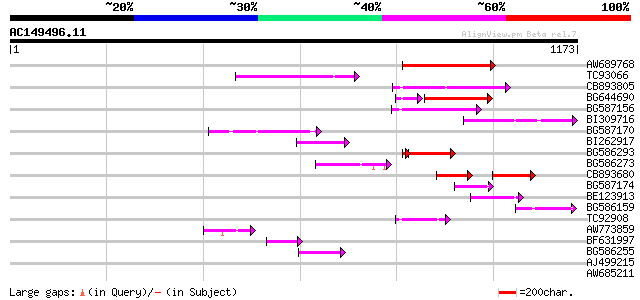
Score E
Sequences producing significant alignments: (bits) Value
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 176 6e-44
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 154 2e-37
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 147 2e-35
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 119 6e-34
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 137 3e-32
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 135 9e-32
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 131 1e-30
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 98 2e-20
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 94 5e-20
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 71 2e-12
CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotia... 68 2e-11
BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse ... 60 5e-09
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 59 8e-09
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 55 2e-07
TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol po... 52 1e-06
AW773859 52 2e-06
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 50 7e-06
BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l ... 45 2e-04
AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 42 0.001
AW685211 weakly similar to GP|4539662|gb| polyprotein {Sorghum b... 41 0.003
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 176 bits (445), Expect = 6e-44
Identities = 88/192 (45%), Positives = 123/192 (63%)
Frame = +1
Query: 813 KWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFK 872
+WL+AMK+E ++ N+ W+L+ P IGCKWV++ K + DG V+ +KARLVAKGF
Sbjct: 94 RWLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFS 273
Query: 873 QIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGF 932
Q G DY ETFSPV +IR++L IA + +EI Q+D+ AFLNG L E+VYM+QP+GF
Sbjct: 274 QTLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGF 453
Query: 933 GDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIVSFL 992
+ VCKL +S+YGLKQA R+W Q+GF K+ +P + +L
Sbjct: 454 -EAANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYL 630
Query: 993 ILYVDDILLIGN 1004
+YVDDIL+ G+
Sbjct: 631 XIYVDDILITGS 666
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 154 bits (389), Expect = 2e-37
Identities = 92/263 (34%), Positives = 144/263 (53%), Gaps = 6/263 (2%)
Frame = +1
Query: 467 ESYETCRSCLL-GKMTKAPFTGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDF 525
+ E C+ L G K F+ R +L IH+D+ GP +T+ GG Y +T+ DDF
Sbjct: 7 DKLEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDF 186
Query: 526 SRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGIL 585
R +VY +++K+E+F FK+++ VE Q GK +K L +D E+ S +F+ GI
Sbjct: 187 PRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIA 366
Query: 586 SQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELPN--FLWGYALLTAAYTLNRVPSKA 643
T P PQ NGV+ER RTLL+ R M+S+A L N LW A TA + +NR P A
Sbjct: 367 RHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSA 546
Query: 644 VE-KTPYEIWNGRKPNVGHFKIWGCEAYIKRLMSTKLEQKSEKCFFVGYPKETRGYYFY- 701
++ K P +IW+G + + +I+GC AY + KL ++ +C F+ Y E++GY +
Sbjct: 547 LDFKVPEDIWSGNLVDYSNLRIFGCPAY-ALVNDGKLAPRAGECIFLSYASESKGYRLWC 723
Query: 702 -KPPEGTIVVAKTGVFLEKDFVS 723
P +++++ F E +S
Sbjct: 724 SDPKSQKLILSRDVTFNEDALLS 792
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 147 bits (372), Expect = 2e-35
