

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.11 - phase: 0
(526 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
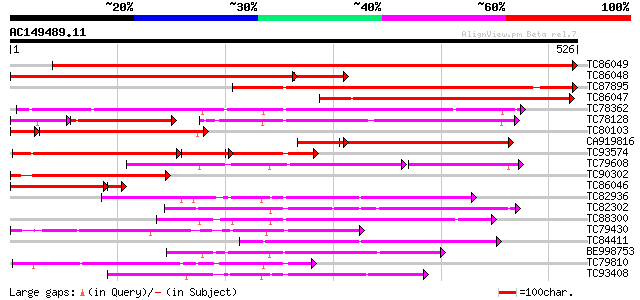
Score E
Sequences producing significant alignments: (bits) Value
TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative tra... 953 0.0
TC86048 weakly similar to PIR|H96561|H96561 probable peptide tra... 463 e-146
TC87895 similar to GP|20466248|gb|AAM20441.1 putative transport ... 478 e-135
TC86047 weakly similar to PIR|T48516|T48516 probable oligopeptid... 355 2e-98
TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter... 289 2e-78
TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T1... 194 4e-77
TC80103 weakly similar to PIR|H96561|H96561 probable peptide tra... 254 2e-76
CA919816 similar to GP|20466248|gb putative transport protein {A... 232 2e-74
TC93574 weakly similar to GP|21536862|gb|AAM61194.1 unknown {Ara... 174 7e-72
TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {... 160 6e-55
TC90302 weakly similar to PIR|H96561|H96561 probable peptide tra... 198 4e-51
TC86046 similar to PIR|H96561|H96561 probable peptide transporte... 186 5e-51
TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptid... 192 3e-49
TC82302 similar to PIR|F84663|F84663 probable nitrate transporte... 182 2e-46
TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affini... 181 8e-46
TC79430 similar to PIR|F86358|F86358 Similar to peptide transpor... 166 3e-41
TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersico... 165 3e-41
BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like... 156 2e-38
TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter... 149 2e-36
TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate tr... 149 2e-36
>TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative transport
protein {Arabidopsis thaliana}, partial (43%)
Length = 1836
Score = 953 bits (2463), Expect = 0.0
Identities = 481/487 (98%), Positives = 484/487 (98%)
Frame = +2
Query: 40 LILIAIGNGGISCSLAFGADQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQ 99
LILIAIGNGGI+CSL+FGA QVNRKDNP+ RVLEIFFSWYYAFTTIAVIIALTGIVYIQ
Sbjct: 2 LILIAIGNGGITCSLSFGAYQVNRKDNPDGYRVLEIFFSWYYAFTTIAVIIALTGIVYIQ 181
Query: 100 DHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPP 159
DHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPP
Sbjct: 182 DHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPP 361
Query: 160 KTSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK 219
KTSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK
Sbjct: 362 KTSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK 541
Query: 220 AIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWI 279
AIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWI
Sbjct: 542 AIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWI 721
Query: 280 IIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLND 339
IIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLND
Sbjct: 722 IIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLND 901
Query: 340 THGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLV 399
THGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLV
Sbjct: 902 THGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLV 1081
Query: 400 SSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD 459
SSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD
Sbjct: 1082SSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD 1261
Query: 460 EVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQR 519
EVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQR
Sbjct: 1262EVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQR 1441
Query: 520 DLKEEDL 526
DLKEEDL
Sbjct: 1442DLKEEDL 1462
>TC86048 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1364
Score = 463 bits (1192), Expect(2) = e-146
Identities = 229/267 (85%), Positives = 245/267 (90%)
Frame = +3
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQ 60
M++LWLTAM+P+A+PPAC++P EGC SA PGQ+AML SALILI IGNGGISCSLAFGADQ
Sbjct: 399 MAMLWLTAMVPAAQPPACSNPPEGCISAKPGQMAMLLSALILIGIGNGGISCSLAFGADQ 578
Query: 61 VNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTV 120
V RK+N NN R LEIFF+WYYA TTIAVI AL GIVYIQDHLGW++GFGVPA LMLISTV
Sbjct: 579 VKRKENSNNNRALEIFFTWYYASTTIAVIAALFGIVYIQDHLGWRIGFGVPAALMLISTV 758
Query: 121 LFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPTDK 180
LFFLASPLYVKI Q+TSL TGFAQVSVAA+KNRKL LPPKTSPEFYH +KDS LVVPTDK
Sbjct: 759 LFFLASPLYVKITQRTSLLTGFAQVSVAAFKNRKLSLPPKTSPEFYHHKKDSVLVVPTDK 938
Query: 181 LRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG 240
LRFLNKACVIKDHEQDIASDGS INRWSLCTVDQVEELKAIIKVIPLWST ITMSINIGG
Sbjct: 939 LRFLNKACVIKDHEQDIASDGSIINRWSLCTVDQVEELKAIIKVIPLWSTGITMSINIGG 1118
Query: 241 SFGLLQAKSLDRHIISSSNFEVPAGSF 267
SFGLLQA SLDRHIIS+SNFEVPAG F
Sbjct: 1119SFGLLQALSLDRHIISNSNFEVPAGIF 1199
Score = 72.8 bits (177), Expect(2) = e-146
Identities = 37/50 (74%), Positives = 40/50 (80%)
Frame = +1
Query: 265 GSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNF 314
GSF++ILI ILIWIIIYDRVLIPLASKIRGKP ISP RMG + F F
Sbjct: 1192 GSFTIILIFVILIWIIIYDRVLIPLASKIRGKPASISPMIRMGTWVVF*F 1341
Score = 28.1 bits (61), Expect = 8.6
Identities = 10/15 (66%), Positives = 13/15 (86%)
Frame = +2
Query: 307 GIGLFFNFLHLITAA 321
G+GLFFNF H++ AA
Sbjct: 1319 GLGLFFNFSHIVVAA 1363
>TC87895 similar to GP|20466248|gb|AAM20441.1 putative transport protein
{Arabidopsis thaliana}, partial (7%)
Length = 1265
Score = 478 bits (1229), Expect = e-135
Identities = 235/320 (73%), Positives = 271/320 (84%)
Frame = +1
Query: 207 WSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGS 266
W LCTVDQVEELKA+++VIPLWST I MS+NIGGSFGLLQAKSLDRHI +SNFEVPAGS
Sbjct: 1 WRLCTVDQVEELKALVRVIPLWSTGIMMSLNIGGSFGLLQAKSLDRHI--TSNFEVPAGS 174
Query: 267 FSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETV 326
