

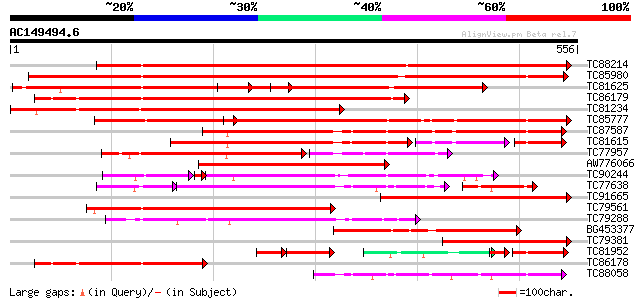
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.6 + phase: 0 /pseudo
(556 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88214 similar to GP|4325339|gb|AAD17339.1| F15P23.1 gene produ... 699 0.0
TC85980 similar to GP|15294146|gb|AAK95250.1 AT4g18030/T6K21_210... 625 e-179
TC81625 similar to PIR|T02472|T02472 hypothetical protein At2g45... 345 e-158
TC86179 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 420 e-118
TC81234 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 365 e-101
TC85777 similar to PIR|F86442|F86442 unknown protein [imported] ... 356 1e-98
TC87587 similar to GP|20259233|gb|AAM14332.1 putative ankyrin pr... 325 3e-89
TC81615 similar to GP|11994314|dbj|BAB02273. ankyrin-like protei... 219 7e-84
TC77957 similar to GP|15450900|gb|AAK96721.1 Unknown protein {Ar... 214 3e-75
AW776066 similar to GP|6752888|gb| unknown {Malus x domestica}, ... 271 4e-73
TC90244 similar to GP|15450749|gb|AAK96646.1 At2g39750/T5I7.5 {A... 207 1e-72
TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protei... 187 3e-71
TC91665 similar to PIR|T02472|T02472 hypothetical protein At2g45... 252 2e-67
TC79561 similar to GP|14194107|gb|AAK56248.1 At1g77260/T14N5_19 ... 229 2e-60
TC79288 similar to GP|8052540|gb|AAF71804.1| F3F9.21 {Arabidopsi... 177 7e-45
BG453377 similar to GP|6752888|gb| unknown {Malus x domestica}, ... 174 1e-43
TC79381 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 172 2e-43
TC81952 similar to GP|6056205|gb|AAF02822.1| unknown protein {Ar... 88 3e-41
TC86178 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x do... 159 3e-39
TC88058 similar to GP|21928175|gb|AAM78114.1 AT5g64030/MBM17_13 ... 157 8e-39
>TC88214 similar to GP|4325339|gb|AAD17339.1| F15P23.1 gene product
{Arabidopsis thaliana}, partial (83%)
Length = 1866
Score = 699 bits (1804), Expect = 0.0
Identities = 317/467 (67%), Positives = 386/467 (81%), Gaps = 1/467 (0%)
Frame = +1
Query: 86 SSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYK 145
S + P C V +EYTPCED RSL++ R +IYRERHCP K EE L+CR+P P+GY+
Sbjct: 7 SERVTHAPVCDVALSEYTPCEDTQRSLKFPRENLIYRERHCPEK-EEVLRCRIPAPYGYR 183
Query: 146 TPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIG 205
P WP SRD AWYANVPH+ELT+EK QNW+ ++GDRF FPGGGTMFP GAGAYIDDIG
Sbjct: 184 VPPRWPESRDWAWYANVPHKELTIEKKNQNWVHFEGDRFRFPGGGTMFPRGAGAYIDDIG 363
Query: 206 KLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
KLINLKDGS+RTALDTGCGVASWGAYL R+I+ +S APRDTHEAQVQFALERGVPALIG
Sbjct: 364 KLINLKDGSVRTALDTGCGVASWGAYLLPRDILAVSFAPRDTHEAQVQFALERGVPALIG 543
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHH 325
V+AS RLP+PSRAFD++HCSRCLIPW + DGI+L EVDRVLRPGGYWILSGPPINW H
Sbjct: 544 VIASIRLPYPSRAFDMAHCSRCLIPWGQNDGIYLTEVDRVLRPGGYWILSGPPINWESHW 723
Query: 326 RGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFC 385
+GW+RT++DLN EQT IE+VAKSLCW KL++K DIAIWQKP NH+ C+ RK+ +RPFC
Sbjct: 724 KGWERTREDLNAEQTSIERVAKSLCWKKLVQKGDIAIWQKPTNHIHCKITRKVFKNRPFC 903
Query: 386 GPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSE 445
++ PD AWYT + TCL P+P+V++ +E +G L NWP+RL SVPPRI G+++G+T+E
Sbjct: 904 DAKD-PDSAWYTKMDTCLTPLPEVTDIKEVSGRGLSNWPERLTSVPPRISSGSLDGITAE 1080
Query: 446 GYSKDNELWKKRIPHYKKVNNQLGTK-RYRNLVDMNANLGGFASALVKNPVWVMNVVPVQ 504
+ ++ ELWKKR+ +YK ++ QL RYRNL+DMNA LGGFA+A++ +PVWVMNVVPV+
Sbjct: 1081MFKENTELWKKRVAYYKTLDYQLAEPGRYRNLLDMNAYLGGFAAAMIDDPVWVMNVVPVE 1260
Query: 505 AKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
A+++TLG +YERGLIGTY +WCEAMSTYPRTYD IHADSLF+LY R
Sbjct: 1261AEINTLGVVYERGLIGTYQNWCEAMSTYPRTYDFIHADSLFTLYEDR 1401
>TC85980 similar to GP|15294146|gb|AAK95250.1 AT4g18030/T6K21_210
{Arabidopsis thaliana}, partial (82%)
Length = 2287
Score = 625 bits (1612), Expect = e-179
Identities = 295/530 (55%), Positives = 372/530 (69%)
Frame = +3
Query: 19 RKPHLYFLIAFLCAAFYLLGAYQQRASFTSLTKKAIITSPSCTIQQVNKPTLDFQSHHNS 78
R+P F + LC FYLLG +Q S + + T P L+F+ H
Sbjct: 357 RRPLSIFAVLALCCLFYLLGTWQSSGSGKGDSLALKVNKMQATTDCNIVPNLNFEPQHKY 536
Query: 79 SDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRV 138
+ + + F C V + +YTPC++ R++ + R MIYRERHCP + EE L+C +
