

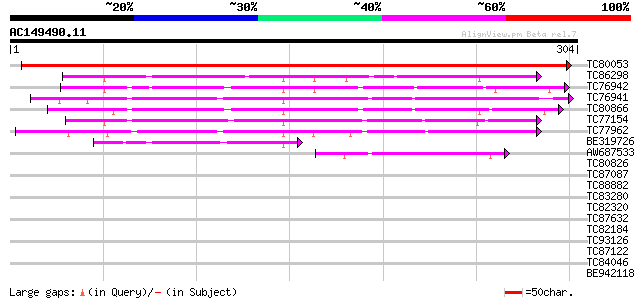
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.11 + phase: 0
(304 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC80053 similar to PIR|JC7335|JC7335 chitinase (EC 3.2.1.14) 1 -... 437 e-123
TC86298 weakly similar to PIR|S49878|S49878 probable narbonin - ... 145 2e-35
TC76942 weakly similar to PIR|S44033|S44033 narbonin - Vicia pan... 139 1e-33
TC76941 similar to PIR|S44033|S44033 narbonin - Vicia pannonica,... 135 3e-32
TC80866 weakly similar to PIR|T12156|T12156 nodulin isoform Nvf... 134 3e-32
TC77154 weakly similar to PIR|S49878|S49878 probable narbonin - ... 124 3e-29
TC77962 similar to PIR|S49848|S49848 probable narbonin - jack be... 122 2e-28
BE319726 weakly similar to PIR|S49848|S49 probable narbonin - ja... 55 3e-08
AW687533 weakly similar to PIR|T12157|T121 nodulin - fava bean, ... 45 3e-05
TC80826 weakly similar to GP|19577787|emb|CAD28575. unnamed prot... 39 0.002
TC87087 similar to GP|20385508|gb|AAM21317.1 auxin-regulated pro... 32 0.30
TC88882 weakly similar to GP|11321444|gb|AAG34171.1 chitinase {A... 31 0.67
TC83280 weakly similar to GP|22136056|gb|AAM91610.1 putative pro... 28 0.85
TC82320 similar to GP|8953735|dbj|BAA98099.1 contains similarity... 30 0.88
TC87632 weakly similar to GP|21536611|gb|AAM60943.1 putative rib... 30 1.5
TC82184 homologue to PIR|D96507|D96507 hypothetical protein T12C... 29 2.0
TC93126 29 2.0
TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltran... 29 2.0
TC84046 29 2.0
BE942118 29 2.6
>TC80053 similar to PIR|JC7335|JC7335 chitinase (EC 3.2.1.14) 1 - cone shell
(Conus tulipa), partial (81%)
Length = 1081
Score = 437 bits (1125), Expect = e-123
Identities = 215/297 (72%), Positives = 253/297 (84%), Gaps = 2/297 (0%)
Frame = +2
Query: 7 KPCFILLILTTSLFA-STILAANSNL-FREYIGAQFKGVKFSDVPINPNVNFHFILSFAI 64
K FILL+ +S +T L++++NL FREYIGA+ +KFSDVPINPNV FHFILSF I
Sbjct: 83 KTFFILLLFISSTHVGATRLSSSTNLIFREYIGAESNNIKFSDVPINPNVEFHFILSFGI 262
Query: 65 DYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPS 124
DYDTSS PSPTNG FNIFWDT+NL+PS+VS IK+QNPNVKVALSLGGD+V+G F PS
Sbjct: 263 DYDTSSSPSPTNGKFNIFWDTKNLNPSQVSSIKSQNPNVKVALSLGGDSVEGGYAYFDPS 442
Query: 125 SIDSWVSNAVSSLTSIIKTYNLDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFAS 184
S++SW+SNAVSSLT IIK YNLDGID+DYEHF G+PNTFAECIGRLI TLK N VI+FAS
Sbjct: 443 SVESWLSNAVSSLTKIIKEYNLDGIDIDYEHFKGNPNTFAECIGRLIKTLKANGVITFAS 622
Query: 185 IAPFDDAEVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVL 244
IAPFDD +VQ+HYLALWKSY ++IDYVNFQFYAYDK T+V+QFIDY+NKQSSNYN KVL
Sbjct: 623 IAPFDDDQVQSHYLALWKSYGHLIDYVNFQFYAYDKGTSVSQFIDYFNKQSSNYNGGKVL 802