FSVI++ +I IWI++YDRVLIPLASK+RGKPV ISPK RMGIGLF NFLHL+TAA FE++
Sbjct: 175 FSVIMVGSIFIWIVLYDRVLIPLASKLRGKPVRISPKIRMGIGLFLNFLHLVTAAAFESI 354
Query: 327 RRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAA 386
RRK+AI+ G+LNDTHGVLKMSA+WLAPQLCL GI+E FN IGQNEFYYKEFP++MSS++A
Sbjct: 355 RRKKAIQAGFLNDTHGVLKMSALWLAPQLCLGGISEAFNAIGQNEFYYKEFPRTMSSISA 534
Query: 387 SLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLY 446
SL GL M GNLVSS V + IE+ T GG EGW+SDNINKG +DKYY VI G++ALNLLY
Sbjct: 535 SLCGLGMAAGNLVSSFVFNTIENVTSRGGKEGWISDNINKGRYDKYYMVIAGVSALNLLY 714
Query: 447 YLVCSWAYGPTVDEVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNG 506
+LVCSWAYGPTVD+VS VS EN SK ++STEFK +NP FD ETSS KELTE KNG
Sbjct: 715 FLVCSWAYGPTVDQVSKVSDENDSKEKDSTEFKKVNPLFD-----ETSSNGKELTEIKNG 879
Query: 507 AQVEKVFKNSEQRDLKEEDL 526
Q EKVFK SE+ KEE+L
Sbjct: 880 GQAEKVFKISEENGSKEEEL 939
>TC86047 weakly similar to PIR|T48516|T48516 probable oligopeptide
transporter protein - Arabidopsis thaliana, partial
(11%)
Length = 983
Score = 355 bits (912), Expect = 2e-98
Identities = 180/238 (75%), Positives = 199/238 (82%), Gaps = 1/238 (0%)
Frame = +3
Query: 288 PLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMS 347
PLASKIRGKP ISP RM G F H + +IFE++RRK+AI+ G+LN+THGVLKMS
Sbjct: 3 PLASKIRGKPASISPMIRMDWGCFLISSHC-SGSIFESIRRKKAIQAGFLNNTHGVLKMS 179
Query: 348 AMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSII 407
AMWLAPQLCLAGIAE FNVIGQNEFYYKEFPK+MSSV+++LSGLAM GNLVSS V S I
Sbjct: 180 AMWLAPQLCLAGIAEAFNVIGQNEFYYKEFPKTMSSVSSALSGLAMAAGNLVSSFVFSTI 359
Query: 408 ESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVS-K 466
E+TT SGGNEGW+SDNINKGHFDKY WVI GIN LNLLYYLVCSWAYGPT D+VS VS +
Sbjct: 360 ENTTSSGGNEGWISDNINKGHFDKYLWVIAGINVLNLLYYLVCSWAYGPTADQVSKVSEE 539
Query: 467 ENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQRDLKEE 524
ENGSK E STEFK++NP DDKVS ETSSKEKELTEFKNG QVEKV K SE+ KEE
Sbjct: 540 ENGSKEENSTEFKNVNPQIDDKVSDETSSKEKELTEFKNGGQVEKVSKVSEENGSKEE 713
>TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter {Cucumis
sativus}, partial (88%)
Length = 1871
Score = 289 bits (740), Expect = 2e-78
Identities = 167/498 (33%), Positives = 277/498 (55%), Gaps = 26/498 (5%)
Frame = +3
Query: 7 TAMIPSARPPACNHPSE-GCESATPGQLAMLFSALILIAIGNGGI-SCSLAFGADQVNRK 64
+A++P RPP C P++ C+ A QL +L+ AL L ++G+GGI SC + F DQ +
Sbjct: 357 SAIVPHFRPPPC--PTQVNCQEAKSSQLWILYLALFLTSLGSGGIRSCVVPFSGDQFDMT 530
Query: 65 DNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFFL 124
R +F +WY+ A + ALT +VYIQDH+GW G G+P I M I+ + F L
Sbjct: 531 KKGVESRKWNLF-NWYFFCMGFASLSALTIVVYIQDHMGWGWGLGIPTIAMFIAIIAFLL 707
Query: 125 ASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVP-----TD 179
S LY +K S F AQV VAA++ R LP + + + DS + + TD
Sbjct: 708 GSRLYKTLKPSGSPFLRLAQVIVAAFRKRNDALPNDPKLLYQNLELDSSISLEGRLSHTD 887
Query: 180 KLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SIN 237
