

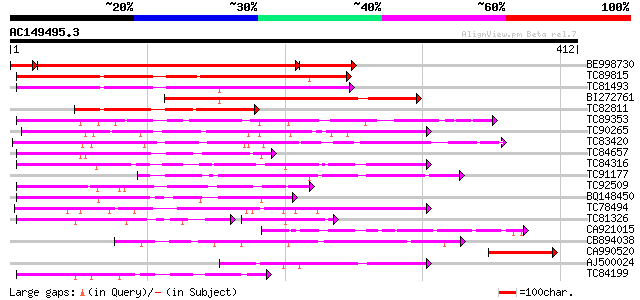
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149495.3 + phase: 0
(412 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE998730 296 1e-96
TC89815 266 1e-71
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 182 2e-46
BI272761 158 4e-39
TC82811 127 6e-30
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 96 3e-20
TC90265 weakly similar to GP|9294666|dbj|BAB03015.1 gene_id:F14O... 96 3e-20
TC83420 weakly similar to GP|5903055|gb|AAD55614.1| May be a pse... 94 1e-19
TC84657 weakly similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.... 94 1e-19
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 88 7e-18
TC91177 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 79 3e-15
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 77 2e-14
BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.1... 77 2e-14
TC78494 76 3e-14
TC81326 weakly similar to PIR|T45651|T45651 hypothetical protein... 57 9e-14
CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-typ... 68 6e-12
CB894038 67 1e-11
CA990520 67 1e-11
AJ500024 67 2e-11
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 66 3e-11
>BE998730
Length = 820
Score = 296 bits (758), Expect(3) = 1e-96
Identities = 145/192 (75%), Positives = 158/192 (81%), Gaps = 1/192 (0%)
Frame = +3
Query: 21 KSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQIEDDFSNLTTA-VPL 79
KSLLRFRSTSKSLKSIIDSHNF NLH KNSLNRS I+R +IYQI+DDFS+LTTA PL
Sbjct: 81 KSLLRFRSTSKSLKSIIDSHNFTNLHLKNSLNRSLIIRRNYDIYQIDDDFSDLTTAEFPL 260
Query: 80 NHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPIT 139
NHPF+ NST I LIGSCNGLLA+SNG IAL PN AN+I IWNPN RKHHIIPFLPLPIT
Sbjct: 261 NHPFSGNSTKITLIGSCNGLLAISNGLIALTQPNDANQIAIWNPNTRKHHIIPFLPLPIT 440
Query: 140 PRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPT 199
P P DM C L +HGFGFD LT DYK+LR+SWLV+ NPFYD HV LFSLKTNSWK IP+
Sbjct: 441 PHPPPDMYCYLYLHGFGFDLLTADYKLLRISWLVTQHNPFYDSHVTLFSLKTNSWKTIPS 620
Query: 200 MPYALVFAQTMG 211
PYAL + Q MG
Sbjct: 621 TPYALQYVQAMG 656
Score = 68.9 bits (167), Expect(3) = 1e-96
Identities = 32/42 (76%), Positives = 37/42 (87%)
Frame = +1
Query: 211 GVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDE 252
GV V++S+HW+MAKKLDG +P LIVAFNLTLEIF EVPLP E
Sbjct: 655 GVFVQNSLHWVMAKKLDGSYPWLIVAFNLTLEIFYEVPLPVE 780
Score = 28.1 bits (61), Expect(3) = 1e-96
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +1
Query: 1 MASDRLPPDTLAEIFSRLPV 20
MA D LP + + EIFSR+PV
Sbjct: 22 MAGDVLPSEIVTEIFSRIPV 81
>TC89815
Length = 807
Score = 266 bits (679), Expect = 1e-71
Identities = 141/251 (56%), Positives = 169/251 (67%), Gaps = 8/251 (3%)
Frame = +1
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
+PP+ +I S LP LLRFRSTSKSLKSIIDSH F NLH KNS N I+R +N+YQ
Sbjct: 46 IPPEIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFTNLHLKNSNNFYLIIRHNANLYQ 225