Sbjct: 537 VEIVEQSEPKAKMFKACDVKYADYTPCQEQDRAMTFPRENMIYRERHCPPQ-EEKLRCLI 713
Query: 139 PPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAG 198
P P GY TPF WP SRD A+YANVP++ LTVEKA QNW++++G+ F FPGGGTMFP GA
Sbjct: 714 PAPEGYTTPFPWPKSRDYAYYANVPYKHLTVEKANQNWVKFEGNVFKFPGGGTMFPQGAD 893
Query: 199 AYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
AYID++ +I +KDG++RTALDTGCGVASWGAYL RN++ +S AP+D HEAQVQFALER
Sbjct: 894 AYIDELASVIPIKDGTVRTALDTGCGVASWGAYLLKRNVLAMSFAPKDNHEAQVQFALER 1073
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPP 318
GVPA+IGVL S RLPFPSRAFD+S CSRCLIPWAE +G++L EVDRVLRPGG+WILSGPP
Sbjct: 1074GVPAIIGVLGSIRLPFPSRAFDMSQCSRCLIPWAENEGMYLMEVDRVLRPGGFWILSGPP 1253
Query: 319 INWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKL 378
INW +++ W+RTK+DL EQ +IE +A+SLCW K EK DIAIW+K IN C+
Sbjct: 1254INWKTYYQTWKRTKEDLKAEQKRIEDLAESLCWEKKYEKGDIAIWRKKINAKSCQRKSLN 1433
Query: 379 ATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGT 438
D +N D WY ++ C P+P+V+++ E AGG LK +P RL +VPPRI
Sbjct: 1434LCD------LDNADDVWYKKMEVCKTPIPEVTSQSEVAGGELKKFPARLFAVPPRIAKDA 1595
Query: 439 IEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVM 498
I GVT E Y +DN+LWKKR+ +YK++N +GT RYRN++DMNA LGGFA+AL WVM
Sbjct: 1596IPGVTVESYQEDNKLWKKRVNNYKRINRLIGTTRYRNIMDMNARLGGFAAALESQKSWVM 1775
Query: 499 NVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLY 548
N VP A +TLG IYERGLIG YHDWCE +TYP TYDLIHA+ LFSLY
Sbjct: 1776NAVPTIAD-NTLGVIYERGLIGIYHDWCEGFATYPXTYDLIHANGLFSLY 1922
>TC81625 similar to PIR|T02472|T02472 hypothetical protein At2g45750
[imported] - Arabidopsis thaliana, partial (45%)
Length = 1431
Score = 345 bits (885), Expect(2) = e-158
Identities = 153/213 (71%), Positives = 182/213 (84%)
Frame = +2
Query: 256 LERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILS 315
L+ GVPA+IGV ASKRLPFPSRAFD++HCSRCLIPW EYDG++LNEVDR+LRPGGYWILS
Sbjct: 803 LKEGVPAVIGVFASKRLPFPSRAFDMAHCSRCLIPWPEYDGLYLNEVDRILRPGGYWILS 982
Query: 316 GPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSA 375
GPPI+W ++ +GW+RTK+DLN+EQTKIE VAKSLCWNK++EK DIAIWQKP NHLDC
Sbjct: 983 GPPIHWKRYWKGWERTKEDLNEEQTKIENVAKSLCWNKIVEKGDIAIWQKPKNHLDC--- 1153
Query: 376 RKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIH 435
K +RPFC Q NPDKAWYT+++TCL P+P+VSNKEE +GG L NWPQRL+S PPRI
Sbjct: 1154 -KFTQNRPFCQEQNNPDKAWYTNMQTCLNPLPKVSNKEEISGGELNNWPQRLKSTPPRIS 1330
Query: 436 MGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQL 468
GTI+ VT +SKDN+LW KR+ +YKKVNNQL
Sbjct: 1331 KGTIKSVTPPTFSKDNQLWNKRVSYYKKVNNQL 1429
Score = 230 bits (587), Expect(2) = e-158
Identities = 123/245 (50%), Positives = 153/245 (62%), Gaps = 8/245 (3%)
Frame = +3
Query: 3 PSSYSRNNKFPRPSSMRKPHLYFLIAFLCAAFYLLGAYQQRASFTS--------LTKKAI 54
P S+ N F + +K +LY IA LC Y LG+YQ S ++ +T K +
Sbjct: 30 PFKPSKYNSF---FNCKKTNLYSFIALLCIISYFLGSYQNNTSTSTSKNIQNKKVTPKCL 200
Query: 55 ITSPSCTIQQVNKPTLDFQSHHNSSDTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRY 114
+P+ T + LDF SHHN ++ ++++ +P C + T+YTPCED TRSL+Y
Sbjct: 201 QKNPTNTKTKTKTTNLDFFSHHNLTNPTTTSTTKSSQYPPCTPSLTDYTPCEDHTRSLKY 380
Query: 115 KRSRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQ 174
R +MIYRERHCP K E LKCRVP P+GYK PF WP SRD+AWYANVP+R LTVEKA Q
Sbjct: 381 TRDKMIYRERHCPKK-HEILKCRVPAPNGYKNPFPWPTSRDMAWYANVPYRHLTVEKAGQ 557
Query: 175 NWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQS 234
NWIR+DGD+F FPGGGTMFPNGA YIDDIGK+ D S R C WG L S
Sbjct: 558 NWIRFDGDKFRFPGGGTMFPNGADKYIDDIGKV----D*S*RWFC*NCC*YWLWGC*LGS 725
Query: 235 RNIIT 239
+ IT
Sbjct: 726 LSTIT 740
Score = 100 bits (250), Expect = 1e-21
Identities = 47/75 (62%), Positives = 60/75 (79%)
Frame = +1
Query: 204 IGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPAL 263
+GKLI+L+DGS+RTA+DTGCGVASWGAYL SR+I+T+S+AP+DTHEAQVQFALER
Sbjct: 646 LGKLIDLEDGSVRTAVDTGCGVASWGAYLLSRDILTVSIAPKDTHEAQVQFALERRCSCS 825
Query: 264 IGVLASKRLPFPSRA 278
K + FP ++
Sbjct: 826 YWCFCFKEITFPFKS 870
>TC86179 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (60%)
Length = 1297
Score = 420 bits (1079), Expect = e-118
Identities = 206/370 (55%), Positives = 259/370 (69%), Gaps = 2/370 (0%)
Frame = +1
Query: 25 FLIAFLCAAFYLLGAYQQRASFTSLTKKAI-ITSPSCTIQQVNKPTLDFQSHHNSSDTII 83