+ ++L+KA ++ D E + N W+L TV +VEELK +++++P+W++ I + + +
Sbjct: 888 QYKWLDKAAIVTDEEAKNLN--KLPNLWNLATVHRVEELKCLVRMLPIWASGILLITASS 1061
Query: 238 IGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKP 297
SF ++QA+++DRH+ S FE+ S ++ ++ ++ +I+Y+R+ +P + P
Sbjct: 1062SQHSFVIVQARTMDRHL--SHTFEISPASMAIFSVLTMMTGVILYERLFVPFIRRFTKNP 1235
Query: 298 VIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCL 357
I+ +RMGIG N + I +A+ E R+K A K L+ ++ +S WL PQ L
Sbjct: 1236AGITCLQRMGIGFVINIIATIVSALLEIKRKKVASKYHLLDSPKAIIPISVFWLVPQYFL 1415
Query: 358 AGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNE 417
G+AE+F +G EF Y + P+SM S A +L +A+ +G+ + +L+++++ T G
Sbjct: 1416HGVAEVFMNVGHLEFLYDQSPESMRSSATALYCIAIAIGHFIGTLLVTLVHKYT--GKER 1589
Query: 418 GWVSD-NINKGHFDKYYWVIVGINALNLLYYLVCSWAYG----------------PTVDE 460
W+ D N+N+G + YY+++ GI +N +YY++C+W Y T E
Sbjct: 1590NWLPDRNLNRGRLEYYYFLVCGIQVINFIYYVICAWIYNYKPLEEINENNQGDLEQTNGE 1769
Query: 461 VSNVSKENGSKVEESTEF 478
+S VS G KV+E EF
Sbjct: 1770LSLVSLNEG-KVDEKREF 1820
>TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T17F3_10
{Arabidopsis thaliana}, partial (77%)
Length = 2022
Score = 194 bits (494), Expect(3) = 4e-77
Identities = 109/303 (35%), Positives = 176/303 (57%), Gaps = 6/303 (1%)
Frame = +2
Query: 177 PTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAIT--M 234
PT+ R L+KA ++ E D+ DG+ +N+W+L ++ QVEE+K + + +P+W+ I
Sbjct: 1046 PTNS-RVLDKAALVM--EGDLNPDGTIVNQWNLVSIQQVEEVKCLARTLPIWAAGILGFT 1216
Query: 235 SINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIR 294
++ G+F + QA +DRH+ F++PAGS VI ++ I +W+ YDR+ +P KI
Sbjct: 1217 AMAQQGTFIVSQAMKMDRHL--GPKFQIPAGSLGVISLIVIGLWVPFYDRICVPSLRKIT 1390
Query: 295 GKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDT-HGVLKMSAMWLAP 353
I+ +R+GIG+ F+ + +I A + E VRR A N T G+ MS MWL P
Sbjct: 1391 KNEGGITLLQRIGIGMVFSIISMIVAGLVEKVRRDVANS----NPTPQGIAPMSVMWLFP 1558
Query: 354 QLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPS 413
QL L G E FN+IG EF+ ++FP M S+A +L + + N VSS+++ + S T +
Sbjct: 1559 QLVLMGFCEAFNIIGLIEFFNRQFPDHMRSIANALFSCSFALANYVSSILVITVHSVTKT 1738
Query: 414 GGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY---GPTVDEVSNVSKENGS 470
+ W+++NIN+G D +Y+++ G+ LNL+Y+L S Y G + + E GS
Sbjct: 1739 HNHPDWLTNNINEGRLDYFYYLLAGVGVLNLVYFLYVSQRYHYKGSVDIQEKPMDVELGS 1918
Query: 471 KVE 473
K E
Sbjct: 1919 KGE 1927
Score = 88.6 bits (218), Expect(3) = 4e-77
Identities = 40/98 (40%), Positives = 61/98 (61%)
Frame = +3
Query: 57 GADQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILML 116
G DQ + ++ + FF+WYY T+ ++ T IVYIQD + WK GF +P + ML
Sbjct: 666 GVDQFD-PTTEKGKKGINSFFNWYYTSFTVVLLFTQTVIVYIQDSISWKFGFAIPTLCML 842
Query: 117 ISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRK 154
S +LFF+ + +YV +K + S+F+ AQV VA++K RK
Sbjct: 843 XSIILFFIGTKIYVHVKPEGSIFSSIAQVLVASFKKRK 956
Score = 45.1 bits (105), Expect(3) = 4e-77
