

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.2 - phase: 0
(365 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
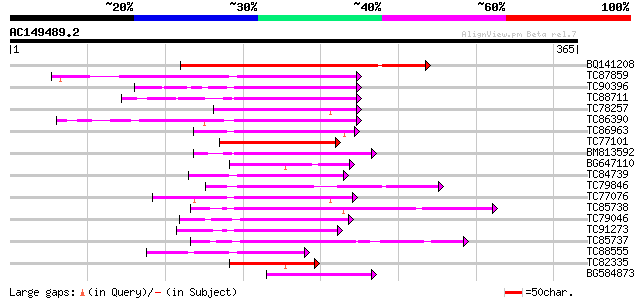
Score E
Sequences producing significant alignments: (bits) Value
BQ141208 similar to PIR|S71477|S71 homeotic protein ovule-speci... 165 2e-41
TC87859 similar to SP|P46604|HT22_ARATH Homeobox-leucine zipper ... 72 3e-13
TC90396 similar to PIR|H96719|H96719 homeobox gene 13 protein 1... 71 6e-13
TC88711 similar to PIR|T07614|T07614 homeobox-leucine zipper pro... 69 2e-12
TC78257 similar to PIR|T47981|T47981 homeobox-leucine zipper pro... 69 4e-12
TC86390 similar to PIR|T52371|T52371 homeobox protein HAT9 [impo... 68 6e-12
TC86963 similar to GP|15148918|gb|AAK84886.1 homeodomain leucine... 66 2e-11
TC77101 similar to GP|15148920|gb|AAK84887.1 homeodomain leucine... 63 2e-10
BM813592 similar to GP|20161555|db putative homeodomain-leucine ... 62 3e-10
BG647110 homologue to PIR|T10695|T10 transcription factor HD-zip... 60 2e-09
TC84739 homologue to PIR|T52367|T52367 homeobox protein HAT14 [i... 60 2e-09
TC79846 similar to GP|8843728|dbj|BAA97276.1 homeodomain transcr... 58 7e-09
TC77076 similar to PIR|T07734|T07734 homeotic protein VAHOX1 - t... 57 9e-09
TC85738 similar to GP|6091551|gb|AAF01764.2| homeodomain-leucine... 57 9e-09
TC79046 similar to PIR|T12634|T12634 homeotic protein - common s... 55 4e-08
TC91273 similar to GP|1435022|dbj|BAA05625.1 DNA-binding protein... 54 7e-08
TC85737 similar to GP|6091551|gb|AAF01764.2| homeodomain-leucine... 54 7e-08
TC88555 similar to PIR|T07614|T07614 homeobox-leucine zipper pro... 54 1e-07
TC82335 homologue to PIR|T10695|T10695 transcription factor HD-z... 54 1e-07
BG584873 similar to GP|20161555|dbj putative homeodomain-leucine... 52 3e-07
>BQ141208 similar to PIR|S71477|S71 homeotic protein ovule-specific -
Phalaenopsis sp., partial (19%)
Length = 661
Score = 165 bits (418), Expect = 2e-41
Identities = 80/161 (49%), Positives = 105/161 (64%)
Frame = +3
Query: 111 DELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQ 170
DE E E G G + + KRK+YHRHT QI+ ME+ FKE PHPD+KQR+
Sbjct: 48 DEFESGTKSCSENHEGGAASGEEQGPRPKRKRYHRHTQHQIQEMESFFKECPHPDDKQRK 227
Query: 171 QLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCP 230
+LS++LGL P QVKFWFQN+RTQ+K ER ENS L+++ EKLR N RE ++ A CP
Sbjct: 228 ELSRELGLEPLQVKFWFQNKRTQMKTQHERSENSQLRADNEKLRADNMRYREALSNASCP 407
Query: 231 NCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKY 271
NCG PT G M+ +E LR+ENA+L+ E++R+ KY
Sbjct: 408 NCGGPTA--IGEMSFDEHHLRLENARLREEIDRISTMAAKY 524
>TC87859 similar to SP|P46604|HT22_ARATH Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22). [Mouse-ear cress]