Query: 245 VSFVSDGSTGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDSKADGFRYEKEAQEVLA 301
VSF+SDGS GL P +GF C+ LKSQQKL+GIFV+SADDS +GFR+EK++Q +LA
Sbjct: 803 VSFLSDGSGGLSPSDGFFKACQRLKSQQKLHGIFVWSADDSMGNGFRFEKQSQALLA 973
>TC86298 weakly similar to PIR|S49878|S49878 probable narbonin - soybean,
partial (71%)
Length = 1281
Score = 145 bits (366), Expect = 2e-35
Identities = 105/281 (37%), Positives = 149/281 (52%), Gaps = 24/281 (8%)
Frame = +1
Query: 29 SNLFREYIGAQFKGVKFSDVP---INPNVN-FHFILSFAID-YDTSSPPSPTNGNFNIFW 83
S +FREYIG + D P I N++ FHFIL FA + YD + + G F W
Sbjct: 133 SKIFREYIGVKPFSTNLRDFPVEIIKTNISEFHFILGFATEEYDAQNKGT---GVFKETW 303
Query: 84 DTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKT 143
+T+ P V +K NPNVKV +S+GG+ P N + +I W++ AVSSL II+
Sbjct: 304 NTRAFGPEAVRNLKGNNPNVKVVISIGGNDTVKTPFNPVEETI--WITRAVSSLKVIIQK 477
Query: 144 YN------LDGIDVDYEHFIGDPNT-----FAECIGRLITTLKNNNV--ISFASIAPFDD 190
Y +DGID++Y + N FA CIG +IT LKN+N I SIAP +
Sbjct: 478 YKDQTGNIIDGIDINYLNVFHTTNDTGKLRFARCIGEVITQLKNDNYLRIKIVSIAPSET 657
Query: 191 AEVQNHYLAL-WKSYSNIIDYVNFQFYAYDKS-TTVAQFIDYYNKQSSNYNSEKVLVSFV 248
E+ HY L W++ +N I++VN+QFY K+ +T+ F+ Y++ S NY VL
Sbjct: 658 NEI--HYRNLFWQNEAN-INWVNYQFYNQSKAVSTLDDFLKLYDQVSRNYKPSIVLPGVS 828
Query: 249 SD----GSTGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDS 285
+D P F+ GCR L L G+F+++ADDS
Sbjct: 829 TDKLHIEPVDKMPREHFIAGCRHLLQIASLPGVFLWNADDS 951
>TC76942 weakly similar to PIR|S44033|S44033 narbonin - Vicia pannonica,
partial (45%)
Length = 1361
Score = 139 bits (350), Expect = 1e-33
Identities = 106/295 (35%), Positives = 152/295 (50%), Gaps = 22/295 (7%)
Frame = +3
Query: 28 NSNLFREYIGAQFKGVKFSDVP---INPNV-NFHFILSFAIDYDTSSPPSPTNGNFNIFW 83
N +FREYIG + ++ P I ++ FHFIL FA ++T T G FN W
Sbjct: 75 NPVIFREYIGVKSYPDSLNNFPADIIGRHIPEFHFILGFA--HETYVDGKGT-GIFNASW 245
Query: 84 DTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKT 143
P V IK + NVKV +S+GG + F P+ W NAV SL I +
Sbjct: 246 KIPFFGPDNVDDIKTNHGNVKVVISIGGRDTK---YPFHPAHKLEWCDNAVESLKKIFQL 416
Query: 144 YN--------LDGIDVDYEHFIGDPNT--FAECIGRLITTLKNNNVISFASIAPFDDAEV 193
YN +DGID++YE+ D + F+ CIG +I LK + I SIAP E
Sbjct: 417 YNRTNSCYNLIDGIDINYEYIHPDVSEEDFSYCIGDVIKRLKKDVGIDVVSIAP--SHET 590
Query: 194 QNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVSDGST 253
Q HY L+ + +N I++VN+QFY D + +F++ + S Y S+K+L +D +
Sbjct: 591 QKHYKTLYLARTNDINWVNYQFYI-DTLKSKDEFVNLFLNLSDEYGSKKLLAGASTDPAD 767
Query: 254 GLGPGN----GFLDGCRMLKSQQKLNGIFVYSADDSKAD----GFRYEKEAQEVL 300
G G FL+GC L S Q L GIF+++A+DS ++ F EK+AQE+L
Sbjct: 768 A-GKGKLSREDFLEGCVDLHSTQSLRGIFIWNANDSASNPNGKPFSLEKKAQEIL 929