Identities = 23/61 (37%), Positives = 35/61 (56%), Gaps = 4/61 (6%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEG---CESATPGQLAMLFSALILIAIGNGGI-SCSLAF 56
M+ + LTA +P +PP+C + C +A Q+ LF LI ++IG+ GI CS+ F
Sbjct: 485 MTAVTLTAWLPKLQPPSCTPQQQALNQCVTANSSQVGFLFMGLIFLSIGSSGIRPCSIPF 664
Query: 57 G 57
G
Sbjct: 665 G 667
>TC80103 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (23%)
Length = 1236
Score = 254 bits (649), Expect(2) = 2e-76
Identities = 121/159 (76%), Positives = 142/159 (89%), Gaps = 2/159 (1%)
Frame = +3
Query: 28 ATPGQLAMLFSALILIAIGNGGISCSLAFGADQVNRKDNPNNRRVLEIFFSWYYAFTTIA 87
AT GQ+AML SA L++IGNGG+SCS+AFGADQVNRKDNPNN RVLEIFFSWYYAFTTI+
Sbjct: 645 ATTGQMAMLLSAFGLMSIGNGGLSCSIAFGADQVNRKDNPNNHRVLEIFFSWYYAFTTIS 824
Query: 88 VIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSV 147
+I+ALT IVYIQ+HLGWK+GFGVPA LML+ST+ FFLASPLYVKI ++T L TGFAQV+
Sbjct: 825 IILALTVIVYIQEHLGWKIGFGVPAALMLLSTICFFLASPLYVKITKRTXLITGFAQVTT 1004
Query: 148 AAYKNRKLPLPPKTSPEFYHQQKDS--ELVVPTDKLRFL 184
AAYKNRKL LPPK SP+FYH +KDS +LV+PTDKLR++
Sbjct: 1005AAYKNRKLQLPPKNSPQFYHHKKDSDLDLVIPTDKLRYI 1121
Score = 50.4 bits (119), Expect(2) = 2e-76
Identities = 19/27 (70%), Positives = 25/27 (92%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCES 27
M +LWLTAMIP+ARPP CN+P++GC+S
Sbjct: 563 MGMLWLTAMIPAARPPPCNNPTKGCKS 643
>CA919816 similar to GP|20466248|gb putative transport protein {Arabidopsis
thaliana}, partial (20%)
Length = 819
Score = 232 bits (592), Expect(2) = 2e-74
Identities = 106/161 (65%), Positives = 128/161 (78%)
Frame = -1
Query: 307 GIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNV 366
G + F+FLHL+TA+I ET+RR+ AI EGY ND H VL MSAMWL PQLCL GIAE FNV
Sbjct: 699 GTRIVFSFLHLVTASIVETIRRRRAINEGYGNDPHSVLNMSAMWLFPQLCLGGIAEAFNV 520
Query: 367 IGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINK 426
IGQNEFYY EFPK+MSS+A+SL GL M VG ++SS + SI+E T SGG +GW+SDNINK
Sbjct: 519 IGQNEFYYTEFPKTMSSIASSLFGLGMAVGYVISSFLFSIVERVTSSGGKDGWISDNINK 340
Query: 427 GHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKE 467
G FDKYYW+I ++ +N+LYY+VCSWAYGPT D S VS +
Sbjct: 339 GRFDKYYWLIATVSGVNILYYVVCSWAYGPTADVESKVSAD 217
Score = 65.9 bits (159), Expect(2) = 2e-74
Identities = 32/47 (68%), Positives = 37/47 (78%)
Frame = -2
Query: 268 SVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNF 314
SVI+I I IWI +YDRV+IPLA K+RGKPV IS K RMG+GLF F
Sbjct: 815 SVIMIFTIFIWIALYDRVVIPLARKLRGKPVRISAKTRMGLGLFSLF 675
>TC93574 weakly similar to GP|21536862|gb|AAM61194.1 unknown {Arabidopsis
thaliana}, partial (35%)
Length = 980
Score = 174 bits (441), Expect(3) = 7e-72
Identities = 87/158 (55%), Positives = 111/158 (70%), Gaps = 1/158 (0%)
Frame = +2
Query: 3 LLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGADQV 61
+LWLTA+I ARPP CN+ E C SAT Q LFS+L L+A G GGI CSLAF ADQ+
Sbjct: 134 VLWLTAIIRHARPPECNN-GESCTSATGMQFLFLFSSLALMAFGAGGIRPCSLAFAADQI 310
Query: 62 NRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVL 121