{Arabidopsis thaliana}, partial (48%)
Length = 1109
Score = 72.0 bits (175), Expect = 3e-13
Identities = 60/203 (29%), Positives = 90/203 (43%), Gaps = 4/203 (1%)
Frame = +2
Query: 28 SNNN---PPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTI 84
SNNN P S+S ++ P+L+L L+ + EE + ST
Sbjct: 146 SNNNVTKPYSSSNHNNYEAEPSLTLGLS------------------TKSYYEEPLDFSTQ 271
Query: 85 GGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGD-GDGDGDGVNKNKKKKRKKY 143
+ S +S + D E+E E E D D DG +KK R
Sbjct: 272 TNSPHHSVVSSFSSGRVKRERDVSSELEMETTEMERVSSRISDEDEDGATAARKKLRL-- 445
Query: 144 HRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHEN 203
T +Q ++E FKE + KQ+Q L+++L L PRQV+ WFQNRR + K Q +
Sbjct: 446 ---TKDQSAMLEESFKEHSTLNPKQKQALARELNLTPRQVEVWFQNRRARTKLKQTEVDC 616
Query: 204 SLLKSEIEKLREKNKTLRETINK 226
LK E L E+N+ L++ + +
Sbjct: 617 EFLKKCCETLTEENRRLQKEVQE 685
>TC90396 similar to PIR|H96719|H96719 homeobox gene 13 protein 11736-10437
[imported] - Arabidopsis thaliana, partial (41%)
Length = 997
Score = 71.2 bits (173), Expect = 6e-13
Identities = 49/146 (33%), Positives = 79/146 (53%)
Frame = +1
Query: 81 GSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKR 140
GS G + + S +SG +S I+ + DEL G++D+ D +G ++ +KK+
Sbjct: 109 GSNGGASFMMKRSMSFSG-IESNHINTNKCDELV--HGDEDQLS---DEEGYSQMGEKKK 270
Query: 141 KKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQER 200
R + EQ++ +E F+ + +++ QL+K LGL PRQV WFQNRR + K Q
Sbjct: 271 ----RLSLEQVKALEKSFEIGNKLEPERKMQLAKALGLQPRQVAIWFQNRRARWKTKQLE 438
Query: 201 HENSLLKSEIEKLREKNKTLRETINK 226
E +LK + + L+ N TL+ NK
Sbjct: 439 KEYEVLKKQFDSLKADNNTLKAQNNK 516
>TC88711 similar to PIR|T07614|T07614 homeobox-leucine zipper protein
homolog h1 - soybean, partial (73%)
Length = 1238
Score = 69.3 bits (168), Expect = 2e-12
Identities = 51/154 (33%), Positives = 78/154 (50%)
Frame = +1
Query: 73 MEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGV 132
+E +E EE V +S S + +S+ E E ++E+ E DE D +
Sbjct: 352 IECQEEEEAG------VSSPNSTVSSVSGKRSLRE-EDHDVENRENISDEEDAE------ 492
Query: 133 NKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRT 192
RKK R + +Q ++E FKE + KQ+ L+KQLGL PRQV+ WFQNRR
Sbjct: 493 -----TARKKL-RLSKDQSAILEETFKEHNTLNPKQKLALAKQLGLRPRQVEVWFQNRRA 654
Query: 193 QIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
+ K Q + +LK E L E+N+ L++ + +
Sbjct: 655 RTKLKQTEVDCEVLKRCCENLTEENRRLQKEVQE 756
>TC78257 similar to PIR|T47981|T47981 homeobox-leucine zipper protein
ATHB-12 - Arabidopsis thaliana, partial (45%)
Length = 994
Score = 68.6 bits (166), Expect = 4e-12
Identities = 37/102 (36%), Positives = 61/102 (59%), Gaps = 7/102 (6%)
Frame = +2
Query: 132 VNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRR 191
++ +KKK K R T EQI+ +E +F+ + +++ QL+++LGL PRQV WFQN+R