>TC76941 similar to PIR|S44033|S44033 narbonin - Vicia pannonica, partial
(23%)
Length = 1119
Score = 135 bits (339), Expect = 3e-32
Identities = 101/300 (33%), Positives = 154/300 (50%), Gaps = 9/300 (3%)
Frame = +3
Query: 12 LLILTTSLFASTIL---AANSNLFREYIGAQF--KGVKFSDVPINPNVNFHFILSFAIDY 66
LL ++T + I+ A + +FREYIG + K KF D+ FH IL+FA +
Sbjct: 42 LLQISTFTMSHPIIDNNAVDRVIFREYIGVKPYPKPFKFPDIDNYKIAEFHIILAFA--H 215
Query: 67 DTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSI 126
+T + G+F+ WD V IK + PNVK+ S+GG +G F P
Sbjct: 216 ETYNEDGKGTGSFSADWDLDICGIQSVKDIKQKCPNVKLVFSIGG---RGTKYPFSPIEK 386
Query: 127 DSWVSNAVSSLTSIIKTYN--LDGIDVDYEHF-IGDPNTFAECIGRLITTLKNNNVISFA 183
+ W NAV SL +IIK YN GID++YEH D N F+ +G +I LKN I
Sbjct: 387 NYWCDNAVDSLKTIIKQYNDIFAGIDINYEHINTNDENDFSNYVGDVINRLKNEVGIDVV 566
Query: 184 SIAPFDDAEVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKV 243
SIAP + N+Y L+ ++++ I++V++QFY T +F+ + + Y EK+
Sbjct: 567 SIAPSHAND--NYYKLLYSAHADDINWVDYQFY-MQPIPTENEFLSLFLSLAREYALEKL 737
Query: 244 LVSFVSDGSTGLG-PGNGFLDGCRMLKSQQKLNGIFVYSADDSKADGFRYEKEAQEVLAS 302
LV +D G P + F+ C L + L+GIF+++A+D YEK A ++L +
Sbjct: 738 LVGASTDPRDGGNVPLDVFVQTCTNLIKHKSLSGIFIWNAND-------YEKIALDILTN 896
>TC80866 weakly similar to PIR|T12156|T12156 nodulin isoform Nvf32-A1 -
fava bean, partial (40%)
Length = 970
Score = 134 bits (338), Expect = 3e-32
Identities = 92/292 (31%), Positives = 147/292 (49%), Gaps = 15/292 (5%)
Frame = +1
Query: 21 ASTILAANSNLFREYIGAQFKGVKFSDVPINPNV--NFHFILSFAIDYDTSSPPSPTNGN 78
+S + + +FREYIG + ++ P + +FHFIL FA D+ + GN
Sbjct: 46 SSVTMXVDPFIFREYIGVKPYPASLNNFPYEIIIAKHFHFILGFA--NDSYNEEGKGTGN 219
Query: 79 FNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLT 138
FN W++ P V +K + P+VKV +S+GG F P++ + W NAV SL
Sbjct: 220 FNANWNSDFFGPQNVMALKRKYPHVKVVISIGGRDAN---FPFFPAAREEWCGNAVDSLK 390
Query: 139 SIIKTYN---------LDGIDVDYEHFIGDPNTFAECIGRLITTLKNNNVISFASIAPFD 189
II++YN +DGID+ Y++ + N F+ +G +I LK I SIAP
Sbjct: 391 EIIRSYNDCSVEDNILIDGIDIFYDYINTNENDFSNYVGDVINRLKKEVRIDVVSIAP-- 564
Query: 190 DAEVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFVS 249
E HY L+ + ++ I++VN+QFY + F++ + + Y+S K+LV S
Sbjct: 565 SHETHKHYKELYLACTDDINWVNYQFY-MQPIPSKNDFLNLFLNLAKEYDSNKLLVGGSS 741
Query: 250 D--GSTGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDS--KADGFRYEKEAQ 297
D + P + F++ C L + L GIF+++A+DS F EK+ Q
Sbjct: 742 DPIDADNFNP-DEFVEACNDLHKTKSLRGIFIWNANDSANNVPPFYLEKKLQ 894
>TC77154 weakly similar to PIR|S49878|S49878 probable narbonin - soybean,
partial (32%)
Length = 1260
Score = 124 bits (312), Expect = 3e-29
Identities = 90/280 (32%), Positives = 145/280 (51%), Gaps = 25/280 (8%)
Frame = +1