Query: 66 IEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNI 125
+ DF NLT +PLNHP S I L GSCNGL+ +SN A++I WNPNI
Sbjct: 226 L--DFPNLTPPIPLNHPLMSYSNRITLFGSCNGLICISN---------IADDIAFWNPNI 372
Query: 126 RKHHIIPFLPLPITPRSPSDMNCSLC-VHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHV 184
RKH IIP+LP TPRS SD VHGFG+D GDYK++R+S+ V LQN +D V
Sbjct: 373 RKHRIIPYLPT--TPRSESDTTLFAARVHGFGYDSFAGDYKLVRISYFVDLQNRSFDSQV 546
Query: 185 RLFSLKTNSWKIIPTMPYALVFAQTMGVLVED-------SIHWIMAKKLDGLHPSLIVAF 237
R+FSLK NSWK +P+M YAL A+TMGV VED S+HW++ +KL+ P LIVAF
Sbjct: 547 RVFSLKMNSWKELPSMNYALCCARTMGVFVEDSNNLNSNSLHWVVTRKLEPFQPDLIVAF 726
Query: 238 NLTLEIFNEVP 248
NLTLEIFNEVP
Sbjct: 727 NLTLEIFNEVP 759
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 182 bits (462), Expect = 2e-46
Identities = 100/248 (40%), Positives = 145/248 (58%), Gaps = 3/248 (1%)
Frame = +1
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP + EI SR+P K LLR RST K +++IDS +FI LH S + ILR S +Y+
Sbjct: 16 LPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHLSKSRDSVIILRQHSRLYE 195
Query: 66 IEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNI 125
+ D +++ L+HP S I ++GSCNGLL + N A++I WNP I
Sbjct: 196 L--DLNSMDRVKELDHPLMCYSNRIKVLGSCNGLLCICN---------IADDIAFWNPTI 342
Query: 126 RKHHIIPFLPLPITPRSPSDMNCSLC---VHGFGFDPLTGDYKILRLSWLVSLQNPFYDP 182
RKH IIP PL + ++ +L V+GFG+D T DYK++ +S V L N YD
Sbjct: 343 RKHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSISNFVDLHNRSYDS 522
Query: 183 HVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLE 242
HV ++++ ++ W +P +PYAL A+ MGV V ++HW++ + L+ LIVAF+L E
Sbjct: 523 HVTIYTMGSDVWMPLPGVPYALCCARPMGVFVSGALHWVVPRALEPDSRDLIVAFDLRFE 702
Query: 243 IFNEVPLP 250
+F EV LP
Sbjct: 703 VFREVALP 726
>BI272761
Length = 600
Score = 158 bits (399), Expect = 4e-39
Identities = 79/190 (41%), Positives = 117/190 (61%), Gaps = 3/190 (1%)
Frame = +2
Query: 113 NAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLC---VHGFGFDPLTGDYKILRL 169
N A++I WNP IRKH IIP PL + ++ +L V+GFG+D T DYK++ +
Sbjct: 5 NIADDIAFWNPTIRKHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSI 184
Query: 170 SWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGL 229
S+ V L N +D HV++++++T+ WK +P+MPYAL A+TMGV V ++HW++ + L+
Sbjct: 185 SYFVDLHNRSFDSHVKIYTMRTDVWKTLPSMPYALCCARTMGVFVSGALHWVVTRDLEPE 364
Query: 230 HPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVW 289
LIVAF+L E+F EV LP + + ++DVA L G LC+ N + DVW
Sbjct: 365 SRDLIVAFDLRFEVFREVALPGTV-------DGKFDMDVALLXGMLCIIENRGSDGFDVW 523
Query: 290 VMKQYGLKDS 299
VM +YG DS
Sbjct: 524 VMXEYGSHDS 553
>TC82811
Length = 961
Score = 127 bits (320), Expect = 6e-30
Identities = 68/135 (50%), Positives = 85/135 (62%), Gaps = 1/135 (0%)
Frame = +1
Query: 48 KNSLNRSFILRLRSNIYQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEI 107