F++ LC FY+LGA+Q R+ F A+ IT + V P L F SHH + I
Sbjct: 199 FIVVGLCCFFYILGAWQ-RSGFGKGDSIALEITKNNAECDVV--PNLSFDSHHAGEVSQI 369
Query: 84 ALS-SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPH 142
S S T F C +T+YTPC+D R++ + R M YRERHCP + EE L C +P P
Sbjct: 370 DESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPE-EEKLHCMIPAPK 546
Query: 143 GYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYID 202
GY TPF WP SRD YAN P++ LTVEKA+QNWI+Y+G+ F FPGGGT FP GA YID
Sbjct: 547 GYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFPQGADKYID 726
Query: 203 DIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPA 262
+ +I + DG++RTALDTGCGVASWGAYL SRN++ +S APRD+HEAQVQFALERGVPA
Sbjct: 727 QLASVIPINDGTVRTALDTGCGVASWGAYLWSRNVVAMSFAPRDSHEAQVQFALERGVPA 906
Query: 263 LIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWN 322
+IGV + +LP+PSRAFD++HCSRCLIPW DG+++ EVDRVLRPGGYW+LSGPPINW
Sbjct: 907 VIGVFGTIKLPYPSRAFDMAHCSRCLIPWGANDGMYMMEVDRVLRPGGYWVLSGPPINWK 1086
Query: 323 KHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDR 382
+++ WQR K++L +EQ KIE+VAK LCW K EK +IAIWQK + CRS R+ +
Sbjct: 1087VNYKPWQRPKEELEEEQRKIEEVAKKLCWEKKSEKAEIAIWQKMTDTESCRS-RQDDSSV 1263
Query: 383 PFCGPQENPD 392
FC + D
Sbjct: 1264EFCESSDPDD 1293
>TC81234 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (50%)
Length = 1149
Score = 365 bits (938), Expect = e-101
Identities = 177/332 (53%), Positives = 224/332 (67%), Gaps = 4/332 (1%)
Frame = +3
Query: 1 MDPSSYSRNNKFPRPSSMRKPHLY---FLIAFLCAAFYLLGAYQQRASFTSLTKKAIITS 57
+ P+SY R SS + + F++ LC FY+LGA+Q+ + +T
Sbjct: 162 LPPTSYDRFYMAKPSSSGSRTRSFVQIFIVVGLCCFFYILGAWQRTGFGKGDLLQLEVTK 341
Query: 58 PSCTIQQVNKPTLDFQSHHNSS-DTIIALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKR 116
V P L F SHH I + S+ F C + +YTPC D R++ + R
Sbjct: 342 KGAGCDIV--PNLSFDSHHGGEVGKIDEVDSKPKVFKPCKARYIDYTPCHDQRRAMTFSR 515
Query: 117 SRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNW 176
MIYRERHCP + EE L C +P P GY TPF WP SRD YAN P++ LTVEKA+QNW
Sbjct: 516 QNMIYRERHCP-REEEKLHCLIPAPKGYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNW 692
Query: 177 IRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRN 236
I+Y+G+ F FPGGGT FP GA YID I +I +++G++RTALDTGCGVASWGAYL SRN
Sbjct: 693 IQYEGNVFRFPGGGTQFPQGADKYIDQIASVIPIENGTVRTALDTGCGVASWGAYLWSRN 872
Query: 237 IITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDG 296
+I +S APRD+HEAQVQFALERGVPA+IGVL + +LP+PS AFD++HCSRCLIPW DG
Sbjct: 873 VIAMSFAPRDSHEAQVQFALERGVPAVIGVLGTIKLPYPSGAFDMAHCSRCLIPWGSNDG 1052
Query: 297 IFLNEVDRVLRPGGYWILSGPPINWNKHHRGW 328
I+L EVDRVLRPGGYW+LSGPPI+W +++ W
Sbjct: 1053IYLMEVDRVLRPGGYWVLSGPPIHWKANYKAW 1148
>TC85777 similar to PIR|F86442|F86442 unknown protein [imported] - Arabidopsis
thaliana, partial (89%)
Length = 2381
Score = 356 bits (913), Expect = 1e-98
Identities = 169/342 (49%), Positives = 231/342 (67%)
Frame = +2
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
LK G + L GVASWG L R ++T+SLAPRD HEAQVQFALERG+PA++GV+++
Sbjct: 713 LKMGQLELQLILVVGVASWGGDLLDRGVLTISLAPRDNHEAQVQFALERGIPAILGVIST 892
Query: 270 KRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQ 329
+RLPFPS +FD++HCSRCLIPW E+ GI+L E+ R+LRPGG+W+LSGPP+N+ + RGW
Sbjct: 893 QRLPFPSNSFDMAHCSRCLIPWTEFGGIYLQEIHRILRPGGFWVLSGPPVNYERRWRGWN 1072
Query: 330 RTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQE 389
T ++ + K++ + S+C+ +KDDI +WQK ++ C T P C
Sbjct: 1073 TTVEEQRTDYEKLQDLLTSMCFKLYNKKDDIYVWQKAKDNA-CYDKLSRDTYPPKCDDSL 1249
Query: 390 NPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSK 449
PD AWYT L+ C + VP K ++ + WPQRL P RI + ++G +S +S
Sbjct: 1250 EPDSAWYTPLRACFV-VPMEKYK-KSGLTYMPKWPQRLNVAPERISL--VQGSSSSTFSH 1417
Query: 450 DNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDT 509
DN WKKRI HYKK+ LGT + RN++DMN GGFA++L+ +P+WVMNVV +T
Sbjct: 1418 DNSKWKKRIQHYKKLLPDLGTNKIRNVMDMNTAYGGFAASLINDPLWVMNVVSSYGP-NT 1594
Query: 510 LGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLYNGR 551
L +++RGLIGT+HDWCEA STYPRTYDL+HAD F+ + R
Sbjct: 1595 LPVVFDRGLIGTFHDWCEAFSTYPRTYDLLHADGFFTAESHR 1720
Score = 157 bits (398), Expect = 8e-39
Identities = 72/142 (50%), Positives = 95/142 (66%), Gaps = 1/142 (0%)
Frame = +1