N NP N R+++ FF+WYY ++V++++ IVYIQ GW +GFG+P LML S+V+
Sbjct: 311 NNPKNPKNERIMKSFFNWYYVSVGVSVMVSMVFIVYIQVKAGWVIGFGIPVGLMLFSSVV 490
Query: 122 FFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPP 159
FFL S +YVK+K SL GFAQV VA++KNR L LPP
Sbjct: 491 FFLGSFMYVKVKPNKSLLAGFAQVIVASWKNRHLTLPP 604
Score = 83.6 bits (205), Expect(3) = 7e-72
Identities = 42/87 (48%), Positives = 61/87 (69%), Gaps = 1/87 (1%)
Frame = +1
Query: 201 GSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG-GSFGLLQAKSLDRHIISSSN 259
G + SLCTV QVEELKA+I+V+PLWST IT+++ I SF ++QA +++R + N
Sbjct: 727 GCQLIHGSLCTVRQVEELKAVIQVLPLWSTGITIAVTISQQSFSVVQATTMNRMV---HN 897
Query: 260 FEVPAGSFSVILIVAILIWIIIYDRVL 286
FE+P SFS I+ + IW+ IYDR++
Sbjct: 898 FEIPPTSFSAFGILTLTIWVAIYDRII 978
Score = 52.8 bits (125), Expect(3) = 7e-72
Identities = 23/48 (47%), Positives = 34/48 (69%)
Frame = +3
Query: 160 KTSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRW 207
+T+ ++ Q S LV PTDK RFLNKAC++K+ E+D+ S+G I+ W
Sbjct: 603 QTTAGLWYFQSGSNLVQPTDKARFLNKACIMKNREKDLDSNGMPIDPW 746
>TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {Arabidopsis
thaliana}, partial (59%)
Length = 1489
Score = 160 bits (406), Expect(2) = 6e-55
Identities = 93/267 (34%), Positives = 150/267 (55%), Gaps = 7/267 (2%)
Frame = +2
Query: 109 GVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQ 168
G+P + MLIS + F PLY + + S FT +V VAA++ +K+P P ++ + +
Sbjct: 5 GIPTLAMLISIIAFIGGYPLYRNLNPEGSPFTRLVKVGVAAFRKKKIPKVPNSTLLYQND 184
Query: 169 QKDSEL-----VVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIK 223
+ D+ + ++ +D+L+FL+KA ++ + + D W L TV +VEELK+II+
Sbjct: 185 ELDASITLGGKLLHSDQLKFLDKAAIVTEEDNTKTPD-----LWRLSTVHRVEELKSIIR 349
Query: 224 VIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIII 281
+ P+W++ I + G+F L QAK+++RH+ S FE+PAGS SV I+ +L +
Sbjct: 350 MGPIWASGILLITAYAQQGTFSLQQAKTMNRHLTKS--FEIPAGSMSVFTILTMLFTTAL 523
Query: 282 YDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTH 341
YDRVLI +A + G I+ RMGIG + A E R+K A++ G + +
Sbjct: 524 YDRVLIRVARRFTGLDRGITFLHRMGIGFVISLFATFVAGFVEMKRKKVAMEHGLIEHSS 703
Query: 342 GVLKMSAMWLAPQLCLAGIAEMFNVIG 368
++ +S WL PQ L G+AE F IG
Sbjct: 704 EIIPISVFWLVPQYSLHGMAEAFMSIG 784
Score = 72.0 bits (175), Expect(2) = 6e-55
Identities = 34/111 (30%), Positives = 65/111 (57%), Gaps = 5/111 (4%)
Frame = +1
Query: 371 EFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDN-INKGHF 429
EF+Y + P+SM+S A + ++ +GN +S+L+++++ T W+ DN +NKG
Sbjct: 793 EFFYDQAPESMTSTAMAFFWTSISLGNYISTLLVTLVHKFTKGPNGTNWLPDNNLNKGKL 972
Query: 430 DKYYWVIVGINALNLLYYLVCSWAYGPTVDEV----SNVSKENGSKVEEST 476
+ +YW+I + +NL+YYL+C+ Y +V N S++N ++ +T
Sbjct: 973 EYFYWLITLLQFINLIYYLICAKMYTYKQIQVHHKGENSSEDNNIELASTT 1125
>TC90302 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (25%)
Length = 933
Score = 198 bits (504), Expect = 4e-51
Identities = 99/149 (66%), Positives = 119/149 (79%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQ 60