Query: 31 LFREYIGAQFKGVKFSDVP---INPNVN-FHFILSFAIDYDTSSPPSPTNGNFNIFWDTQ 86
+FREYIG + D P IN ++ FHFIL + S G F W+
Sbjct: 94 IFREYIGVKDIPKNLKDFPAEMINDDIEEFHFILGTL--REVYSGDGKGKGEFYRTWNFN 267
Query: 87 NLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN- 145
N SP++V+ +K + NVKV +S+GG + F P I+SW + A S+ +I Y
Sbjct: 268 NFSPAKVAKLKKDHKNVKVIISIGGFGAENP---FNPKEIESWSTKAKQSIKKLINEYQE 438
Query: 146 ----------------LDGIDVDYEHFIGDPNTFAECIGRLITTL-KNNNVISFASIAPF 188
+DGID++YE+ +P+ F+ CIG LI L K++ I SIAP
Sbjct: 439 YSKDSSSTDECHCDDIIDGIDINYEYSNCNPDEFSSCIGELIRKLKKSSKSIKLVSIAPT 618
Query: 189 DDAEVQNHYLALWKSYSNIIDYVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSEKVLVSFV 248
+ ++ HY L+ + +II++V+++FY S + + ++ YNK + Y ++ L+ V
Sbjct: 619 E--LLKPHYHKLYWANKDIINWVDYKFYNQTVS-SADELVNLYNKLLNEYGTDVKLLPGV 789
Query: 249 S---DGSTGLGPGNGFLDGCRMLKSQQKLNGIFVYSADDS 285
S D +T + + F+ GC+ L + L GIFV++A+DS
Sbjct: 790 STDPDSNTNM-TRDVFIKGCKSLLESESLPGIFVWNANDS 906
>TC77962 similar to PIR|S49848|S49848 probable narbonin - jack bean, partial
(18%)
Length = 1239
Score = 122 bits (305), Expect = 2e-28
Identities = 98/317 (30%), Positives = 155/317 (47%), Gaps = 35/317 (11%)
Frame = +2
Query: 4 HIPKPCFILLILTTSLFASTILAANSN----------LFREYIGAQFKGVKFSDVPIN-- 51
H KP + LT S+F+ +I ++ + +FREY+G + + D P+N
Sbjct: 2 HQIKPTLSINFLTFSIFSFSITMSDHSQNIKTVLRPKIFREYVGVKDEPETLDDFPVNII 181
Query: 52 -PNVN-FHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSL 109
+VN FHFIL FA + + G+F W+ SP +V K + N+KV +++
Sbjct: 182 HDDVNQFHFILGFATE---AYKDGKGTGHFIRDWNFDYFSPEKVFEHKKKYKNMKVMITI 352
Query: 110 GGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN--------------LDGIDVDYEH 155
GG G F P W+ NA SS+ II+ Y +DGID++YE+
Sbjct: 353 GG---HGPKYPFNPKEKKVWIFNATSSIRHIIQDYENYLVNDNSCHCTSIIDGIDINYEY 523
Query: 156 FIGDPN--TFAECIGRLITTLKNNNVIS----FASIAPFDDAEVQNHYLALWKSYSNIID 209
F+ CIG +I LK + +S + SIAP + +Q HY L+ + I+
Sbjct: 524 IDSSVTGADFSNCIGEVIKRLKKDKHVSKSMEYVSIAPTE--LLQAHYRTLFWDHKMNIN 697
Query: 210 YVNFQFYAYDKSTTVAQFIDYYNKQSSNYNSE-KVLVSFVSDGSTGLGPGNGFLDGCRML 268
YV+++FY ST +F + YN+ ++Y E K+LV +D S F++G L
Sbjct: 698 YVDYKFYNQTISTE-NEFDELYNQLVTDYGGELKLLVGVSTDPSDTKMKRQVFIEGVTRL 874
Query: 269 KSQQKLNGIFVYSADDS 285
+ + L G+FV+SA+DS
Sbjct: 875 INNKSLPGLFVWSANDS 925
>BE319726 weakly similar to PIR|S49848|S49 probable narbonin - jack bean,
partial (42%)
Length = 448
Score = 55.1 bits (131), Expect = 3e-08
Identities = 38/119 (31%), Positives = 55/119 (45%), Gaps = 7/119 (5%)
Frame = +3
Query: 46 SDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKV 105
S PI+ N + L A YD + G F W+ + P +V +K N NVKV
Sbjct: 111 SSTPISQNSTSFWALQ-ARRYDDKKRGT---GVFKTTWNVEFFGPEDVKRLKENNKNVKV 278