Query: 84 ALSSETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHG 143
+L + F+FP C ++ +YTPC DP R +Y R+ ERHCP E +C VPPP G
Sbjct: 334 SLQIKPFSFPECSNDYQDYTPCTDPKRWRKYGTYRLTLLERHCPPIFERK-ECLVPPPPG 510
Query: 144 YKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDD 203
YK P WP SRD WY NVP+ + +K+ Q+W+ +G++F FPGGGTMFPNG G Y+D
Sbjct: 511 YKPPIRWPKSRDECWYRNVPYDWINKQKSNQHWLIKEGEKFQFPGGGTMFPNGVGEYVDL 690
Query: 204 IGKLI-NLKDGSIRTALDTGCG 224
+ LI +KDGS+RTA+DTGCG
Sbjct: 691 MQDLIPGIKDGSVRTAIDTGCG 756
>TC87587 similar to GP|20259233|gb|AAM14332.1 putative ankyrin protein
{Arabidopsis thaliana}, partial (70%)
Length = 1641
Score = 325 bits (833), Expect = 3e-89
Identities = 168/364 (46%), Positives = 230/364 (63%), Gaps = 7/364 (1%)
Frame = +3
Query: 190 GTMFPNGAGAYIDDIGKLINLKD------GSIRTALDTGCGVASWGAYLQSRNIITLSLA 243
GT F +GA YI I ++N D G +RT LD GCGVAS+GAYL + +II +SLA
Sbjct: 24 GTHFHHGADKYIAAIANMLNFTDNNLNNEGRLRTVLDVGCGVASFGAYLLASDIIAMSLA 203
Query: 244 PRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVD 303
P D H+ Q+QFALERG+PA +GVL +KRLP+PSR+F+++HCSRC I W + DGI L E+D
Sbjct: 204 PNDVHQNQIQFALERGIPAYLGVLGTKRLPYPSRSFELAHCSRCRIDWLQRDGILLLELD 383
Query: 304 RVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIW 363
RVLRPGGY+ S P + + ++DL + ++ + +CW ++D IW
Sbjct: 384 RVLRPGGYFAYSSP--------EAYAQDEEDL-RIWREMSALVGRMCWRIASKRDQTVIW 536
Query: 364 QKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNW 423
QKP+ + DC R+ T P C E+PD W +++ C+ P N+ + G L W
Sbjct: 537 QKPLTN-DCYMEREPGTRPPLCQSDEDPDAVWGVNMEVCITPYSDHDNRAQ--GSELAPW 707
Query: 424 PQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHY-KKVNNQLGTKRYRNLVDMNAN 482
P RL S PPR+ G++SE + KD ELW+KR+ +Y + ++ + RN++DM AN
Sbjct: 708 PSRLTSPPPRL---ADLGLSSEVFEKDTELWQKRVENYWNLMGPKISSNTVRNVMDMKAN 878
Query: 483 LGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHAD 542
LG FA+AL VWVMNVVP TL +++RGLIGT HDWCEA STYPRTYDL+HA
Sbjct: 879 LGSFAAALKDKDVWVMNVVPPYGP-STLRIVFDRGLIGTTHDWCEAFSTYPRTYDLLHAW 1055
Query: 543 SLFS 546
++FS
Sbjct: 1056TVFS 1067
>TC81615 similar to GP|11994314|dbj|BAB02273. ankyrin-like protein
{Arabidopsis thaliana}, partial (69%)
Length = 1489
Score = 219 bits (558), Expect(3) = 7e-84
Identities = 104/244 (42%), Positives = 148/244 (60%), Gaps = 6/244 (2%)
Frame = +3
Query: 158 WYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLKD----- 212
W AN+PH L EK+ QNW+ G++ FPGGGT F GA YI I ++N +
Sbjct: 3 WRANIPHTHLATEKSDQNWMVVKGEKIVFPGGGTHFHRGADKYIASIANMLNFPNNIINN 182
Query: 213 -GSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKR 271
G +R D GCGVAS+G YL S +++ +SLAP D HE Q+QFALERG+PA +GVL + R
Sbjct: 183 GGRLRNVFDVGCGVASFGGYLLSSDVLAMSLAPNDVHENQIQFALERGIPAYLGVLGTFR 362
Query: 272 LPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRT 331
LP+PSR+F+++HCSRC I W + DGI L E+DR+LRPGGY++ S P + +
Sbjct: 363 LPYPSRSFELAHCSRCRIDWLQRDGILLLELDRILRPGGYFVYSSP--------EAYAQD 518
Query: 332 KKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENP 391
++D + ++ + +CW +K+ IW KP+ + DC R+ T P C ++P
Sbjct: 519 EED-RRIWREMSALVGRMCWKVAAKKNQTVIWVKPLTN-DCYLKREPRTQPPLCSSDDDP 692
Query: 392 DKAW 395
D W
Sbjct: 693 DTVW 704
Score = 75.5 bits (184), Expect(3) = 7e-84
Identities = 34/51 (66%), Positives = 41/51 (79%)
Frame = +2
Query: 496 WVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFS 546
WVMNVVP +TL IY+RGL+GT H+WCEA STYPRTYDL+HA ++FS
Sbjct: 995 WVMNVVPENGP-NTLKIIYDRGLLGTVHNWCEAFSTYPRTYDLLHAWTIFS 1144
Score = 56.2 bits (134), Expect(3) = 7e-84
Identities = 32/93 (34%), Positives = 52/93 (55%), Gaps = 1/93 (1%)
Frame = +1
Query: 399 LKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRI 458
+K C+ +K G L WP RL + PPR+ ++E + KD ++W++R+
Sbjct: 715 MKACITRYSDQVHKAR--GSDLSPWPARLTTPPPRL---ADFNYSNEMFVKDMDVWQRRV 879
Query: 459 PHY-KKVNNQLGTKRYRNLVDMNANLGGFASAL 490
+Y K + N++ RN++DM ANLG FA+AL
Sbjct: 880 SNYWKLLGNKIKPDTIRNVMDMKANLGSFAAAL 978
>TC77957 similar to GP|15450900|gb|AAK96721.1 Unknown protein {Arabidopsis
thaliana}, partial (61%)
Length = 1689
Score = 214 bits (546), Expect(2) = 3e-75
Identities = 107/213 (50%), Positives = 135/213 (63%), Gaps = 12/213 (5%)
Frame = +1
Query: 91 NFPRCGVNFTEYTPCEDPTRSLRYKR------SRMIYRERHCPVKGEEDLKCRVPPPHGY 144