Identities = 89/249 (35%), Positives = 142/249 (56%), Gaps = 4/249 (1%)
Frame = +3
Query: 792 LLMDQDEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKK 851
+L +P T+ EA+ EKW +M +EM++ N W L D G IG KW+FK
Sbjct: 21 MLTMTSDPTTFEEAVKS---EKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKT 191
Query: 852 KIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDV 911
K++ +G++ YKARLVAKG+ Q +GVDY E F+PVA +IR+++A+AA +I + V
Sbjct: 192 KLNENGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA----QIKRDGV 359
Query: 912 KTAFL-NGNLLEDVYMTQPEGFGDPKATKKVC--KLQRSIYGLKQASRSWNLRFDETVQQ 968
+ + + + M + ++V +++R++YGLKQA R+W R + +
Sbjct: 360 CIS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS*RVKRALYGLKQAPRAWYSRIEAYFTK 539
Query: 969 YGFIKNEDEPCVYKKVS-GSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSMKDLGE 1027
GF K E ++ K+S G + + LYVDD++ IGND +E K ++K F+M DLG+
Sbjct: 540 EGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGK 719
Query: 1028 ASYILGIRI 1036
Y LG+ +
Sbjct: 720 MHYFLGVEV 746
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 119 bits (299), Expect(2) = 6e-34
Identities = 57/141 (40%), Positives = 96/141 (67%)
Frame = -2
Query: 859 VSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNG 918
++ K++LV +G+ Q G+DYDE FSPVA +++IRIL+A AA+ ++++QMDVK+AF+NG
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 919 NLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEP 978
+L E+V++ QP GF D + V +L +++YGLKQA R+W R + + + GF + + +
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 979 CVYKKVSGSIVSFLILYVDDI 999
++ + + +YVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 44.3 bits (103), Expect(2) = 6e-34
Identities = 19/57 (33%), Positives = 32/57 (55%)
Frame = -3
Query: 798 EPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKID 854
EP EA+ + W+ +M+ E+ ++VW L+ PEG IG +WVF+ K++
Sbjct: 588 EPKNVKEALRDAD---WINSMQEELHQFERSKVWYLVPRPEGKTVIGTRWVFRNKLE 427
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 137 bits (344), Expect = 3e-32
Identities = 72/185 (38%), Positives = 108/185 (57%)
Frame = -1
Query: 791 VLLMDQDEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFK 850
V L + P +Y EA+ E ++W E++ +E +M N W + P+G + +W+F
Sbjct: 582 VNLNENHIPRSYEEAM---EDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWIFT 412
Query: 851 KKIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMD 910
K DG + K RLVA+GF +G DY ETF+PVA + +IRI+L++A + +WQMD
Sbjct: 411 IKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQMD 232
Query: 911 VKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQYG 970
VK AFL G L ++VYM P G V +L+++IYGLKQ+ R+W + T+ G
Sbjct: 231 VKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNGRG 52
Query: 971 FIKNE 975
F K+E
Sbjct: 51 FRKSE 37
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 135 bits (340), Expect = 9e-32
Identities = 78/234 (33%), Positives = 125/234 (53%)
Frame = +2
Query: 940 KVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIVSFLILYVDDI 999
KVC+LQ+SIYGLKQASR W + E++ +G++++ + ++ K S + L++YVDDI
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 1000 LLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLRR 1059
+L GNDI +Q +K +L F +KDLG Y LG+ + R + L L+Q +Y ++L
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGIL--LNQRKYTLELLED 370
Query: 1060 FNMHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYA 1119
K P L L + SP +DE Y IG ++Y + TRPD+S+A
Sbjct: 371 SGNLAVKSTLTPYDISLKLHNSDSPLYNDETQ------YRRLIGKLIY-LTTTRPDISFA 529
