

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
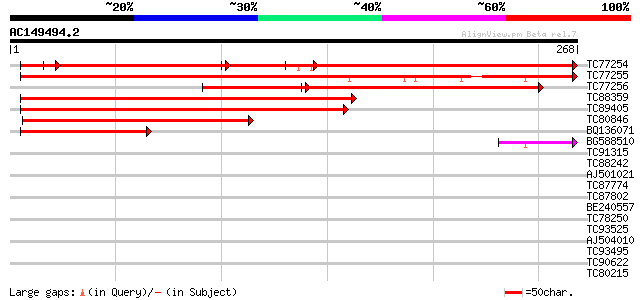
Query= AC149494.2 - phase: 0
(268 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77254 similar to GP|10334497|emb|CAC10210. hypothetical protei... 268 e-118
TC77255 homologue to GP|10334497|emb|CAC10210. hypothetical prot... 327 2e-90
TC77256 similar to GP|10334497|emb|CAC10210. hypothetical protei... 223 8e-59
TC88359 weakly similar to GP|6624711|emb|CAB51831.2 hypothetical... 154 3e-38
TC89405 weakly similar to GP|20160649|dbj|BAB89594. hypothetical... 138 2e-33
TC80846 weakly similar to GP|20160649|dbj|BAB89594. hypothetical... 76 1e-14
BQ136071 similar to PIR|T48609|T48 hypothetical protein F18O22.1... 60 1e-09
BG588510 41 6e-04
TC91315 weakly similar to GP|8777369|dbj|BAA96959.1 gene_id:MJE7... 33 0.12
TC88242 similar to GP|15100049|gb|AAK84220.1 transcription facto... 32 0.26
AJ501021 similar to PIR|C84587|C84 hypothetical protein At2g2028... 30 0.98
TC87774 similar to GP|7578871|gb|AAF64163.1| plastid-specific ri... 29 1.7
TC87802 similar to GP|11994617|dbj|BAB02754. contains similarity... 29 2.2
BE240557 similar to GP|11994240|dbj gb|AAF40458.1~gene_id:MUJ8.1... 29 2.2
TC78250 similar to GP|17380924|gb|AAL36274.1 unknown protein {Ar... 28 2.8
TC93525 weakly similar to GP|16902292|dbj|BAB71851. kinesin-rela... 28 2.8
AJ504010 GP|20516699|gb hypothetical protein {Thermoanaerobacter... 28 3.7
TC93495 similar to GP|21553756|gb|AAM62849.1 zinc finger protein... 28 4.8
TC90622 weakly similar to PIR|C86366|C86366 protein F26F24.2 [im... 28 4.8
TC80215 similar to GP|7547106|gb|AAF63778.1| unknown protein {Ar... 28 4.8
>TC77254 similar to GP|10334497|emb|CAC10210. hypothetical protein {Cicer
arietinum}, partial (62%)
Length = 1985
Score = 268 bits (685), Expect(3) = e-118
Identities = 135/139 (97%), Positives = 135/139 (97%), Gaps = 1/139 (0%)
Frame = +1
Query: 131 GDVKLQRKMQV-KRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGN 189
GDVKLQRK KRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGN
Sbjct: 1252 GDVKLQRKNAG*KRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGN 1431
Query: 190 HKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASS 249
HKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASS
Sbjct: 1432 HKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASS 1611
Query: 250 SSSSTNNSVHPKFKWEHFE 268
SSSSTNNSVHPKFKWEHFE
Sbjct: 1612 SSSSTNNSVHPKFKWEHFE 1668
Score = 150 bits (379), Expect(3) = e-118
Identities = 79/88 (89%), Positives = 83/88 (93%)
Frame = +2
Query: 17 GLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAK 76
G + +KIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAK
Sbjct: 908 GYTTKTKKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAK 1087
Query: 77 SISEISLEVDRLAGQVSALETIISKGGK 104
SIS+ISLEVD LAGQVSAL+TIISKGGK
Sbjct: 1088SISKISLEVDSLAGQVSALKTIISKGGK 1171
Score = 60.1 bits (144), Expect = 9e-10
Identities = 34/47 (72%), Positives = 38/47 (80%), Gaps = 1/47 (2%)
Frame = +3
Query: 101 KGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKL-QRKMQVKRVQK 146