+FP C +E PC D R L Y+ S M + ERHCP E C +PPP GY
Sbjct: 616 SFPVCDDRHSELIPCLD--RHLIYQLRMKLDLSVMEHYERHCP-PAERRYNCLIPPPAGY 786
Query: 145 KTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDI 204
K P WP SRD W AN+PH L EK+ QNW+ G++ FPGGGT F GA YI +
Sbjct: 787 KVPVKWPKSRDEVWKANIPHTHLAHEKSDQNWMVEKGEKIAFPGGGTHFHYGADKYIASM 966
Query: 205 GKLINLK------DGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
++N +G +RT LD GCGVAS+G YL S +IIT+SLAP D H+ Q+QFALER
Sbjct: 967 ANMLNFSNNNLNNEGRLRTVLDVGCGVASFGGYLLSSDIITMSLAPNDVHQNQIQFALER 1146
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRCLIPW 291
G+PA +GVL +KRLP+PSR+F+++HCSRC I W
Sbjct: 1147GIPAYLGVLGTKRLPYPSRSFELAHCSRCRIDW 1245
Score = 86.3 bits (212), Expect(2) = 3e-75
Identities = 48/140 (34%), Positives = 74/140 (52%)
Frame = +2
Query: 295 DGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKL 354
DGI L E+DRVLRPGGY+ S P + + +++L + + V + +CW
Sbjct: 1256 DGILLLELDRVLRPGGYFAYSSP--------EAYAQDEENLRIWKEMSDLVGR-MCWRIA 1408
Query: 355 IEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEE 414
+K+ IWQKP+ + DC R+ T P C +PD + +++ C+ P + NK +
Sbjct: 1409 SKKEQTVIWQKPLTN-DCYKKREPGTRPPLCQSDADPDAVFGVNMEVCITPYSEHDNKAK 1585
Query: 415 TAGGVLKNWPQRLESVPPRI 434
+G L WP RL S PPR+
Sbjct: 1586 GSG--LAPWPARLTSPPPRL 1639
>AW776066 similar to GP|6752888|gb| unknown {Malus x domestica}, partial
(31%)
Length = 583
Score = 271 bits (694), Expect = 4e-73
Identities = 123/187 (65%), Positives = 148/187 (78%)
Frame = +1
Query: 186 FPGGGTMFPNGAGAYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPR 245
FPGGGT FP GA AYI+ + +I L +G IRTALDTGCGVASWGAYL +N+I +S+APR
Sbjct: 4 FPGGGTQFPQGADAYINQLASVIPLDNGMIRTALDTGCGVASWGAYLFKKNVIAMSIAPR 183
Query: 246 DTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRV 305
D+HEAQVQFALERGVPA+IGVL + LPFPS AFD++HCSRCLIPW G+++ EVDRV
Sbjct: 184 DSHEAQVQFALERGVPAIIGVLGTIMLPFPSGAFDMAHCSRCLIPWGANGGLYMKEVDRV 363
Query: 306 LRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQK 365
LRPGGYWILSGPPINW + R WQR + +L +EQ +IE AK LCW K EK +IAIW+K
Sbjct: 364 LRPGGYWILSGPPINWKNNFRAWQRPENELEEEQRQIENTAKLLCWEKKHEKGEIAIWRK 543
Query: 366 PINHLDC 372
+N +C
Sbjct: 544 ALNIDEC 564
>TC90244 similar to GP|15450749|gb|AAK96646.1 At2g39750/T5I7.5 {Arabidopsis
thaliana}, partial (49%)
Length = 1319
Score = 207 bits (528), Expect(3) = 1e-72
Identities = 118/298 (39%), Positives = 176/298 (58%), Gaps = 11/298 (3%)
Frame = +3
Query: 193 FPNGAGAYIDDIGKLI-NLKDGSIRTA--LDTGCGVASWGAYLQSRNIITLSLAPRDTHE 249
F +GA Y+D I K+I + G T+ L GCGVAS+GAYL RN+IT+S+AP+D HE
Sbjct: 477 FIHGADEYLDHISKMIPEITFGRHITSCSLMLGCGVASFGAYLLQRNVITMSVAPKDVHE 656
Query: 250 AQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPG 309
Q+QFALERGVPA++ A++RL +PS+AFD+ HCSRC I W DGI L EV+R+LR G
Sbjct: 657 NQIQFALERGVPAMVAAFATRRLLYPSQAFDLIHCSRCRINWTRDDGILLLEVNRMLRAG 836
Query: 310 GYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINH 369
GY++ + P+ KH ++ L ++ ++ + LCW L + IA+WQKP ++
Sbjct: 837 GYFVWAAQPV--YKH-------EEALEEQWEEMLNLTTRLCWKFLKKDGYIAVWQKPFDN 989
Query: 370 LDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLES 429
C R+ T P C P ++PD WY DLK C+ +P+ N+ E + +WP RL++
Sbjct: 990 -SCYLNREAGTKPPLCDPSDDPDNVWYVDLKACISELPK--NEYEAN---ITDWPARLQT 1151
Query: 430 VPPRIHMGTIEGVTS--EGYSKDNELWKK------RIPHYKKVNNQLGTKRYRNLVDM 479
P R+ ++ S E + +++ W + R H+KK+ R RN++DM
Sbjct: 1152PPNRLQSIKVDAFISRKELFKAESKYWNEIIEAYVRALHWKKI-------RLRNVMDM 1304
Score = 80.5 bits (197), Expect(3) = 1e-72
Identities = 39/91 (42%), Positives = 49/91 (52%), Gaps = 2/91 (2%)
Frame = +1
Query: 92 FPRCGVNFTEYTPCEDPTRSLRYKRSRMIYR--ERHCPVKGEEDLKCRVPPPHGYKTPFT 149
F C +EY PC D +++ S ERHCP G++ L C VP P GY+ P
Sbjct: 169 FGLCSRGMSEYIPCLDNVEAIKKLPSTEKGERFERHCPEDGKK-LNCLVPAPKGYRAPIP 345
Query: 150 WPASRDVAWYANVPHRELTVEKAVQNWIRYD 180
WP SRD W++NVPH L +K QNWI D
Sbjct: 346 WPKSRDEVWFSNVPHTRLVEDKGGQNWISRD 438
Score = 25.0 bits (53), Expect(3) = 1e-72
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = +2
Query: 182 DRFFFPGGGTMF 193
D+F FPGGGT F
Sbjct: 443 DKFKFPGGGTQF 478
>TC77638 similar to GP|20161455|dbj|BAB90379. ankyrin-like protein {Oryza
sativa (japonica cultivar-group)}, partial (62%)
Length = 2345