Query: 1120 LSATSRYQSNPGNEYWIAVKNILKYLRRTKDTFLIYGG*EELSVIGYIDASFQT 1173
+ S++ S P ++ A +L+YL+ L Y L + + D+ + T
Sbjct: 530 VQQLSQFVSKPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWAT 691
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 131 bits (330), Expect = 1e-30
Identities = 81/233 (34%), Positives = 122/233 (51%)
Frame = -3
Query: 412 LYVLDLDMPIYNINAKRLKPNELNPTYLWHCRLGHINENRISKLHKDGFLDSFDFESYET 471
LY+L+ P+ N + LN LWH RLGH + ++ + L FE+ +
Sbjct: 665 LYMLEKLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRALNLM-----LPGVVFEN-KN 504
Query: 472 CRSCLLGKMTKAPFTGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFGYV 531
C +C+LGK K F + LI+TD+ +++ R YF+T D+ S++ ++
Sbjct: 503 CEACILGKHCKNVFPRTSTVYENCFDLIYTDLWTAPSLS-RDNHKYFVTFIDEKSKYTWL 327
Query: 532 YLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPP 591
L+ K FK FQ V N KIK+LRSD GGEY S F +HL GIL Q + P
Sbjct: 326 TLIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCP 147
Query: 592 GTPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAV 644
TPQ NGV++R+N+ L+++ RS+M A + TA Y +N +P+K +
Sbjct: 146 YTPQQNGVAKRKNKHLMEVARSLMFQANVS---------TACYLINWIPTKVL 15
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 98.2 bits (243), Expect = 2e-20
Identities = 47/114 (41%), Positives = 66/114 (57%), Gaps = 3/114 (2%)
Frame = +1
Query: 593 TPQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVE-KTPYEI 651
TPQ NGV+ER NRTLL+ R+M+ A + W A+ TA Y +NR PS ++ KTP E+
Sbjct: 85 TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEM 264
Query: 652 WNGRKPNVGHFKIWGCEAYI--KRLMSTKLEQKSEKCFFVGYPKETRGYYFYKP 703
W G+ + ++GC Y+ TKL+ KS KC F+GY +GY + P
Sbjct: 265 WKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWDP 426
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 94.0 bits (232), Expect(2) = 5e-20
Identities = 45/96 (46%), Positives = 65/96 (66%)
Frame = +2
Query: 826 YTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSP 885
Y Q L+ P G+ PIG +W++K K + DG + YKARLVAKG+ + G+D+DE F+P
Sbjct: 44 YQKQTLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAP 223
Query: 886 VAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLL 921
V I++I +LLA+AA + I +DVK AFLNG+ +
Sbjct: 224 VVRIETI*LLLALAATNGC*IHHIDVKIAFLNGHFV 331
Score = 23.1 bits (48), Expect(2) = 5e-20
Identities = 7/16 (43%), Positives = 14/16 (86%)
Frame = +3
Query: 814 WLEAMKSEMDSMYTNQ 829
W++AMK+E++S+ N+
Sbjct: 9 WIDAMKAEIESIIKNR 56
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 71.2 bits (173), Expect = 2e-12
Identities = 45/180 (25%), Positives = 92/180 (51%), Gaps = 21/180 (11%)
Frame = -2
Query: 632 AAYTLNRVPSKAV-EKTPYEIWNGRKPNVGHFKIWGCEAYI--KRLMSTKLEQKSEKCFF 688
A Y +NR+P++ + ++ P+E+ N RKP++ + +++GC Y+ + KLE +S K F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 689 VGYPKETRGYYFYKPPEGTIVVAKTGVFL-EKDFVSRRISGSKVDLKEIQDPQSTEIPVE 747
+GY +GY Y P ++V++ F+ E+ + + ++ DL+++ ++ + V
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEK---NQEDLRDLTSDKAGVLRVI 354
Query: 748 EQG-----QDTQTVMTENPAPVTQEPRRSSRI------------RQEPERYGYLISQEGD 790
+G Q+ + P + EPRR+++ QE + G +S+EG+
Sbjct: 353 LEGLGIKMNQDQSTRSRQPEESSNEPRRAAQTPHLDHEGGSEPETQENGQEGVGLSEEGE 174
>CB893680 weakly similar to GP|1167523|db ORF(AA 1-1338) {Nicotiana tabacum},
partial (7%)
Length = 780
Score = 68.2 bits (165), Expect = 2e-11
Identities = 36/74 (48%), Positives = 50/74 (66%)
Frame = -2
Query: 883 FSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVC 942