Sbjct: 215 ISSMRKKKNKNTKRFTDEQIKSLETMFETETRLEPRKKLQLARELGLQPRQVAIWFQNKR 394
Query: 192 TQIKAIQ-ERHENSL------LKSEIEKLREKNKTLRETINK 226
+ K+ Q ER N L L S+ E ++++ +TL + K
Sbjct: 395 ARWKSKQLEREYNKLQNSYNNLASKFESMKKERQTLLIQLQK 520
>TC86390 similar to PIR|T52371|T52371 homeobox protein HAT9 [imported] -
Arabidopsis thaliana, partial (58%)
Length = 1288
Score = 67.8 bits (164), Expect = 6e-12
Identities = 62/203 (30%), Positives = 90/203 (43%), Gaps = 7/203 (3%)
Frame = +2
Query: 31 NPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVE 90
NP STS P+L+L L+G IS+ + +G G E
Sbjct: 341 NPYSTSN------EPSLTLGLSG-----------ESYNLISHKQATKG-----YGEELCR 454
Query: 91 EISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGD-------GDGDGDGVNKNKKKKRKKY 143
+ SS +S S S + E DE E++ + D D D N KK
Sbjct: 455 QTSSPHSVVNSSFSSGRVLQVKRERDEEEEEVEEERVSSRVSDEDEDATNARKKL----- 619
Query: 144 HRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHEN 203
R T EQ ++E FK + KQ+Q L++QL L PRQV+ WFQNRR + K Q +
Sbjct: 620 -RLTKEQSLLLEESFKLHSTLNPKQKQALARQLNLRPRQVEVWFQNRRARTKLKQTEVDC 796
Query: 204 SLLKSEIEKLREKNKTLRETINK 226
LK E L ++N+ L++ + +
Sbjct: 797 EFLKKCCETLTDENRRLKKELQE 865
>TC86963 similar to GP|15148918|gb|AAK84886.1 homeodomain leucine zipper
protein HDZ2 {Phaseolus vulgaris}, partial (87%)
Length = 1519
Score = 65.9 bits (159), Expect = 2e-11
Identities = 39/114 (34%), Positives = 61/114 (53%), Gaps = 7/114 (6%)
Frame = +1
Query: 119 EDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGL 178
E +E GD D + + KKR R +SEQ++ +E F+ + ++ QL+K+LGL
Sbjct: 676 EKEENCGDEDYEACYHQQGKKR----RLSSEQVQFLEKSFEVENKLEPDRKVQLAKELGL 843
Query: 179 APRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLR-------EKNKTLRETIN 225
PRQV WFQNRR + K Q + LK+ + L+ ++N L+E +N
Sbjct: 844 QPRQVAIWFQNRRARFKTKQLEKDYGTLKASFDSLKDDYDNLLQENDKLKEEVN 1005
>TC77101 similar to GP|15148920|gb|AAK84887.1 homeodomain leucine zipper
protein HDZ3 {Phaseolus vulgaris}, complete
Length = 1532
Score = 62.8 bits (151), Expect = 2e-10
Identities = 31/78 (39%), Positives = 49/78 (62%)
Frame = +1
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+++ +K R TSEQ+ ++E F+E + +++ QL+K+LGL PRQV WFQNRR + K
Sbjct: 613 EEQSPEKKRRLTSEQVHMLEKSFEEENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWK 792
Query: 196 AIQERHENSLLKSEIEKL 213
Q + +LKS + L
Sbjct: 793 TKQLERDYDVLKSSYDSL 846
>BM813592 similar to GP|20161555|db putative homeodomain-leucine zipper
{Oryza sativa (japonica cultivar-group)}, partial (42%)
Length = 683
Score = 62.4 bits (150), Expect = 3e-10
Identities = 41/118 (34%), Positives = 60/118 (50%)
Frame = +2
Query: 119 EDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGL 178
E++E +GD RKK R T EQ ++E F+++ + KQ++ L+ QL L