KNS N I+R +N+YQ+ DF NLT +PLNHP S I L GSCNGL+ +S
Sbjct: 4 KNSNNFYLIIRHNANLYQL--DFPNLTPPIPLNHPLMSYSNRITLFGSCNGLICIS---- 165
Query: 108 ALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNC-SLCVHGFGFDPLTGDYKI 166
N A++I WNPNIRKH IIP+ LP TPRS SD + VHGFG+D GDYK+
Sbjct: 166 -----NIADDIAFWNPNIRKHRIIPY--LPTTPRSESDTTLFAARVHGFGYDSFAGDYKL 324
Query: 167 LRLSWLVSLQNPFYD 181
+R+S+ V LQN +D
Sbjct: 325 VRISYFVDLQNRSFD 369
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 95.9 bits (237), Expect = 3e-20
Identities = 97/369 (26%), Positives = 164/369 (44%), Gaps = 20/369 (5%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNS-----LNRSFILRLR 60
+P + + EI RLPV+SLL+FR K K++I F H S L F+ +
Sbjct: 105 MPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTAYPQLVSVFVSIAK 284
Query: 61 SNI--YQIEDDFSNLTT--AVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAAN 116
N+ Y ++ N + P + ++T++ +IGSCNGLL +S+
Sbjct: 285 CNLVSYPLKPLLDNPSAHRVEPADFEMI-HTTSMTIIGSCNGLLCLSD----------FY 431
Query: 117 EITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQ 176
+ T+WNP+I+ + P P S + GFG+D + YK+L + +Q
Sbjct: 432 QFTLWNPSIK----LKSKPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVLAV-----VQ 584
Query: 177 NPFY--DPHVRLFSLKTNSWKIIPTMP----YALVFAQTMGVLVEDSIHWIMAKKLDGLH 230
N + + +++ W I P + +G V +++WI++KK
Sbjct: 585 NCYNLDETKTLIYTFGGKDWTTIQKFPCDPSRCDLGRLGVGKFVSGNLNWIVSKK----- 749
Query: 231 PSLIVAFNLTLEIFNEVPLPDEIGEEE----VNSND-SVEIDVAALGGCLCMTVNYETTK 285
+IV F++ E + E+ LP + G++ V+SN V D + T
Sbjct: 750 --VIVFFDIEKETYGEMSLPQDYGDKNTVLYVSSNRIYVSFD------------HSNKTH 887
Query: 286 IDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKK 345
VW+MK+YG+ +SW KL + + +T S P C SD + G+ +L+ H K
Sbjct: 888 WVVWMMKEYGVVESWTKLMIIPQDKLT----SPGPYCL-SDALFISEHGV-LLMRPQHSK 1049
Query: 346 LFWYDLKTE 354
L Y+L +
Sbjct: 1050LSVYNLNND 1076
>TC90265 weakly similar to GP|9294666|dbj|BAB03015.1
gene_id:F14O13.16~pir||T05437~similar to unknown protein
{Arabidopsis thaliana}, partial (6%)
Length = 1373
Score = 95.5 bits (236), Expect = 3e-20
Identities = 90/344 (26%), Positives = 159/344 (46%), Gaps = 46/344 (13%)
Frame = +3
Query: 9 DTLA-EIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNR--------SFILRL 59
D LA I S+LP+KSL RFR KS + ++H+F++ KN ++ S +L++
Sbjct: 90 DELAFSILSKLPLKSLKRFRCVRKSWTLLFENHHFMSTFCKNLISNHHYYYGDVSLLLQI 269
Query: 60 --RSNIYQIEDDFSNLTTAVPLNHPFTRNSTNIALI--GSCNGLLAVSNGEIALRHPNAA 115
R + D + N+ + +PF ++ GS G+L + N N
Sbjct: 270 GDRDLLSLSSDRYENM-VKLDWPNPFQEEDPWFCILDSGSITGILCLYN------RNNRN 428
Query: 116 NEITI-WNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWL-- 172
NE T+ WNP ++ +IP PL P+ +HGFG+D DYK++R +
Sbjct: 429 NERTVFWNPATKEFKVIPPSPLEAV---PTYQGFGTVLHGFGYDHARDDYKLIRYLYYFL 599
Query: 173 ----------VSLQNPFY-----DPHVRLFSLKTNSWKIIPTMPY---------ALVFAQ 208
+SLQ+ + D ++SL++NSWK + Y +