Score = 187 bits (474), Expect(3) = 3e-71
Identities = 108/276 (39%), Positives = 164/276 (59%), Gaps = 8/276 (2%)
Frame = +1
Query: 164 HRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLI-NLKDGS-IRTALDT 221
H L + ++ + G+ FPGGGT F +GA YID I + + ++ G R LD
Sbjct: 1291 HLSLLKLRGIKIG*KLPGEYLTFPGGGTQFKHGALHYIDFIQETLPDIAWGKRTRVILDV 1470
Query: 222 GCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLASKRLPFPSRAFDI 281
GCGVAS+G +L R+++ +SLAP+D HEAQVQFALERG+PA+ V+ +KRLPFP R FD
Sbjct: 1471 GCGVASFGGFLFDRDVLAMSLAPKDEHEAQVQFALERGIPAISAVMGTKRLPFPGRVFDA 1650
Query: 282 SHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTK 341
HC+RC +PW G L E++RVLRPGG+++ S PI +Q+ +D+ + +
Sbjct: 1651 VHCARCRVPWHIEGGKLLLELNRVLRPGGFFVWSATPI--------YQKLPEDV-EIWNE 1803
Query: 342 IEKVAKSLCWNKL-IEKD-----DIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAW 395
++ + KS+CW + I KD +AI++KP+++ DC R + P C ++ + AW
Sbjct: 1804 MKALTKSICWELVSISKDKVNGVGVAIYKKPLSN-DCYEQRS-KNEPPMCQKSDDHNAAW 1977
Query: 396 YTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVP 431
Y L+ C+ VP VS+ E + WP++ + P
Sbjct: 1978 YIKLQACIHKVP-VSSSERGS-----QWPEKWPARP 2067
Score = 62.8 bits (151), Expect(3) = 3e-71
Identities = 37/83 (44%), Positives = 45/83 (53%), Gaps = 4/83 (4%)
Frame = +2
Query: 86 SSET-FNFPRCGVNF-TEYTPCEDPTRSLRYKRSRMIY--RERHCPVKGEEDLKCRVPPP 141
SS+T +N+ C V ++ PC D + +R RS Y RERHCP EE C V P
Sbjct: 1052 SSKTGYNWKVCNVTAGPDFIPCLDNWKVIRSLRSTKHYEHRERHCP---EEPPTCLVSLP 1222
Query: 142 HGYKTPFTWPASRDVAWYANVPH 164
GYK WP SR+ WY NVPH
Sbjct: 1223 EGYKCSIEWPKSREKIWYYNVPH 1291
Score = 58.9 bits (141), Expect(3) = 3e-71
Identities = 34/75 (45%), Positives = 48/75 (63%), Gaps = 2/75 (2%)
Frame = +2
Query: 445 EGYSKDNELWKKRIPHYKKVNNQLGTK--RYRNLVDMNANLGGFASALVKNPVWVMNVVP 502
E ++ DN+ WK+ + K N LG + RN++DMN+ GGFA+AL + +WVMNVV
Sbjct: 2129 EDFAADNKHWKRVVS--KSYLNGLGIQWSNVRNVMDMNSIYGGFAAALKELNIWVMNVVS 2302
Query: 503 VQAKVDTLGAIYERG 517
+ + DTL IYERG
Sbjct: 2303 IDS-ADTLPIIYERG 2344
>TC91665 similar to PIR|T02472|T02472 hypothetical protein At2g45750
[imported] - Arabidopsis thaliana, partial (35%)
Length = 991
Score = 252 bits (644), Expect = 2e-67
Identities = 114/189 (60%), Positives = 147/189 (77%), Gaps = 1/189 (0%)
Frame = +1
Query: 364 QKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNW 423
QKP NH+ C+ RK+ RPFC ++PD AWYT L TCL P+P V+N +E AGG + W
Sbjct: 1 QKPTNHMHCKVKRKIFKTRPFCEEAQDPDMAWYTKLDTCLTPLPDVNNVKEVAGGEISKW 180
Query: 424 PQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLG-TKRYRNLVDMNAN 482
P+RL S+PPRI +++G+T E + ++ ELWKKR +YK +++QL T RYRNL+DMN+
Sbjct: 181 PKRLTSIPPRISGESLKGITPEMFKENTELWKKRESYYKTLDSQLAETGRYRNLLDMNSY 360
Query: 483 LGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHAD 542
LGGFA+ALV +PVWVMN+VPV+A+++TLG IYERGLIGTY +WCEAMSTYPRTYD IH D
Sbjct: 361 LGGFAAALVDDPVWVMNIVPVEAEINTLGVIYERGLIGTYQNWCEAMSTYPRTYDFIHGD 540
Query: 543 SLFSLYNGR 551
S+FSLY R
Sbjct: 541 SVFSLYQHR 567
>TC79561 similar to GP|14194107|gb|AAK56248.1 At1g77260/T14N5_19
{Arabidopsis thaliana}, partial (33%)
Length = 1092
Score = 229 bits (585), Expect = 2e-60
Identities = 119/251 (47%), Positives = 161/251 (63%), Gaps = 7/251 (2%)
Frame = +2
Query: 76 HNSSDT--IIALSSETFNFPRC-GVNFTEYTPCEDPTRSL-RYKRSRMIYR-ERHCPVKG 130
+N+SDT ++ + + C V +Y PC D + ++ S + ERHCP +
Sbjct: 341 NNNSDTNFVVEEVKKVKKYKICKDVRMVDYIPCLDNFEEIAKFNGSERGEKYERHCPEQ- 517
Query: 131 EEDLKCRVPPPHGYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGG 190
E+ L C VP P GY+ P WP SRD W++NVPH L +K QNWI D+F FPGGG
Sbjct: 518 EKGLNCVVPRPKGYRKPILWPKSRDEIWFSNVPHTRLVEDKGGQNWISRKKDKFVFPGGG 697
Query: 191 TMFPNGAGAYIDDIGKLI-NLKDG-SIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTH 248
T F +GA Y+D I +++ ++ G + R ALD GCGVAS+GA+L RN+ TLS+AP+D H
Sbjct: 698 TQFIHGADKYLDQISEMVPDIAFGYNTRVALDIGCGVASFGAFLMQRNVTTLSIAPKDVH 877
Query: 249 EAQVQFALERGVPALIGVLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRP 308
E Q+QFALERGVPAL+ V A+ RL FPS+AFD+ HCSRC I W DGI L E +R+LR
Sbjct: 878 ENQIQFALERGVPALVAVFATHRLLFPSQAFDLIHCSRCRINWTRDDGILLLEANRLLRA 1057
Query: 309 GGYWILSGPPI 319
G Y++ + P+
Sbjct: 1058GRYFVWAAQPV 1090
>TC79288 similar to GP|8052540|gb|AAF71804.1| F3F9.21 {Arabidopsis thaliana},