K GKVVETDVLSLIEKLMNQLLKLDGIVADG + + K +VK+ K
Sbjct: 1161 KVGKVVETDVLSLIEKLMNQLLKLDGIVADGGC*IAEEKCRVKKSTK 1301
Score = 44.7 bits (104), Expect(3) = e-118
Identities = 19/19 (100%), Positives = 19/19 (100%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQK 24
GELKKMLTGPTGLHHQDQK
Sbjct: 874 GELKKMLTGPTGLHHQDQK 930
>TC77255 homologue to GP|10334497|emb|CAC10210. hypothetical protein {Cicer
arietinum}, partial (87%)
Length = 2000
Score = 327 bits (839), Expect = 2e-90
Identities = 184/278 (66%), Positives = 215/278 (77%), Gaps = 15/278 (5%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G+LKKML+GPTGLHHQDQK+FYK+KERDSKAFLD+VGVKDKSKLV++EDPI+QEKR LE+
Sbjct: 886 GDLKKMLSGPTGLHHQDQKLFYKDKERDSKAFLDVVGVKDKSKLVLVEDPISQEKRILEL 1065
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RKN KME+AAKSIS+ISLEVDRLAG+VSA E+IISKGGKVVE+D+L LIE LMNQLLKLD
Sbjct: 1066 RKNAKMEKAAKSISQISLEVDRLAGRVSAFESIISKGGKVVESDMLGLIELLMNQLLKLD 1245
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS-------NGGHVPKKKPQQKVKLPPI 178
GI+ADGDVKLQRKMQV+RVQKYVETLDMLK KNS N P++K +L I
Sbjct: 1246 GIIADGDVKLQRKMQVRRVQKYVETLDMLKAKNSMTNSNDVNAQVQPQQKHSNGKRLETI 1425
Query: 179 DEQLEGM-SIGNH----KLQPSLEQQSQRNSNGNSQVFQ-QLQHKPSTNSTSEVVVTTKW 232
EQ + + S G H +Q ++Q ++N N S VFQ Q QH+P S VVVTTKW
Sbjct: 1426 QEQPQQIHSNGYHGSIVPIQEQQQEQPRKNPNELSLVFQEQQQHQP-----SGVVVTTKW 1590
Query: 233 ETFDSLPPLI--PVTSASSSSSSTNNSVHPKFKWEHFE 268
ETFDS PPLI P TS ++ S + NNS PKF WE FE
Sbjct: 1591 ETFDSTPPLITFPSTSTTTPSMTNNNSGPPKFSWEFFE 1704
>TC77256 similar to GP|10334497|emb|CAC10210. hypothetical protein {Cicer
arietinum}, partial (34%)
Length = 879
Score = 223 bits (567), Expect = 8e-59
Identities = 111/114 (97%), Positives = 113/114 (98%)
Frame = +2
Query: 139 MQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQPSLEQ 198
++VKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLE MSIGNHKLQPSLEQ
Sbjct: 536 LKVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLERMSIGNHKLQPSLEQ 715
Query: 199 QSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSS 252
QSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSS
Sbjct: 716 QSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSS 877
Score = 93.6 bits (231), Expect = 7e-20
Identities = 49/51 (96%), Positives = 50/51 (97%)
Frame = +3
Query: 92 VSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVK 142
VSALETIISKGGKVVE DVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQV+
Sbjct: 54 VSALETIISKGGKVVEKDVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVR 206
>TC88359 weakly similar to GP|6624711|emb|CAB51831.2 hypothetical protein
{Oryza sativa}, partial (68%)
Length = 1084
Score = 154 bits (390), Expect = 3e-38
Identities = 77/159 (48%), Positives = 117/159 (73%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G +KK+++ TGL ++Q++ YK KER++ FLD+ GVKDKSKLV+++D + E+R+++M
Sbjct: 295 GVVKKIMSDETGLKVEEQRVIYKGKERENGEFLDVCGVKDKSKLVLIQDASSIERRFIQM 474
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
R N K++ A ++I+ +SL++D+LA QVSA+E IS G KV E +++LIE LM Q ++L+