Sbjct: 600 PSSRDFEDLGISLQDVPWGDISNDSFWEIYSLRSNSWKKLDINMYLGDIRCSFSGFDCVK 779
Query: 209 TMGVLVEDSIHWIMAKKLDGLHPS---LIVAFNLTLEIF--NEVPLPDEIGEEEVNSNDS 263
+ + ++ HW +D HP + +F+L E+F +PL + +++ S S
Sbjct: 780 SQRLYLDGRCHW--WHLID--HPDAKRALASFDLVNEVFFTTLIPLDPPLDVDDIFSVFS 947
Query: 264 VEIDVAALGGCLCMTV-NYETTKIDVWVMKQYGLKDSWCKLFTM 306
+ + AL G + + + ++ T D++V+ + G+K+SW KLFT+
Sbjct: 948 RPLYLVALSGSIALILWDFGTPTFDIYVLGEVGVKESWTKLFTI 1079
>TC83420 weakly similar to GP|5903055|gb|AAD55614.1| May be a pseudogene of
F6D8.29. {Arabidopsis thaliana}, partial (10%)
Length = 1210
Score = 94.0 bits (232), Expect = 1e-19
Identities = 95/415 (22%), Positives = 171/415 (40%), Gaps = 56/415 (13%)
Frame = +1
Query: 3 SDRLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSL---------NR 53
S +P D I S+LP+KSL RF KS + + F+N+ R N L N
Sbjct: 79 SSHIPDDLTFSILSKLPLKSLTRFTCAQKSWSLLFQNSIFMNMFRTNFLLSKHDEDDENT 258
Query: 54 SFILR-------LRSNIYQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSC--NGLLAVSN 104
+L+ +++Y + + + + L P N + I ++ S NG+L + N
Sbjct: 259 CVLLKQMEGRRPYHTSLYTLSGEKFENSVRLDLPTPLLENDSFIRILCSAGINGVLCLYN 438
Query: 105 GEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDY 164
+ NA I +WNP + ++P P ++ + FG+DP+T DY
Sbjct: 439 DSL-----NANKTIVLWNPATEEFKVVPHSHQPY-----ENIEFNPRPFAFGYDPVTNDY 588
Query: 165 KILRL---------SWL---------------VSLQNPFYDPH-------------VRLF 187
K++R+ +W+ + Q FYD H + ++
Sbjct: 589 KVIRVANYHIYFEDNWIYLPRKESLLWRKDDPLWKQYMFYDSHFWEGHELQVYEPFLEIY 768
Query: 188 SLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEV 247
SL++NSW+ + + + M + + + HW + ++ +F+ T EIF E
Sbjct: 769 SLRSNSWRKLDMDLSVFLHGRCM-MNLNELCHWQVNDQM--------ASFDFTNEIFFET 921
Query: 248 PLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVN-YETTKIDVWVMKQYGLKDSWCKLFTM 306
LP ++ S + D+A L G + N T + +W++ + + +SW KLF +
Sbjct: 922 TLP----SFDLKDGMSTKKDLAVLHGSVAFIHNVVNTGYLHIWILGELAVNESWTKLFVL 1089
Query: 307 MKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKKLFWYDLKTEQVSYEEI 361
P+ V I+ + L+ ++L W DL T+++ EEI
Sbjct: 1090------------GPVPCIGRPIGVRIKSVMFYLK-EDEELAWIDLSTQRI--EEI 1209
>TC84657 weakly similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 884
Score = 93.6 bits (231), Expect = 1e-19
Identities = 62/201 (30%), Positives = 103/201 (50%), Gaps = 12/201 (5%)
Frame = +1
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNS----LNR-----SFI 56
LP + + +I RLPVKSL+RF+ KSL ++I HNF H + S NR +
Sbjct: 328 LPHELIIQIMLRLPVKSLIRFKCVCKSLLALISDHNFAKSHFELSTATHTNRIVFMSTLA 507
Query: 57 LRLRSNIYQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAAN 116
L RS ++ + + +T++ LN + +++ + SC G + ++ ++
Sbjct: 508 LETRSIDFEASLNDDSASTSLNLNFMPPESYSSLEIKSSCRGFIVLT----------CSS 657
Query: 117 EITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQ 176
I +WNP+ H IPF P S D S C++GFG+D L DY ++ +S+ S+