partial (40%)
Length = 1665
Score = 177 bits (450), Expect = 7e-45
Identities = 109/319 (34%), Positives = 165/319 (51%), Gaps = 10/319 (3%)
Frame = +3
Query: 95 CGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPHGYKTPFTWPASR 154
C + PC + + + ER C E+ C V PP YK P WP +
Sbjct: 768 CSPELENFVPCFNVSDGNEF--------ERKCEY--EQSQNCLVLPPVNYKVPLRWPTGK 917
Query: 155 DVAWYANVP---HRELTVEKAVQNWIRYDGDRFFFPGGGTMFPNGAGAYIDDIGKLINLK 211
DV W ANV L+ + + D ++ F MF +G Y I ++I L+
Sbjct: 918 DVIWVANVKITAQEVLSSGSLTKRMMMLDEEQISFRSASHMF-DGVEDYSHQIAEMIGLR 1094
Query: 212 DGS------IRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIG 265
+ S IRT LD GCG S+GA+L I+TL +A + +QVQ LERG+PA+I
Sbjct: 1095 NESSFIQAGIRTVLDIGCGYGSFGAHLFDSQILTLCIANYEPSGSQVQLTLERGLPAMIA 1274
Query: 266 VLASKRLPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPPINWNKHH 325
SK+LP+PS +FD+ HC+RC I W + DG L E DR+LRPGGY++ + P N
Sbjct: 1275 SFTSKQLPYPSLSFDMLHCARCGIDWDQKDGNLLIEADRLLRPGGYFVWTSPLTN----- 1439
Query: 326 RGWQRTKKDLNQEQTKI-EKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARKLATDRPF 384
R K+ NQ++ KI ++LCW L ++D+ +++K + +C ++RK + RP
Sbjct: 1440 ---ARNKE--NQKRWKIVHDFTENLCWEMLSQQDETVVFKK-ASKKNCYTSRKKGS-RPL 1598
Query: 385 CGPQENPDKAWYTDLKTCL 403
CG + + +Y +L+ C+
Sbjct: 1599 CGRGLDVESPYYRELQNCI 1655
>BG453377 similar to GP|6752888|gb| unknown {Malus x domestica}, partial
(29%)
Length = 534
Score = 174 bits (440), Expect = 1e-43
Identities = 86/185 (46%), Positives = 120/185 (64%)
Frame = +1
Query: 318 PINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKLIEKDDIAIWQKPINHLDCRSARK 377
PI+W +++ WQR K+DL +EQ KIE VAK LCW K EK++IA+WQK ++ CR R+
Sbjct: 1 PIHWKANYKAWQRPKEDLEEEQRKIEDVAKLLCWEKKSEKNEIAVWQKTVDSETCRR-RQ 177
Query: 378 LATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNKEETAGGVLKNWPQRLESVPPRIHMG 437
+ FC + D WY ++ C+ P +V G LK +PQRL +VPP+I G
Sbjct: 178 EDSGVKFCESTDAND-VWYKKMEACVTPNRKVH-------GDLKPFPQRLYAVPPKIASG 333
Query: 438 TIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLGGFASALVKNPVWV 497
++ GV++E Y DN+ WKK + YKK+N LG+ RYRN++DMNA LG FA+A+ +WV
Sbjct: 334 SVPGVSAETYQDDNKRWKKHVNAYKKINKLLGSGRYRNIMDMNAGLGSFAAAIQSPKLWV 513
Query: 498 MNVVP 502
MNVVP
Sbjct: 514 MNVVP 528
>TC79381 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (31%)
Length = 981
Score = 172 bits (437), Expect = 2e-43
Identities = 78/127 (61%), Positives = 97/127 (75%)
Frame = +2
Query: 425 QRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLGTKRYRNLVDMNANLG 484
QRL ++PPR+ G+I GV+SE Y DN++WKK + YKK+N+ L + RYRN++DMNA LG
Sbjct: 5 QRLYAIPPRVSSGSIPGVSSETYQNDNKMWKKHVNAYKKINSLLDSGRYRNIMDMNAGLG 184
Query: 485 GFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSL 544
FA+A+ + WVMNVVP A+ TLG IYERGLIG YHDWCE STYPRTYDLIHA+ L
Sbjct: 185 SFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCEGFSTYPRTYDLIHANGL 364
Query: 545 FSLYNGR 551
FSLY +
Sbjct: 365 FSLYQDK 385
Score = 35.4 bits (80), Expect = 0.058
Identities = 33/153 (21%), Positives = 62/153 (39%), Gaps = 3/153 (1%)
Frame = +2
Query: 210 LKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALERGVPALIGVLAS 269
L G R +D G+ S+ A + S +++ P ++ + ERG+ +
Sbjct: 134 LDSGRYRNIMDMNAGLGSFAAAIHSSKSWVMNVVPTIAEKSTLGVIYERGLIGIYHDWCE 313
Query: 270 KRLPFPSRAFDISHCSRCLIPWAEYDGI--FLNEVDRVLRPGGYWILSGPPINWNKHHRG 327
+P R +D+ H + + + L E+DR+LRP G I+
Sbjct: 314 GFSTYP-RTYDLIHANGLFSLYQDKCNTEDILLEMDRILRPEGAVII------------- 451
Query: 328 WQRTKKDLNQEQTKIEKVAKSLCWN-KLIEKDD 359
+D K++K+ + WN KL++ +D
Sbjct: 452 -----RDEVDVLIKVKKLIGGMRWNMKLVDHED 535
>TC81952 similar to GP|6056205|gb|AAF02822.1| unknown protein {Arabidopsis
thaliana}, partial (52%)
Length = 1126
Score = 87.8 bits (216), Expect(5) = 3e-41
Identities = 39/55 (70%), Positives = 45/55 (80%)
Frame = +1
Query: 494 PVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCEAMSTYPRTYDLIHADSLFSLY 548
PVWVMNVVP K +TL IY+RGLIGT+HDWCE S+YPRTYDL+HA+ LFS Y
Sbjct: 709 PVWVMNVVPASMK-NTLSGIYDRGLIGTFHDWCEPFSSYPRTYDLLHANYLFSQY 870
Score = 63.2 bits (152), Expect(5) = 3e-41
Identities = 27/47 (57%), Positives = 35/47 (74%)
Frame = +1
Query: 272 LPFPSRAFDISHCSRCLIPWAEYDGIFLNEVDRVLRPGGYWILSGPP 318