Query: 106 ALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYN-------LDGIDVDYEHFI 157
+S GG + F P+ + W AV+SL II + +DGID++YEH +
Sbjct: 279 VISFGGCD---EKTPFNPAEDNIWTEKAVASLKVIILRFKDQSGRSIIDGIDINYEHIL 446
Score = 32.3 bits (72), Expect = 0.23
Identities = 19/46 (41%), Positives = 25/46 (54%), Gaps = 4/46 (8%)
Frame = +2
Query: 22 STILAANSNLFREYIGAQFKGVKFSDVP---INPNVN-FHFILSFA 63
S ++ +FREYIG + D P IN N++ FHFIL FA
Sbjct: 20 SFLIVRMPKIFREYIGVKPYSKSLRDFPINIINSNISEFHFILGFA 157
>AW687533 weakly similar to PIR|T12157|T121 nodulin - fava bean, partial
(30%)
Length = 387
Score = 45.1 bits (105), Expect = 3e-05
Identities = 35/114 (30%), Positives = 59/114 (51%), Gaps = 10/114 (8%)
Frame = +3
Query: 165 ECIGRLITTLKNNN--VISFASIAPFDDAEVQNHYLALWKSYSNIIDYVNFQFYAYDKST 222
EC+G++IT LK + I+ SIAP + + +HY L+ + I+ V+++ Y K
Sbjct: 36 ECLGQVITDLKKDRDLNINVVSIAPSEQND--SHYRKLYWENKDNINLVDYKLYNQTKIV 209
Query: 223 -TVAQFIDYYNKQSSNYNSEKVLVSFVSD-GSTGLG------PGNGFLDGCRML 268
T +F+ Y+K +++Y+ EK L +D G T P F+ GC+ L
Sbjct: 210 QTSEEFVKLYSKIANDYSPEKFLPGISTDPGDTEPADKIIKMPREIFIAGCKHL 371
>TC80826 weakly similar to GP|19577787|emb|CAD28575. unnamed protein product
{Helianthus annuus}, partial (24%)
Length = 1285
Score = 39.3 bits (90), Expect = 0.002
Identities = 36/153 (23%), Positives = 67/153 (43%), Gaps = 9/153 (5%)
Frame = +3
Query: 11 ILLILTTSLFASTILAANSNLFREYIGAQFK------GVKFSDVPINPNVNFHFILSFAI 64
IL+ L T F + ++++N +++ + G FS I+ N H +L+F
Sbjct: 36 ILVFLVTIFFNVSSSSSSNNSQYQFLNHGVRSAYWPAGDDFSPSLIDTNYFTHILLAFI- 212
Query: 65 DYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGDPVNFIPS 124
P P + I + + +++++P VK LS+GG G + + S
Sbjct: 213 ------QPEPISFKLEITKSGIKWGQNFIKALRHRSPPVKTLLSIGG----GGSNSTLFS 362
Query: 125 SIDSWVSNA---VSSLTSIIKTYNLDGIDVDYE 154
I S N ++S + + Y DG+D+D+E
Sbjct: 363 EIASTKQNREIFINSTIEVARKYRFDGVDLDWE 461
>TC87087 similar to GP|20385508|gb|AAM21317.1 auxin-regulated protein
{Populus tremula x Populus tremuloides}, partial (69%)
Length = 1157
Score = 32.0 bits (71), Expect = 0.30
Identities = 26/102 (25%), Positives = 46/102 (44%), Gaps = 8/102 (7%)
Frame = +3
Query: 176 NNNVISFASIAPFDDAEVQNHYLALWKSYSNIIDYVNFQFYAYD----KSTTVAQFIDYY 231
NNN ISF ++ ++ L ++KSY + D + F ++ +S + F++
Sbjct: 483 NNNPISFVKVSMDGAPYLRKVDLKMYKSYPELSDALAKMFNSFTTGNCESQGIKDFMNES 662
Query: 232 NKQSSNYNSEKVLVSFV-SDGS---TGLGPGNGFLDGCRMLK 269
NK NS + ++ DG G P F+D C+ L+
Sbjct: 663 NKLMDLLNSSDYVPTYEDKDGDWMLVGDVPWEMFIDSCKRLR 788
>TC88882 weakly similar to GP|11321444|gb|AAG34171.1 chitinase {Aspergillus
nidulans}, partial (10%)
Length = 1510
Score = 30.8 bits (68), Expect = 0.67
Identities = 28/99 (28%), Positives = 44/99 (44%), Gaps = 13/99 (13%)