Sbjct: 658 NIYLWNPSTGHHKQIPF------PASNLDAKYSCCLYGFGYDHLRDDYLVVSVSYNTSI- 816
Query: 177 NPFYD---PHVRLFSLKTNSW 194
+P D H++ FSL+ N+W
Sbjct: 817 DPVDDNISSHLKFFSLRANTW 879
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 87.8 bits (216), Expect = 7e-18
Identities = 76/313 (24%), Positives = 140/313 (44%), Gaps = 12/313 (3%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRS---- 61
LP + + I RLPV+SLLRF+ KS K++ +F N H S ++ S
Sbjct: 42 LPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQLVACESVSAY 221
Query: 62 -----NIYQIEDDFSNL-TTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAA 115
Y IE N TT +P+++ + T ++GSCNG L + + N
Sbjct: 222 RTWEIKTYPIESLLENSSTTVIPVSNTGHQRYT---ILGSCNGFLCL--------YDNYQ 368
Query: 116 NEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSL 175
+ +WNP+I + +S ++ +GFG+D + YK+L + +
Sbjct: 369 RCVRLWNPSIN-----------LKSKSSPTID-RFIYYGFGYDQVNHKYKLLAVKAFSRI 512
Query: 176 QNPFYDPHVRLFSLKTNSWKIIPT--MPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSL 233
+++ NS K + P + +G V +++WI+ ++ DG +
Sbjct: 513 TETM------IYTFGENSCKNVEVKDFPRYPPNRKHLGKFVSGTLNWIVDER-DGR--AT 665
Query: 234 IVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMKQ 293
I++F++ E + +V LP + + L C+C+ ++ T+ +W+MK+
Sbjct: 666 ILSFDIEKETYRQVLLPQ-------HGYAVYSPGLYVLSNCICVCTSFLDTRWQLWMMKK 824
Query: 294 YGLKDSWCKLFTM 306
YG+ +SW KL ++
Sbjct: 825 YGVAESWTKLMSI 863
>TC91177 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1298
Score = 79.0 bits (193), Expect = 3e-15
Identities = 67/242 (27%), Positives = 109/242 (44%), Gaps = 5/242 (2%)
Frame = +2
Query: 94 GSCNGLLAVSNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVH 153
GSC G + + N + + IWNP+ H IPF + P D N +
Sbjct: 356 GSCRGFILL----------NCYSSLCIWNPSTGFHKRIPFTTIDSNP----DANY---FY 484
Query: 154 GFGFDPLTGDYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVL 213
GFG+D T DY ++ +S+ S + H+ +FSL+ N W + L ++ L
Sbjct: 485 GFGYDESTDDYLVISMSYEPSPSSDGMLSHLGIFSLRANVWTRVEGGNLLLYSQNSLLNL 664
Query: 214 VED----SIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVA 269
VE +IHW+ + + + +IVAF+L E+ LP+EI + S D+
Sbjct: 665 VESLSNGAIHWLAFR--NDISMPVIVAFHLMERKLLELRLPNEI-----INGPSRAYDLW 823
Query: 270 ALGGCLCM-TVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGS 328
GCL + + + +WVM++Y ++ SW K T++ S + S P Y+ G
Sbjct: 824 VYRGCLALWHILPDRVTFQIWVMEKYNVQSSWTK--TLVLSFDGNPAHSFWPKYYTKSGD 997
Query: 329 KV 330
V
Sbjct: 998 IV 1003
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 76.6 bits (187), Expect = 2e-14
Identities = 64/230 (27%), Positives = 99/230 (42%), Gaps = 14/230 (6%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSN--- 62
LP + +AEI RLPVK L + R KS S+I F H +S ++ LRSN
Sbjct: 174 LPFELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSSTTPHHLI-LRSNNGS 350
Query: 63 ------IYQIEDDFSNLTTAVP---LNHP--FTRNSTNIALIGSCNGLLAVSNGEIALRH 111