LPFPS +F++ HCSRC I + E DGI L E+DR+LR GY++ S PP
Sbjct: 94 LPFPSGSFEMIHCSRCRIDFHENDGILLKELDRLLRSNGYFVYSAPP 234
Score = 40.0 bits (92), Expect(5) = 3e-41
Identities = 16/29 (55%), Positives = 24/29 (82%)
Frame = +3
Query: 243 APRDTHEAQVQFALERGVPALIGVLASKR 271
AP+D HE Q+QFALERG+ A+I +++K+
Sbjct: 6 APKDGHENQIQFALERGIGAMISAMSTKQ 92
Score = 32.3 bits (72), Expect(5) = 3e-41
Identities = 28/135 (20%), Positives = 54/135 (39%), Gaps = 5/135 (3%)
Frame = +2
Query: 348 SLCWNKLIEKDDIAIWQKPINHLDC---RSARKLATDRPFCGPQENPDKAWYTDLKTCLM 404
++CW + K AIW K N C + +KL C + +W LK C+
Sbjct: 296 AMCWRLIARKVQTAIWIKENNQPSCLLQNAEQKLIN---VCDVDDESKPSWNIPLKNCIQ 466
Query: 405 --PVPQVSNKEETAGGVLKNWPQRLESVPPRIHMGTIEGVTSEGYSKDNELWKKRIPHYK 462
S K + L + ++L + G+ + ++ D+ W+ + H+
Sbjct: 467 VRSANAESYKLPPSHERLSVFSEKLNKI----------GIHRDEFTSDSIFWQDQTRHHW 616
Query: 463 KVNNQLGTKRYRNLV 477
++ N T+ RN++
Sbjct: 617 RLMNVKETE-IRNVI 658
Score = 24.3 bits (51), Expect(5) = 3e-41
Identities = 11/20 (55%), Positives = 14/20 (70%)
Frame = +3
Query: 471 KRYRNLVDMNANLGGFASAL 490
++Y +DMNA GGFA AL
Sbjct: 639 QKYAT*LDMNAFYGGFAVAL 698
>TC86178 similar to GP|6752888|gb|AAF27920.1| unknown {Malus x domestica},
partial (29%)
Length = 691
Score = 159 bits (401), Expect = 3e-39
Identities = 84/172 (48%), Positives = 106/172 (60%), Gaps = 2/172 (1%)
Frame = +3
Query: 25 FLIAFLCAAFYLLGAYQQRASFTSLTKKAI-ITSPSCTIQQVNKPTLDFQSHHNSSDTII 83
F++ LC FY+LGA+Q R+ F A+ IT + V P L F SHH + I
Sbjct: 186 FIVVGLCCFFYILGAWQ-RSGFGKGDSIALEITKNNAECDVV--PNLSFDSHHAGEVSQI 356
Query: 84 ALS-SETFNFPRCGVNFTEYTPCEDPTRSLRYKRSRMIYRERHCPVKGEEDLKCRVPPPH 142
S S T F C +T+YTPC+D R++ + R M YRERHCP + EE L C +P P
Sbjct: 357 DESNSNTKVFKPCEARYTDYTPCQDQRRAMTFPRENMNYRERHCPPE-EEKLHCMIPAPK 533
Query: 143 GYKTPFTWPASRDVAWYANVPHRELTVEKAVQNWIRYDGDRFFFPGGGTMFP 194
GY TPF WP SRD YAN P++ LTVEKA+QNWI+Y+G+ F FPGGGT FP
Sbjct: 534 GYVTPFPWPKSRDYVPYANAPYKSLTVEKAIQNWIQYEGNVFRFPGGGTQFP 689
>TC88058 similar to GP|21928175|gb|AAM78114.1 AT5g64030/MBM17_13
{Arabidopsis thaliana}, partial (39%)
Length = 1335
Score = 157 bits (398), Expect = 8e-39
Identities = 95/259 (36%), Positives = 145/259 (55%), Gaps = 11/259 (4%)
Frame = +2
Query: 299 LNEVDRVLRPGGYWILSGPPINWNKHHRGWQRTKKDLNQEQTKIEKVAKSLCWNKL---- 354
L E++R+LRPGG+++ S PI +Q+ +D+ + +++ + K++CW +
Sbjct: 5 LLELNRLLRPGGFFVWSATPI--------YQKLPEDV-EIWNEMKALTKAMCWEVVSISR 157
Query: 355 --IEKDDIAIWQKPINHLDCRSARKLATDRPFCGPQENPDKAWYTDLKTCLMPVPQVSNK 412
+ K IA+++KP ++ +C R + C ++P+ AW L+TC+ P S
Sbjct: 158 DKLNKVGIAVYKKPTSN-ECYEKRS-KNEPSICQDYDDPNAAWNIPLQTCMHKAPVSST- 328
Query: 413 EETAGGVLKNWPQRLESVP---PRIHMGTIEGVTSEGYSKDNELWKKRIPHYKKVNNQLG 469
E WP+RL P +G E ++ D+E WK+ + K N +G
Sbjct: 329 -ERGSQWPGEWPERLSKSPYWLSNSEVGVYGKPAPEDFTADHEHWKRVVS--KSYLNGIG 499
Query: 470 TK--RYRNLVDMNANLGGFASALVKNPVWVMNVVPVQAKVDTLGAIYERGLIGTYHDWCE 527
+ RN++DM + GGFA+AL+ +WVMNVVPV + DTL IYERGL G YHDWCE
Sbjct: 500 IQWSNVRNVMDMRSVYGGFAAALMDLKIWVMNVVPVDSP-DTLPIIYERGLFGIYHDWCE 676
Query: 528 AMSTYPRTYDLIHADSLFS 546
+ STYPR+YDL+HAD LFS
Sbjct: 677 SFSTYPRSYDLVHADHLFS 733
Score = 29.3 bits (64), Expect = 4.1
Identities = 27/119 (22%), Positives = 55/119 (45%), Gaps = 3/119 (2%)
Frame = +2
Query: 199 AYIDDIGKLINLKDGSIRTALDTGCGVASWGAYLQSRNIITLSLAPRDTHEAQVQFALER 258
+Y++ IG ++ ++R +D + A L I +++ P D+ + + ER
Sbjct: 479 SYLNGIG----IQWSNVRNVMDMRSVYGGFAAALMDLKIWVMNVVPVDSPDT-LPIIYER 643
Query: 259 GVPALIGVLASKRLPFPSRAFDISHCSRC---LIPWAEYDGIFLNEVDRVLRPGGYWIL 314
G+ + +P R++D+ H L +++ + + EVDR+LRP G I+
Sbjct: 644 GLFGIYHDWCESFSTYP-RSYDLVHADHLFSKLKKRCKFEAV-VAEVDRILRPEGKLIV 814
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.137 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,522,315
Number of Sequences: 36976
Number of extensions: 326121
Number of successful extensions: 1631
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 1524
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1560
length of query: 556
length of database: 9,014,727
effective HSP length: 101
effective length of query: 455
effective length of database: 5,280,151
effective search space: 2402468705
effective search space used: 2402468705
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149494.6