Frame = +3
Query: 131 SNAVSSLTSIIKTYNLDGIDVDYEHFIGDPNTFAECIGRLITTL--------KN---NNV 179
S + +L S ++ Y DG+D D+E+ D AE G+ T L KN V
Sbjct: 1002 SKFIGNLLSFMRQYGFDGVDFDWEYPGADDRGGAEEDGKSFTLLLEELKAAIKNQPLEYV 1181
Query: 180 ISFASIAPFDDAEVQNHYLALW--KSYSNIIDYVNFQFY 216
+SF + F YL + K+ ++ +D+VN Y
Sbjct: 1182 VSFTTPTSF-------WYLRHFDLKASTDAVDFVNIMSY 1277
>TC83280 weakly similar to GP|22136056|gb|AAM91610.1 putative protein
{Arabidopsis thaliana}, partial (52%)
Length = 1230
Score = 27.7 bits (60), Expect(2) = 0.85
Identities = 13/29 (44%), Positives = 16/29 (54%)
Frame = +1
Query: 137 LTSIIKTYNLDGIDVDYEHFIGDPNTFAE 165
L+ I LDG++VD E I DP AE
Sbjct: 583 LSRAIYMLKLDGVEVDEEQTISDPELLAE 669
Score = 21.2 bits (43), Expect(2) = 0.85
Identities = 7/21 (33%), Positives = 11/21 (52%)
Frame = +2
Query: 78 NFNIFWDTQNLSPSEVSFIKN 98
N+ + W QNL E+ + N
Sbjct: 479 NYILIWCIQNLRKKEIMLLNN 541
>TC82320 similar to GP|8953735|dbj|BAA98099.1 contains similarity to unknown
protein~emb|CAB85560.1~gene_id:K10D11.2 {Arabidopsis
thaliana}, partial (30%)
Length = 875
Score = 30.4 bits (67), Expect = 0.88
Identities = 45/161 (27%), Positives = 73/161 (44%), Gaps = 19/161 (11%)
Frame = -1
Query: 15 LTTSLFASTILA--------ANSNLFREYIGAQF------KGVKFSDV--PINPNVNFHF 58
L T LF+ST+L+ AN N + + F KG + + P N N+N
Sbjct: 515 LGTKLFSSTLLSLCHLIIFCANGNNQQSILPKAFFHSLANKGSQAENPIDPFNLNLNDDL 336
Query: 59 ILSFAIDYDTSSPPSPTN-GNFNIFWDTQNLSPSEVSFIKNQNPNVKVALSLGGDTVQGD 117
IL I Y +S +PTN G+F IF +++ +++ K++ + K+ G +T Q D
Sbjct: 335 ILFGNIAYKSSLASTPTN*GDFIIF----SITIIKLA*CKDKPESPKLPFG-GFNTPQFD 171
Query: 118 PVNFIPSS--IDSWVSNAVSSLTSIIKTYNLDGIDVDYEHF 156
N P S + S + + S S T +G D ++ F
Sbjct: 170 SKNPFPKSTFVSSSFKHFLFSTDSDTDT*LFNGPDTNF*SF 48
>TC87632 weakly similar to GP|21536611|gb|AAM60943.1 putative ribosomal
protein S2 {Arabidopsis thaliana}, partial (81%)
Length = 1572
Score = 29.6 bits (65), Expect = 1.5
Identities = 35/165 (21%), Positives = 70/165 (42%), Gaps = 7/165 (4%)
Frame = +1
Query: 48 VPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNI--FWDTQNLSPSEVSFIKNQNPNVKV 105
+P+NP+V F ++ I + N N NI D +L + + K+ +
Sbjct: 595 IPVNPSVQFVYLFCNLITKTLLLEKNNNNNNSNIKLLDDDHHLREKQAAKEKS-----NL 759
Query: 106 ALSLGGDTVQGDPVNFIPSSIDSWVSNAVSSLTSIIKTYNLDGIDVDYEH-----FIGDP 160
A LG + D ++ +P ++ N + L ++K + G ++ +++ I D
Sbjct: 760 AAKLGVTVIPYDAISPLPQDVEQ-TKNLLDKLV-VLKLNHASGTNMGFQNPKSAMDICDG 933
Query: 161 NTFAECIGRLITTLKNNNVISFASIAPFDDAEVQNHYLALWKSYS 205
TF + I I TL ++ S + F+ + + L + + YS
Sbjct: 934 QTFLDLIINQIETL-DSKYGSRVPLLIFNKDDTHDSTLKVLEKYS 1065
>TC82184 homologue to PIR|D96507|D96507 hypothetical protein T12C22.10