+ I+ S T VP L +P T + SC+G++ ++
Sbjct: 351 GRFALIVSPIQSVLSTSTVPVPQTQLTYPTCLTEEFASPYEWCSCDGIICLTTD------ 512
Query: 112 PNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSW 171
+ +WNP I K +P L RSPS C+ FG+DP +YK+ +++
Sbjct: 513 ---YSSAVLWNPFINKFKTLPPLKYISLKRSPS------CLFTFGYDPFADNYKVFAITF 665
Query: 172 LVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWI 221
V V + ++ T+SW+ I P + F G+ V +HW+
Sbjct: 666 CVKRTT------VEVHTMGTSSWRRIEDFP-SWSFIPDSGIFVAGYVHWL 794
>BQ148450 similar to GP|10177464|dbj gb|AAD21700.1~gene_id:MQB2.18~similar to
unknown protein {Arabidopsis thaliana}, partial (9%)
Length = 666
Score = 76.6 bits (187), Expect = 2e-14
Identities = 61/220 (27%), Positives = 98/220 (43%), Gaps = 16/220 (7%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNI-- 63
LP + EI SRLPVK L++F+ K KS I +F+ H + S R L S +
Sbjct: 3 LPFEIQVEILSRLPVKYLMQFQCVCKLWKSQISKPDFVKKHLRVSNTRHLFLLTFSKLSP 182
Query: 64 ------YQIEDDFSNLT-TAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAAN 116
Y + F+ +T T L +P + +++GSC+G+L + + L P
Sbjct: 183 ELVIKSYPLSSVFTEMTPTFTQLEYPLNNRDESDSMVGSCHGILCI---QCNLSFP---- 341
Query: 117 EITIWNPNIRKHHIIPFLPLP----ITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWL 172
+WNP+IRK +P P I P + FG+D + YK++ +
Sbjct: 342 --VLWNPSIRKFTKLPSFEFPQNKFINP-----------TYAFGYDHSSDTYKVVAVFCT 482
Query: 173 VSLQNPFYD--PHVRLFSLKTNSWKIIPT-MPYALVFAQT 209
++ N Y V + ++ TN W+ I T P+ + F T
Sbjct: 483 SNIDNGVYQLKTLVNVHTMGTNCWRRIQTEFPFKIPFTGT 602
>TC78494
Length = 1450
Score = 75.9 bits (185), Expect = 3e-14
Identities = 81/342 (23%), Positives = 149/342 (42%), Gaps = 39/342 (11%)
Frame = +1
Query: 4 DRLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSII-DSH---NFINLHRKNS--------L 51
+ LP + ++ I SRLP + LL+ + K+ ++I DSH N+ + H + +
Sbjct: 181 ENLPHELVSNILSRLPSRELLKNKLVCKTWYNLITDSHFTNNYYSFHNQLQNQEEHLLVI 360
Query: 52 NRSFILRLRSNIYQIEDDFS----NLTTAVPLNHPFTRNSTN---IALIGSCNGLLAVSN 104
R FI L++ I F+ N+ +++ LN P NS + ++G CNG+ +
Sbjct: 361 RRPFISSLKTTISLHSSTFNDPKKNVCSSL-LNPPEQYNSEHKYWSEIMGPCNGIYLLQG 537
Query: 105 GEIALRHPNAANEITIWNPNIRKHHIIP-FLPLPITPRSPSDMNCSLCVHG-FGFDPLTG 162
NPN+ + + F LP + + S+ SL + FGFDP T
Sbjct: 538 -----------------NPNVLMNASLQQFKALPESHLTDSNGIYSLTDYASFGFDPKTN 666
Query: 163 DYKILRLS--WLVSL---QNPFYDPHVRLFSLKTNSWKII--PTMPYALVF-------AQ 208
DYK++ L WL Q ++ L+SL +NSWK + T+P + +
Sbjct: 667 DYKVIVLKDLWLKETDERQKGYWTG--ELYSLNSNSWKKLDAETLPLPIEICGSSSSSSS 840
Query: 209 TMGVLVEDSIHWIM----AKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSV 264
+ V + HW + G++ +++F++ E+F ++ +P +
Sbjct: 841 RVYTYVNNCCHWWSFVNNHDESQGMNQDFVLSFDIVNEVFRKIKVPRICESSQETFVTLA 1020
Query: 265 EIDVAALGGCLCMTVNYETTKIDVWVMKQYGLKDSWCKLFTM 306
+ ++ G + + DVWVM+ Y + SW K +++
Sbjct: 1021PFEESSTIGFIVNPIRGNVKHFDVWVMRDYWDEGSWIKQYSV 1146
>TC81326 weakly similar to PIR|T45651|T45651 hypothetical protein F13I12.200