Sbjct: 475 RINAKIQTANRAINNVSLQLDQLAEQVSAIEKSISDGVKVPEVQIITLIEMLMKQAIRLE 654
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHV 164
I A+GD Q+ +Q KRVQK VETLD+LK N+ HV
Sbjct: 655 SISAEGDASAQKILQGKRVQKCVETLDILKTSNATMKHV 771
>TC89405 weakly similar to GP|20160649|dbj|BAB89594. hypothetical
protein~similar to Arabidopsis thaliana chromosome3
At3g51780, partial (39%)
Length = 930
Score = 138 bits (348), Expect = 2e-33
Identities = 74/155 (47%), Positives = 107/155 (68%)
Frame = +3
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G+LKK+L TGL + Q++F++ E+D K L GVK KSK+++ME ++EK+ E
Sbjct: 204 GDLKKLLVPKTGLEPEQQRLFFRGIEKDDKEQLHQEGVKGKSKILLMERAASKEKKLEET 383
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RK +M +A ++I+ + EVD+L+ +V+ALE I+ G K E + L L E LM+QLLKLD
Sbjct: 384 RKLNEMSKATEAINGVRAEVDKLSDRVAALEAAINAGKKASEKEFLLLTELLMSQLLKLD 563
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSN 160
GI A+G+ KLQRK +V+RVQ V+ +D LK KNSN
Sbjct: 564 GIEAEGEAKLQRKAEVRRVQGLVDMVDSLKAKNSN 668
>TC80846 weakly similar to GP|20160649|dbj|BAB89594. hypothetical
protein~similar to Arabidopsis thaliana chromosome3
At3g51780, partial (13%)
Length = 670
Score = 76.3 bits (186), Expect = 1e-14
Identities = 38/109 (34%), Positives = 70/109 (63%)
Frame = +1
Query: 7 ELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMR 66
++KK+L TGL + Q +F+ KE++++ L + GVKD SKL+++ED ++E E+R
Sbjct: 340 DVKKLLAHKTGLKPEQQILFFNGKEKENEENLHMEGVKDMSKLLLLEDAASKESNIEEVR 519
Query: 67 KNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIE 115
K +M +A ++++ + EVD+L +VSAL+ ++ G KV + + + E
Sbjct: 520 KQNEMLKAFQAVAVVGSEVDKLCDRVSALDVAVNGGTKVSDKEFVVTTE 666
>BQ136071 similar to PIR|T48609|T48 hypothetical protein F18O22.150 -
Arabidopsis thaliana, partial (65%)
Length = 673
Score = 59.7 bits (143), Expect = 1e-09
Identities = 26/62 (41%), Positives = 43/62 (68%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELK +L+ T L ++Q++ YK KERD FL ++GV+DK K++++EDP +E + L +
Sbjct: 301 GELKMVLSLVTSLEPREQRLLYKGKERDDNEFLHMIGVRDKDKVLLLEDPAIKEMKLLGL 480
Query: 66 RK 67
+
Sbjct: 481 AR 486
>BG588510
Length = 132
Score = 40.8 bits (94), Expect = 6e-04
Identities = 21/39 (53%), Positives = 22/39 (55%), Gaps = 2/39 (5%)
Frame = +3
Query: 232 WETFDSLPPLI--PVTSASSSSSSTNNSVHPKFKWEHFE 268
WET D PPLI P T SS S + N S KF WE FE
Sbjct: 3 WETLDCTPPLITFPSTCTSSPSMTNNTSGAAKFTWEFFE 119
>TC91315 weakly similar to GP|8777369|dbj|BAA96959.1
gene_id:MJE7.2~pir||C71412~similar to unknown protein
{Arabidopsis thaliana}, partial (3%)
Length = 1044
Score = 33.1 bits (74), Expect = 0.12
Identities = 31/112 (27%), Positives = 53/112 (46%), Gaps = 8/112 (7%)
Frame = +2
Query: 31 ERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM---RKNIKMERAAKSISEISLEVDR 87
E+ ++AFL++ K S+ V D I Q LEM +KN +++ + E LE++
Sbjct: 164 EQLAQAFLELEAQKGASEDKVQWDEIKQHFSDLEMALNKKNEELQAKEREYEEKQLEMNT 343
Query: 88 LAGQVSALET-----IISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVK 134
L Q A T ++ + ++ +T V ++IE N V DG+ K
Sbjct: 344 LLTQRKAAVTSKEQDLLDRLQELKDTAVAAIIEARPNDETTTFDFVYDGEDK 499