F P+ + +I LL+I A + + +DVKTAFL G+L+ED+YM QPEGF + K V
Sbjct: 554 FVPIVKLNTIMFLLSIVAIENLYLE*LDVKTAFLRGDLVEDIYMHQPEGF-S*EVGKMVG 378
Query: 943 KLQRSIYGLKQASR 956
KL++S+YGLKQ R
Sbjct: 377 KLKKSMYGLKQGPR 336
Score = 60.1 bits (144), Expect = 5e-09
Identities = 34/89 (38%), Positives = 54/89 (60%)
Frame = -3
Query: 999 ILLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLR 1058
+L++G++I ++ +KT K MKDLG A I+G++I D+ + +L LSQ YI +VL+
Sbjct: 271 LLVVGSNIDEIKNLKTRFSKEIDMKDLGPAKKIIGMQIMIDKQKGVL*LSQVEYITRVLQ 92
Query: 1059 RFNMHESKKGFIPMSSGLNLSKTQSPSTD 1087
FNM + ++S LS QSP T+
Sbjct: 91 IFNMGNAILVSTTLASHFCLSHEQSPQTE 5
>BG587174 similar to PIR|A47759|A4775 retrovirus-related reverse transcriptase
homolog - rape retrotransposon copia-like (fragment),
partial (84%)
Length = 249
Score = 60.1 bits (144), Expect = 5e-09
Identities = 36/84 (42%), Positives = 48/84 (56%), Gaps = 3/84 (3%)
Frame = -1
Query: 920 LLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQY---GFIKNED 976
L E +YMTQPEGF P VCKL++S+YGLKQ+ R W RFD + G +
Sbjct: 246 LEEKIYMTQPEGFLFPGKEDHVCKLRKSLYGLKQSPRQWYKRFDSYRSSWATTGVLMTVV 67
Query: 977 EPCVYKKVSGSIVSFLILYVDDIL 1000
++S I +L+LYVDD+L
Sbjct: 66 ST*TR*RMSRYI--YLVLYVDDML 1
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 59.3 bits (142), Expect = 8e-09
Identities = 35/111 (31%), Positives = 63/111 (56%), Gaps = 1/111 (0%)
Frame = +1
Query: 953 QASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIV-SFLILYVDDILLIGNDIPTLQE 1011
Q+ R W RF V+++G+I+ + + ++ K S ++ + LI+YVDDI L G+ ++
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1012 IKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNM 1062
+K L + F +KDLG Y LG+ + R + +SQ +Y+ +L+ M
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGS--SISQRKYVLDLLKETRM 327
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 55.1 bits (131), Expect = 2e-07
Identities = 38/127 (29%), Positives = 63/127 (48%), Gaps = 2/127 (1%)
Frame = +1
Query: 1047 LSQGRYIDKVLRRFNMHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIM 1106
+ Q +Y+ +L RF M +S P++ L K D+ ++ Y +G +M
Sbjct: 34 ICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIK------DENGVKVDATKYKQIVGCLM 195
Query: 1107 YAMICTRPDVSYALSATSRYQSNPGNEYWIAVKNILKYLRRTKDTFLIY--GG*EELSVI 1164
Y + TRPD+ Y LS SR+ + P + AVK +L+YL T + ++Y G E+L
Sbjct: 196 Y-LAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEKLE-- 366
Query: 1165 GYIDASF 1171
Y D+ +
Sbjct: 367 AYTDSDY 387
>TC92908 similar to GP|10140704|gb|AAG13538.1 putative gag-pol polyprotein
{Oryza sativa}, partial (1%)
Length = 638
Score = 52.0 bits (123), Expect = 1e-06
Identities = 39/115 (33%), Positives = 58/115 (49%), Gaps = 1/115 (0%)
Frame = +2
Query: 798 EPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDG 857
EP Y +A + ++WL AM+ E+ + N L EG + C+ V+K +G
Sbjct: 278 EPKNYVQAA---Q*QEWLAAMEQEIQVLEENNTSTLEPLREGKKWVDCRPVYKIIHKANG 448
Query: 858 KVSTYKARLVAKGFKQIHGVDY-DETFSPVAMIKSIRILLAIAAYHDYEIWQMDV 911
++ YKA+LVAK F Q+ G D+ D S + R LL IAA ++ MDV
Sbjct: 449 EIEKYKAQLVAKDFVQVEGEDF*DLCLS--NKDDNCRCLLTIAAAKG*QLHLMDV 607
>AW773859
Length = 538
Score = 51.6 bits (122), Expect = 2e-06
Identities = 35/113 (30%), Positives = 51/113 (44%), Gaps = 6/113 (5%)
Frame = -3
Query: 402 LYANANLSNGLYVLDL-DMPIYNINAKRLKPNELNPTYL-----WHCRLGHINENRISKL 455
+ + L GLY L D P I + +++P PT+L WH RLGH++ ++ L
Sbjct: 356 MIGSGELFEGLYFLTTQDTPSATIASSQVQPQP-QPTFLPQEALWHFRLGHLSNRKLLSL 180