- Arabidopsis thaliana, partial (12%)
Length = 838
Score = 57.4 bits (137), Expect(2) = 9e-14
Identities = 56/171 (32%), Positives = 77/171 (44%), Gaps = 12/171 (7%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLH-----RKNSLNR-SFILRL 59
LP + + +I RLPVKSL RF+S KS S+I + +F N H K++ +R FI L
Sbjct: 168 LPHELIFQILLRLPVKSLTRFKSVRKSWFSLISAPHFANSHFQLTSAKHAASRIMFISTL 347
Query: 60 RSNIYQIE-DDFSNLTTAVPLNHPF--TRNSTNIALIGSCNGLLAVSNGEIALRHPNAAN 116
I+ F N LN F TR+ + + GSC G + + R P
Sbjct: 348 SHETRSIDFKAFLNDDDPASLNITFSLTRSHFPVEIRGSCRGFI------LLYRPP---- 497
Query: 117 EITIWNPN--IRKH-HIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDY 164
+I IWNP+ +KH H+ SP D GFG+D DY
Sbjct: 498 DIYIWNPSTGFKKHIHL-----------SPVDSKSVAQCQGFGYDQSRDDY 617
Score = 37.0 bits (84), Expect(2) = 9e-14
Identities = 25/75 (33%), Positives = 44/75 (58%), Gaps = 4/75 (5%)
Frame = +1
Query: 169 LSWLVSLQ-NPF-YDPHVRLFSLKTNSWKIIPT--MPYALVFAQTMGVLVEDSIHWIMAK 224
++ +VSL NP + H++ FS++ N+WK I PY ++ + G+L IHW+ +
Sbjct: 613 ITLVVSLSYNPSAFSTHLKFFSVRDNTWKEIEGNYFPYGVLSSCREGLLFNGVIHWLALR 792
Query: 225 KLDGLHPSLIVAFNL 239
+ GL ++I AF+L
Sbjct: 793 RDLGL--TVIGAFDL 831
>CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-type (EC
2.7.7.7) (PolIII). {Listeria monocytogenes}, partial
(0%)
Length = 824
Score = 68.2 bits (165), Expect = 6e-12
Identities = 59/200 (29%), Positives = 101/200 (50%), Gaps = 6/200 (3%)
Frame = -1
Query: 184 VRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEI 243
V +++L T+ WK I +PY +F + GV V +++W+ + S I++ ++ E
Sbjct: 761 VSVYTLGTDYWKRIEDIPYN-IFEE--GVFVSGTVNWLASDD------SFILSLDVEKES 609
Query: 244 FNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMKQYGLKDSWCKL 303
+ +V LPD ND + V L CLC+ +DVW+M +YG ++SW KL
Sbjct: 608 YQQVLLPD-------TENDLWILGV--LRNCLCILAT-SNLFLDVWIMNEYGNQESWTKL 459
Query: 304 FTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKKLFWYDLKTEQVSYEEIPD 363
+++ + H + + YSS+ ++L+E E + KL YD KT ++ IP+
Sbjct: 458 YSVPN--MQDHGLEAYTVLYSSEDDQLLLEFNE--MRSDKVKLVVYDSKTGTLN---IPE 300
Query: 364 F--NGAKIC----VGSLVPP 377
F N +IC + SL+ P
Sbjct: 299 FQNNYDQICQNFYIESLISP 240
>CB894038
Length = 855
Score = 67.4 bits (163), Expect = 1e-11
Identities = 70/294 (23%), Positives = 120/294 (40%), Gaps = 39/294 (13%)
Frame = +1
Query: 77 VPLNHPFTRNSTNIALIG--SCNGLLAVSNGEIALRHPNAANEITIWNPNIRKHHIIPFL 134
V L +P + + +IG S NG+L + GE I++WNP + +IP
Sbjct: 31 VSLINPIQSDCEALQIIGFGSVNGILCLQYGE---------TRISLWNPTTNEFKVIP-- 177
Query: 135 PLPITPRSPSDMN-----------CSLCVHGFGFDPLTGDYKIL---------------R 168
P R P ++ +HGFG+D + DYK++ R
Sbjct: 178 --PAGTRLPHIVHKFKSKLVDPFYIQTTIHGFGYDSVADDYKLICLQSFEPYYFYNDKQR 351
Query: 169 LSWLVSLQNPFYDPHVRLFSLKTNSW-KIIPTMP-----YALVFAQTMGVLVEDSI-HWI 221
+ + LQ+ P ++SL +NSW K+ MP + L + L D + HW+
Sbjct: 352 MKQSLLLQHKSLQPFWMIYSLTSNSWKKLYVNMPRSSPTFQLEYYHGNHRLYMDGVCHWL 531