>TC88242 similar to GP|15100049|gb|AAK84220.1 transcription factor bZIP29
{Arabidopsis thaliana}, partial (39%)
Length = 1930
Score = 32.0 bits (71), Expect = 0.26
Identities = 30/114 (26%), Positives = 46/114 (40%)
Frame = +2
Query: 145 QKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQPSLEQQSQRNS 204
Q + T+ N GG +P P Q +PPI S N+++ S Q ++
Sbjct: 206 QSQMRTVTRNHHHNQRGGGIPPSHPHQ---IPPISPY----SHMNNQIPVSRPQMPSHST 364
Query: 205 NGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSV 258
+ T S + + + + DSLPPL P T SSS+S + V
Sbjct: 365 S-------------PTPSHTRSLSQPSFFSLDSLPPLSPCTFRESSSTSDHADV 487
>AJ501021 similar to PIR|C84587|C84 hypothetical protein At2g20280 [imported]
- Arabidopsis thaliana, partial (42%)
Length = 611
Score = 30.0 bits (66), Expect = 0.98
Identities = 21/79 (26%), Positives = 40/79 (50%), Gaps = 1/79 (1%)
Frame = +2
Query: 123 KLDGIVADGDVKLQRKMQVKRVQKYVETL-DMLKIKNSNGGHVPKKKPQQKVKLPPIDEQ 181
K +V D L+ K + K VQKYV+ L ++ + KKK ++K K +++
Sbjct: 158 KKQKVVEDKTFGLKNKNKSKAVQKYVQNLKSSVQPRTDPKQDAKKKKEEEKAKEKELND- 334
Query: 182 LEGMSIGNHKLQPSLEQQS 200
L +++ K+ P ++ +S
Sbjct: 335 LFKIAVSQPKVPPGVDPKS 391
>TC87774 similar to GP|7578871|gb|AAF64163.1| plastid-specific ribosomal
protein 3 precursor {Spinacia oleracea}, partial (52%)
Length = 864
Score = 29.3 bits (64), Expect = 1.7
Identities = 17/53 (32%), Positives = 24/53 (45%)
Frame = +1
Query: 211 FQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSVHPKFK 263
F LQH +N T+ ++ + T S+ L+P TS S NN K K
Sbjct: 4 FHSLQHSTESNPTTMNMLASSSSTTSSMSVLLPTTSKPLFSIRPNNVHMNKLK 162
>TC87802 similar to GP|11994617|dbj|BAB02754. contains similarity to kinesin
protein~gene_id:MGL6.9 {Arabidopsis thaliana}, partial
(17%)
Length = 846
Score = 28.9 bits (63), Expect = 2.2
Identities = 23/88 (26%), Positives = 45/88 (51%), Gaps = 12/88 (13%)
Frame = -3
Query: 100 SKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQ-------RKMQVK---RVQKYVE 149
S+GG + T ++L L Q + D G ++ + RK+++K R++K +E
Sbjct: 421 SRGGSIALTTHVAL*ATLGRQFSRSDYTRMRGKLESEEEHESDERKVELK*NGRMKKRME 242
Query: 150 T--LDMLKIKNSNGGHVPKKKPQQKVKL 175
+M+K ++ +GG K+KP+ K+
Sbjct: 241 NGEFEMMKFRSESGGKKEKQKPETMGKV 158
>BE240557 similar to GP|11994240|dbj gb|AAF40458.1~gene_id:MUJ8.11~similar to
unknown protein {Arabidopsis thaliana}, partial (6%)
Length = 349
Score = 28.9 bits (63), Expect = 2.2
Identities = 22/64 (34%), Positives = 29/64 (44%), Gaps = 6/64 (9%)
Frame = +1
Query: 207 NSQVFQQLQHKPSTNSTSEVVVT-TKWETFDSL-----PPLIPVTSASSSSSSTNNSVHP 260
NS +F +Q T V+VT TK+E L PP P S S ++TN S
Sbjct: 154 NSLLFLHIQVAIPVVPTIRVLVTFTKFEELQPLYEFATPPTSPSMSREESPATTNFSASS 333
Query: 261 KFKW 264
F+W
Sbjct: 334 WFQW 345
>TC78250 similar to GP|17380924|gb|AAL36274.1 unknown protein {Arabidopsis
thaliana}, partial (52%)
Length = 1148
Score = 28.5 bits (62), Expect = 2.8
Identities = 19/49 (38%), Positives = 25/49 (50%), Gaps = 2/49 (4%)
Frame = +3
Query: 207 NSQVFQQLQHKPSTNSTS--EVVVTTKWETFDSLPPLIPVTSASSSSSS 253
+SQ F Q K S TS E++VTT +T L+ + S SSSS