Query: 456 HKDGFLDSFDFESYETCRSCLLGKMTKAPFTGQGERASDLLGLIHTDVCGPLN 508
H + + D S C C + K PF RAS L H D+ GP +
Sbjct: 179 HSNFPFITIDQNS--VCDICHYSRHKKLPFQLSTNRASKCYELFHFDIWGPFS 27
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 49.7 bits (117), Expect = 7e-06
Identities = 29/75 (38%), Positives = 42/75 (55%)
Frame = -2
Query: 532 YLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPP 591
++MKHKSE+F F + + + G++ + + + G E S EF+ KE I Q T
Sbjct: 643 FMMKHKSEAFYFSRNGRFWSRVKQGRR*NVFKLNNGLEICSAEFNELCKEEHITRQYTVR 464
Query: 592 GTPQWNGVSERRNRT 606
TPQ NGV+ER N T
Sbjct: 463 NTPQKNGVAERMNIT 419
Score = 35.4 bits (80), Expect = 0.13
Identities = 46/157 (29%), Positives = 70/157 (44%), Gaps = 6/157 (3%)
Frame = -1
Query: 540 SFTFFKEFQNEVENQLGKKIKMLRSD-RGGEYL-----SLEFDNHLKECGILSQLTPPGT 593
S FFKE++ VE+Q GKK+K L+++ R G +L+ H K I + T
Sbjct: 620 SILFFKEWKILVESQTGKKVKRLQTE*RLGNLFCRVQ*TLQGRTHYK--AIYGE--KYST 453
Query: 594 PQWNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVEKTPYEIWN 653
+W + N L+ R M S+A L A+ T Y +N +P + +
Sbjct: 452 KEWCC*ANEYN--FLERARCMFSNAGLNRSFQAEAISTKCYLVNVLPLLL*TVRLHLRYG 279
Query: 654 GRKPNVGHFKIWGCEAYIKRLMSTKLEQKSEKCFFVG 690
N+ +I GC AY + KLE +S+K F G
Sbjct: 278 --LINLLITQILGCPAYY-HVNEGKLEPRSKKGLFWG 177
>BG586255 similar to GP|7682800|gb Hypothetical protein T15F17.l {Arabidopsis
thaliana}, partial (12%)
Length = 436
Score = 45.1 bits (105), Expect = 2e-04
Identities = 27/101 (26%), Positives = 50/101 (48%), Gaps = 2/101 (1%)
Frame = -1
Query: 597 NGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVEKTPYEIWNGRK 656
NG++E + + + R ++ ++ P WG+A+L AA + PS + +P ++ +G
Sbjct: 373 NGLAESFIKRIQLITRPLLMRSKQPVSAWGHAVLHAAELIRIRPSSEHKYSPSQLLSGHV 194
Query: 657 PNVGHFKIWGCEAY--IKRLMSTKLEQKSEKCFFVGYPKET 695
P+V H K +G Y I TK+ + +VG+ T
Sbjct: 193 PDVSHIKTFGYPVYVPIAPPHRTKMGPQRRMGIYVGFDSPT 71
>AJ499215 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (18%)
Length = 567
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/120 (25%), Positives = 59/120 (48%), Gaps = 2/120 (1%)
Frame = +3
Query: 300 STSWVLDTGCGSHICTDVQGLRKSRALAKGEVD-LRVGNGAKVAALAVGTYVLTLPSGLL 358
S W++D+GC H+ D ++ L K + +R+ NGA + +GT ++ SG
Sbjct: 99 SKYWLIDSGCTHHMTHDRDLFKE---LNKSTISKVRMLNGAHIEVEGIGTVLVKSHSG-Y 266
Query: 359 LNLENCYYVPAISRNIISISCLDKAGFEFTIKN-KCCSVYLDNILYANANLSNGLYVLDL 417
+ N Y P ++++++S+ L G++ ++ KC +N A + +VLDL
Sbjct: 267 KQISNVLYAPKLNQSLLSVPQLLTKGYKVLFEHEKCVIKDQNNKEVLRAQMKARKFVLDL 446
>AW685211 weakly similar to GP|4539662|gb| polyprotein {Sorghum bicolor},
partial (2%)
Length = 388
Score = 40.8 bits (94), Expect = 0.003
Identities = 20/70 (28%), Positives = 39/70 (55%)
Frame = -2
Query: 816 EAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIH 875
+AM EM ++ +N+ W +I P+ + +K ++ K R+ AKG+ +++
Sbjct: 222 QAMIGEMTALESNRTWTII-------PLPLEMPYKHRL---------KVRMAAKGYSKVY 91
Query: 876 GVDYDETFSP 885
G+ Y++TFSP
Sbjct: 90 GLHYNDTFSP 61
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,247,260
Number of Sequences: 36976
Number of extensions: 488878
Number of successful extensions: 2282
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 2250
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2268
length of query: 1173
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1066
effective length of database: 5,058,295
effective search space: 5392142470
effective search space used: 5392142470
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149496.11