Query: 222 MAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNY 281
+ +V+F+L E F P+P I + + + ++ N
Sbjct: 532 SLP----TSGACMVSFDLNNETFFVTPIPSYILRVRRRAWQQLMVVNHSIALVSLPYHNT 699
Query: 282 ETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSH----LKSSSPLCYSSDGSKVL 331
+T I +W + G+K+SW KLFT C L + L ++++ +K+L
Sbjct: 700 QTYHISIW--GEVGVKESWIKLFTGENPCTLVEYPIGLGMNGELVFANEDNKLL 855
>CA990520
Length = 592
Score = 67.0 bits (162), Expect = 1e-11
Identities = 33/51 (64%), Positives = 38/51 (73%), Gaps = 1/51 (1%)
Frame = +2
Query: 349 YDLKTEQVSYEE-IPDFNGAKICVGSLVPPSFPVDNSSKKENHTCKSKKRY 398
YD ++ QVSY E IPDF+ VGSLVPPSFPVDNS KKEN T KSK+ +
Sbjct: 2 YDFRSAQVSYVEGIPDFHDVMFSVGSLVPPSFPVDNSRKKENRTSKSKRSF 154
>AJ500024
Length = 538
Score = 66.6 bits (161), Expect = 2e-11
Identities = 44/159 (27%), Positives = 76/159 (47%), Gaps = 5/159 (3%)
Frame = -1
Query: 153 HGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKII--PTMPYALVFAQT- 209
+ FG+D L DY ++ +S+ +L N + + FSL+ N WK + P PY F +
Sbjct: 496 YSFGYDQLKDDYLVVLMSYDPALDNIY--SCLEFFSLRDNVWKEMDCPYCPYRSNFDEMP 323
Query: 210 -MGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDV 268
G L +IHW+ + + IVAF+L E+P PD+ +D+ + +
Sbjct: 322 KAGCLYNGAIHWLAFHRDFPWRDNFIVAFDLNERKLFEMPPPDDF------EHDAGDCGL 161
Query: 269 AALGGCLCM-TVNYETTKIDVWVMKQYGLKDSWCKLFTM 306
G + + +Y ++WVMK+Y + SW K+ +
Sbjct: 160 WVFGEFFSLWSTDYPNDSFEIWVMKEYKVNSSWTKILVL 44
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 65.9 bits (159), Expect = 3e-11
Identities = 58/198 (29%), Positives = 87/198 (43%), Gaps = 13/198 (6%)
Frame = +1
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLH----RKNSLNRSFILR--- 58
LP + + +I LPVKSLLRF+ KS S+I +F N H K+S F+L
Sbjct: 109 LPLELIIQILLWLPVKSLLRFKCVCKSWFSLISDTHFANSHFQITAKHSRRVLFMLNHVP 288
Query: 59 ----LRSNIYQIEDDFSNLTTAVP--LNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHP 112
L ++ S + +P + P TN + SC G + + N
Sbjct: 289 TTLSLDFEALHCDNAVSEIPNPIPNFVEPPCDSLDTNSS---SCRGFIFLHNDP------ 441
Query: 113 NAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWL 172
++ IWNP+ R + IP SP+D N C++GFG+D L DY ++
Sbjct: 442 ----DLFIWNPSTRVYKQIPL--------SPNDSNSFHCLYGFGYDQLRDDYLVVS---- 573
Query: 173 VSLQNPFYDPHVRLFSLK 190
V+ Q P +R FS +
Sbjct: 574 VTCQELMDYPCLRFFSFE 627
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,693,403
Number of Sequences: 36976
Number of extensions: 255824
Number of successful extensions: 1659
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 1578
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1593
length of query: 412
length of database: 9,014,727
effective HSP length: 98
effective length of query: 314
effective length of database: 5,391,079
effective search space: 1692798806
effective search space used: 1692798806
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149495.3