Sbjct: 522 SSQRFSSSQKKTSKQLTSCGEIIVTTNLDTVSKRNYLVRLKKTSPSSSS 668
>TC93525 weakly similar to GP|16902292|dbj|BAB71851. kinesin-related protein
{Arabidopsis thaliana}, partial (3%)
Length = 798
Score = 28.5 bits (62), Expect = 2.8
Identities = 14/33 (42%), Positives = 18/33 (54%)
Frame = +3
Query: 235 FDSLPPLIPVTSASSSSSSTNNSVHPKFKWEHF 267
F P P +S SSSSS TN + P+F H+
Sbjct: 438 FSHRKPSTPYSSTSSSSSLTNGRIIPRFFIFHY 536
>AJ504010 GP|20516699|gb hypothetical protein {Thermoanaerobacter
tengcongensis}, partial (9%)
Length = 285
Score = 28.1 bits (61), Expect = 3.7
Identities = 23/81 (28%), Positives = 35/81 (42%), Gaps = 3/81 (3%)
Frame = +3
Query: 183 EGMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNST---SEVVVTTKWETFDSLP 239
EG I P L ++ N + + + PS+N+ + +T T SLP
Sbjct: 36 EGSVIPTFGFSP-LTTSVRKQENNGATCWIWVPPSPSSNTLVTKTTTSITESSSTTLSLP 212
Query: 240 PLIPVTSASSSSSSTNNSVHP 260
P P T SSS S + +S+ P
Sbjct: 213 PCFPFTFPSSSISISRSSLIP 275
>TC93495 similar to GP|21553756|gb|AAM62849.1 zinc finger protein-like
{Arabidopsis thaliana}, partial (39%)
Length = 819
Score = 27.7 bits (60), Expect = 4.8
Identities = 12/37 (32%), Positives = 24/37 (64%)
Frame = -3
Query: 222 STSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSV 258
S +++T K T ++PP +P+ S++SSS + ++V
Sbjct: 310 SRLSLILTIKPSTVVTIPPRVPLPSSNSSSGTFTHAV 200
>TC90622 weakly similar to PIR|C86366|C86366 protein F26F24.2 [imported] -
Arabidopsis thaliana, partial (10%)
Length = 537
Score = 27.7 bits (60), Expect = 4.8
Identities = 22/101 (21%), Positives = 42/101 (40%)
Frame = +1
Query: 98 IISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVKRVQKYVETLDMLKIK 157
++ K E ++ + + N LDG+ G V+ + + +TL +
Sbjct: 1 MVEKSSYTKEDAIIWVSAMMTNHKTCLDGLEEKGYVQANQVLDRNLTSLLGQTL----VL 168
Query: 158 NSNGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQPSLEQ 198
SN K++PQ+ L D L+ S+ NH+ ++ Q
Sbjct: 169 YSNNKIKVKEQPQRSTILEENDGLLDSWSLANHRADFTVAQ 291
>TC80215 similar to GP|7547106|gb|AAF63778.1| unknown protein {Arabidopsis
thaliana}, partial (50%)
Length = 1486
Score = 27.7 bits (60), Expect = 4.8
Identities = 22/85 (25%), Positives = 34/85 (39%), Gaps = 5/85 (5%)
Frame = -1
Query: 8 LKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSK-----LVVMEDPIAQEKRY 62
L K + H Q+ K FY +SK I+ + K K L +M + R
Sbjct: 268 LSKWAISRSSQHQQNGK*FYSKLLENSKFLYQILKLNSKMKVDK*ILKLMNASVKSNLRL 89
Query: 63 LEMRKNIKMERAAKSISEISLEVDR 87
+ RK + ER + + + VDR
Sbjct: 88 VRRRKQSERERWMIIVFLVRVVVDR 14
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.311 0.129 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,829,165
Number of Sequences: 36976
Number of extensions: 84048
Number of successful extensions: 721
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 674
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 710
length of query: 268
length of database: 9,014,727
effective HSP length: 94
effective length of query: 174
effective length of database: 5,538,983
effective search space: 963783042
effective search space used: 963783042
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC149494.2