Sbjct: 143 EEEESSVNGD---------TPRKKL-RLTKEQSHLLEESFRKNHTLNPKQKECLAMQLKL 292
Query: 179 APRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPT 236
PRQV+ WFQNRR + K Q E LK L E+N+ L+ + + G PT
Sbjct: 293 RPRQVEVWFQNRRARSKLKQTEMECEYLKRWFGSLTEQNRRLQREVEELRAMKVGPPT 466
>BG647110 homologue to PIR|T10695|T10 transcription factor HD-zip -
Arabidopsis thaliana, partial (16%)
Length = 844
Score = 59.7 bits (143), Expect = 2e-09
Identities = 34/85 (40%), Positives = 48/85 (56%), Gaps = 4/85 (4%)
Frame = +1
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL----GLAPRQVKFWFQNRRTQIKAI 197
KY R+T EQ+ +E L+ E P P +RQQL ++ + P+Q+K WFQNRR + K
Sbjct: 148 KYVRYTPEQVEALERLYHECPKPTSLRRQQLIRECPILSHIEPKQIKVWFQNRRCREK-- 321
Query: 198 QERHENSLLKSEIEKLREKNKTLRE 222
+R E L++ KL NK L E
Sbjct: 322 -QRKEAGRLQAVNRKLTAMNKLLME 393
>TC84739 homologue to PIR|T52367|T52367 homeobox protein HAT14 [imported] -
Arabidopsis thaliana (fragment), partial (70%)
Length = 737
Score = 59.7 bits (143), Expect = 2e-09
Identities = 37/103 (35%), Positives = 51/103 (48%)
Frame = +1
Query: 116 DEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQ 175
D+ + D GV +KK R + +Q +E FKE + KQ+ L+KQ
Sbjct: 13 DQRNSSRVSDEDDNCGVRNTRKKLRL-----SKDQSAFLEESFKEHHTLNPKQKLALAKQ 177
Query: 176 LGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNK 218
L L PRQV+ WFQNRR + K Q + LK E L E+N+
Sbjct: 178 LNLRPRQVEVWFQNRRARTKLKQTEVDCEYLKRCCETLTEENR 306
>TC79846 similar to GP|8843728|dbj|BAA97276.1 homeodomain transcription
factor-like {Arabidopsis thaliana}, partial (42%)
Length = 679
Score = 57.8 bits (138), Expect = 7e-09
Identities = 41/153 (26%), Positives = 73/153 (46%)
Frame = +2
Query: 127 GDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFW 186
G+ + NKK+K + EQ+ ++E F + +++ +L+ +LGL PRQV W
Sbjct: 233 GENSANDMNKKRKL------SDEQVNILEENFGNEHKLESEKKDRLAMELGLDPRQVAVW 394
Query: 187 FQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPTTNRDGTMATE 246
FQNRR + K +KL E+ +L++ C + T R+ +E
Sbjct: 395 FQNRRARWKN--------------KKLEEEYFSLKKNHESTILEKCLLETKLRE--QHSE 526
Query: 247 EQQLRIENAKLKAEVERLRAALGKYASGTMSPS 279
+LR + ++ + E++RLR + S + S S
Sbjct: 527 ALKLREQLSEAEKEIQRLREPFERVTSSSSSTS 625
>TC77076 similar to PIR|T07734|T07734 homeotic protein VAHOX1 - tomato,
partial (43%)
Length = 1664
Score = 57.4 bits (137), Expect = 9e-09
Identities = 41/142 (28%), Positives = 74/142 (51%), Gaps = 10/142 (7%)
Frame = +1
Query: 93 SSEYSGPAKSKSIDEYEGDELEDDE---GEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSE 149
S+ + G S ++ +G + + G D + +G+ + D +KKR R + +
Sbjct: 496 SASFFGSRSMVSFEDVQGRKRRNRSFFGGFDLDENGEDEMDEYFHQSEKKR----RLSVD 663
Query: 150 QIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQ-ERHENSL--- 205