[imported] - Arabidopsis thaliana, partial (37%)
Length = 723
Score = 29.3 bits (64), Expect = 2.0
Identities = 26/84 (30%), Positives = 40/84 (46%), Gaps = 9/84 (10%)
Frame = +1
Query: 71 PPSPTNGNFNIFWDTQNLSPSEV---------SFIKNQNPNVKVALSLGGDTVQGDPVNF 121
P + ++ ++NI Q++SP + SFI N N NV V ++ +T +
Sbjct: 283 PLTSSSSHYNI---PQDMSPKSIQRVAAAAANSFIDNNNNNVNVNVNDNANTPSSSSLVS 453
Query: 122 IPSSIDSWVSNAVSSLTSIIKTYN 145
PSS+ S S+ VSSL S N
Sbjct: 454 SPSSMVS--SDDVSSLMSSFDQVN 519
>TC93126
Length = 670
Score = 29.3 bits (64), Expect = 2.0
Identities = 20/61 (32%), Positives = 26/61 (41%)
Frame = +1
Query: 42 GVKFSDVPINPNVNFHFILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPSEVSFIKNQNP 101
G F D P+ P F+L T P+P F + D N+S E SF+K N
Sbjct: 46 GDVFWDEPLLPTNQSSFVL-------TQPCPTPNLSAFVQYRDQPNISFGEQSFLKGSNS 204
Query: 102 N 102
N
Sbjct: 205 N 207
>TC87122 weakly similar to GP|19911203|dbj|BAB86928. glucosyltransferase-10
{Vigna angularis}, partial (41%)
Length = 1588
Score = 29.3 bits (64), Expect = 2.0
Identities = 20/90 (22%), Positives = 39/90 (43%), Gaps = 6/90 (6%)
Frame = +2
Query: 59 ILSFAIDYDTSSPPSPTNGNFNIFWDTQNLSPS---EVSFIKNQNPNVKVALSLGGDTVQ 115
+LS++ P P G+ N F + ++F+ N+ + ++ SLG D ++
Sbjct: 104 VLSYSQKPHVVCVPFPAQGHVNPFMQLAKILNKLGFHITFVNNEFNHKRLIKSLGHDFLK 283
Query: 116 GDP---VNFIPSSIDSWVSNAVSSLTSIIK 142
G P IP + NA S+ +++
Sbjct: 284 GQPDFRFETIPDGLPPSDENATQSIGGLVE 373
>TC84046
Length = 691
Score = 29.3 bits (64), Expect = 2.0
Identities = 13/18 (72%), Positives = 14/18 (77%)
Frame = -3
Query: 48 VPINPNVNFHFILSFAID 65
V INPNV HF LSFAI+
Sbjct: 497 VLINPNVQLHFTLSFAIN 444
Score = 27.7 bits (60), Expect = 5.7
Identities = 13/23 (56%), Positives = 16/23 (69%)
Frame = -1
Query: 171 ITTLKNNNVISFASIAPFDDAEV 193
I T K N V +FASI PFD+ +V
Sbjct: 445 INTCKINVVTTFASICPFDEDQV 377
>BE942118
Length = 693
Score = 28.9 bits (63), Expect = 2.6
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Frame = +3
Query: 207 IIDYVNFQFYAYDK-STTVAQFIDYYNKQSSNYNSEKVLVS 246
I+ Y FQF+AY K S +V FI Y YN E V ++
Sbjct: 279 ILPYELFQFFAYKKNSASVMYFISYQFSSCICYN*EHVSIA 401
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,730,701
Number of Sequences: 36976
Number of extensions: 135384
Number of successful extensions: 760
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 734
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 737
length of query: 304
length of database: 9,014,727
effective HSP length: 96
effective length of query: 208
effective length of database: 5,465,031
effective search space: 1136726448
effective search space used: 1136726448
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149490.11