Q++ +E F+E + +++ +L+K LGL PRQV WFQNRR + K Q E+ +SL
Sbjct: 664 QVQFLEKSFEEDNKLEPERKTKLAKDLGLQPRQVAIWFQNRRARWKTKQLEKDYDSLNDG 843
Query: 206 ---LKSEIEKLREKNKTLRETI 224
LK+E + L ++ L+ +
Sbjct: 844 YESLKTEYDNLLKEKDRLQSEV 909
>TC85738 similar to GP|6091551|gb|AAF01764.2| homeodomain-leucine zipper
protein 56 {Glycine max}, partial (77%)
Length = 2144
Score = 57.4 bits (137), Expect = 9e-09
Identities = 52/206 (25%), Positives = 91/206 (43%), Gaps = 8/206 (3%)
Frame = +3
Query: 117 EGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL 176
EG D+EG D G +KKR R + +Q++ +E F+ + ++ +L+++L
Sbjct: 1008 EGLDEEGCVDEPG-------QKKR----RLSVDQVKALEKNFEVENKLEPDRKVKLAQEL 1154
Query: 177 GLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKL--------REKNKTLRETINKAC 228
GL PRQV WFQNRR + K Q + +LK+ + L R+K L+E
Sbjct: 1155 GLQPRQVAVWFQNRRARWKTKQLERDYGVLKANYDALKLNCEDLQRDKETLLKEVKELKS 1334
Query: 229 CPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQEN 288
T + + +E+ + ++ ++ A E A+ + S ++ C + D
Sbjct: 1335 RLQIQEENTTTESDVCVKEELITLQESENTASDE---TAILRSDSKDLNNDCFKNGDGVA 1505
Query: 289 IKSSLDFYTGIFCLDESRIMDVVNQA 314
DF G D S I++ + A
Sbjct: 1506 SLFPADFKDGSSDSDSSAILNEESNA 1583
>TC79046 similar to PIR|T12634|T12634 homeotic protein - common sunflower,
partial (56%)
Length = 1136
Score = 55.1 bits (131), Expect = 4e-08
Identities = 34/112 (30%), Positives = 57/112 (50%)
Frame = +2
Query: 110 GDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQR 169
G EL ++ +E D DG +KK+R EQ++ +E F+ + +++
Sbjct: 329 GIELGEEANIPEEDLSD---DGSQAGEKKRRLNM-----EQVKTLEKSFELGNKLEPERK 484
Query: 170 QQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLR 221
QL++ L L PRQV WFQNRR + K Q + +LK + + ++ N L+
Sbjct: 485 MQLARALNLQPRQVAIWFQNRRARWKTKQLEKDYDVLKRQYDAIKLDNDALQ 640
>TC91273 similar to GP|1435022|dbj|BAA05625.1 DNA-binding protein {Daucus
carota}, partial (33%)
Length = 722
Score = 54.3 bits (129), Expect = 7e-08
Identities = 32/107 (29%), Positives = 56/107 (51%)
Frame = +1
Query: 108 YEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEK 167
+ G EL ++ ++E DG G +KKR R EQ++ +E F+ + +
Sbjct: 427 FSGIELGEEANVEEELSDDGSQLG-----EKKR----RLNMEQVKTLEKSFELGNKLEPE 579
Query: 168 QRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLR 214
++ QL++ LGL PRQ+ WFQNRR + K Q + +L + + ++
Sbjct: 580 RKMQLARALGLQPRQIAIWFQNRRARWKTKQLEKDYDVLXRQYDTIK 720
>TC85737 similar to GP|6091551|gb|AAF01764.2| homeodomain-leucine zipper
protein 56 {Glycine max}, partial (63%)
Length = 1622
Score = 54.3 bits (129), Expect = 7e-08
Identities = 45/179 (25%), Positives = 85/179 (47%)
Frame = +2
Query: 117 EGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL 176
+G ++EG + G + ++KK+R + +Q++ +E F+ + +++++L+ +L
Sbjct: 503 DGFEEEGCVEETG---HHSEKKRRLRV-----DQVKALEKNFEVENKLEPERKEKLAIEL 658
Query: 177 GLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCPNCGVPT 236
GL PRQV WFQNRR + K Q + +LK+ + L+ K + + NKA
Sbjct: 659 GLQPRQVAVWFQNRRARWKTKQLERDYGVLKANYDALKLKFDAIAQD-NKAFHKEI---- 823
Query: 237 TNRDGTMATEEQQLRIENAKLKAEVERLRAALGKYASGTMSPSCSTSHDQENIKSSLDF 295
++ E++ N +K E+ L + +PS TS+ K LD+
Sbjct: 824 --KELKSKLGEEEKSTINVLVKEELTMLESC----DEDKHNPSSETSNPSSESKDHLDY 982
>TC88555 similar to PIR|T07614|T07614 homeobox-leucine zipper protein
homolog h1 - soybean, partial (47%)
Length = 704
Score = 53.5 bits (127), Expect = 1e-07
Identities = 37/105 (35%), Positives = 54/105 (51%)
Frame = +3
Query: 89 VEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTS 148
V +S S + +S E G++L+ E D G D + ++KK R T
Sbjct: 411 VSSPNSTVSSVSGKRSEREVTGEDLDM---ERDCSRGISDEEDAETSRKKLRL-----TK 566
Query: 149 EQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQ 193
+Q ++E FKE + KQ+ L+KQLGL RQV+ WFQNRR +
Sbjct: 567 DQSIILEESFKEHNTLNPKQKLALAKQLGLRARQVEVWFQNRRAR 701
>TC82335 homologue to PIR|T10695|T10695 transcription factor HD-zip -
Arabidopsis thaliana, partial (20%)
Length = 720
Score = 53.5 bits (127), Expect = 1e-07
Identities = 26/62 (41%), Positives = 38/62 (60%), Gaps = 4/62 (6%)
Frame = +1
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL----GLAPRQVKFWFQNRRTQIKAI 197
KY R+T EQ+ +E L+ E P P +RQQL ++ + P+Q+K WFQNRR + K
Sbjct: 211 KYVRYTPEQVEALERLYHECPKPTSLRRQQLIRECPILSHIEPKQIKVWFQNRRCREKQR 390
Query: 198 QE 199
+E
Sbjct: 391 KE 396
>BG584873 similar to GP|20161555|dbj putative homeodomain-leucine zipper
{Oryza sativa (japonica cultivar-group)}, partial (31%)
Length = 737
Score = 52.4 bits (124), Expect = 3e-07
Identities = 28/71 (39%), Positives = 39/71 (54%)
Frame = +1
Query: 166 EKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETIN 225
+KQ++ L+ QL L PRQV+ WFQNRR + K Q E LK L E+N+ L+ +
Sbjct: 295 KKQKECLAMQLKLRPRQVEVWFQNRRARSKLKQTEMECEYLKRWFGSLTEQNRRLQREVE 474
Query: 226 KACCPNCGVPT 236
+ G PT
Sbjct: 475 ELRAMKVGPPT 507
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.309 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,513,930
Number of Sequences: 36976
Number of extensions: 141153
Number of successful extensions: 1665
Number of sequences better than 10.0: 189
Number of HSP's better than 10.0 without gapping: 1253
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1471
length of query: 365
length of database: 9,014,727
effective HSP length: 97
effective length of query: 268
effective length of database: 5,428,055
effective search space: 1454718740
effective search space used: 1454718740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149489.2