M+LLWLTAMI P+EG +S T ++AML SA ILI+I +GG+SCS+AFGADQ
Sbjct: 512 MALLWLTAMIK---------PTEGDQSPTSWEMAMLISAFILISIASGGVSCSMAFGADQ 664
Query: 61 VNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTV 120
VN KDNPNN RVLE+FFSWYYAF +I+ IIALT IVYI DHLGWK+GFGVPA LM +ST+
Sbjct: 665 VNIKDNPNNNRVLEMFFSWYYAFASISAIIALTVIVYIPDHLGWKIGFGVPAALMFLSTL 844
Query: 121 LFFLASPLYVKIKQKTSLFTGFAQVSVAA 149
LFFLASPLYVKI+++ +LF F QV VA+
Sbjct: 845 LFFLASPLYVKIQKRNNLFASFGQVIVAS 931
>TC86046 similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (22%)
Length = 710
Score = 186 bits (473), Expect(2) = 5e-51
Identities = 90/91 (98%), Positives = 91/91 (99%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQ 60
MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQ
Sbjct: 386 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQ 565
Query: 61 VNRKDNPNNRRVLEIFFSWYYAFTTIAVIIA 91
VNRKDNPNNRRVLEIFFSWYYAFTTIAVII+
Sbjct: 566 VNRKDNPNNRRVLEIFFSWYYAFTTIAVIIS 658
Score = 33.1 bits (74), Expect(2) = 5e-51
Identities = 14/18 (77%), Positives = 14/18 (77%)
Frame = +3
Query: 91 ALTGIVYIQDHLGWKVGF 108
ALTGIVY QDHLGW F
Sbjct: 657 ALTGIVYXQDHLGWXSWF 710
>TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptide
transporter [imported] - Arabidopsis thaliana, partial
(53%)
Length = 1112
Score = 192 bits (487), Expect = 3e-49
Identities = 116/357 (32%), Positives = 197/357 (54%), Gaps = 9/357 (2%)
Frame = +3
Query: 86 IAVIIALTGIVYIQDHLGWKVGFGVPAILMLISTVLFFLASPLYV-KIKQKTSLFTGFAQ 144
+ +IA G+VYIQ++LGW +G+G+P +++S V+F++ +P+Y K++ S +
Sbjct: 75 LGALIATLGLVYIQENLGWGLGYGIPTAGLILSLVIFYIGTPIYRHKVRTSKSPAKDIIR 254
Query: 145 VSVAAYKNRKLPLP--PKTSPEFYHQQ---KDSELVVPTDKLRFLNKACVIKDHEQDIAS 199
V + A+K+RKL LP P EF + + V T LRFL+KA + +D +
Sbjct: 255 VFIVAFKSRKLQLPSNPSELHEFQMEHCVIRGKRQVYHTPTLRFLDKAAIKED-----PT 419
Query: 200 DGSAINRWSLCTVDQVEELKAIIKVIPLWSTAI---TMSINIGGSFGLLQAKSLDRHIIS 256
GS+ R TV+QVE K I+ ++ +W + T+ I F + Q +LDR++
Sbjct: 420 TGSS--RRVPMTVNQVEGAKLILGMLLIWLVTLIPSTIWAQINTLF-VKQGTTLDRNL-- 584
Query: 257 SSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLH 316
+F++PA S + +++L+ + +YDR+ +P + G P I+ +R+GIG +
Sbjct: 585 GPDFKIPAASLGSFVTLSMLLSVPMYDRLFVPFMRQKTGHPRGITLLQRLGIGFSIQIIA 764
Query: 317 LITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKE 376
+ A E VRR IKE ++ ++ MS WL PQ L GIA++FN IG EF+Y +
Sbjct: 765 IAIAYAVE-VRRMHVIKENHIFGPKDIVPMSIFWLLPQYVLIGIADVFNAIGLLEFFYDQ 941
Query: 377 FPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYY 433
P+ M S+ + +GVGN ++S ++++ + T G + W++DN+N H YY
Sbjct: 942 SPEDMQSLGTTFFTSGIGVGNFLNSFLVTMTDKITGRGDRKSWIADNLNDSHLXYYY 1112
>TC82302 similar to PIR|F84663|F84663 probable nitrate transporter
[imported] - Arabidopsis thaliana, partial (51%)
Length = 1133
Score = 182 bits (463), Expect = 2e-46
Identities = 108/333 (32%), Positives = 187/333 (55%), Gaps = 2/333 (0%)
Frame = +3
Query: 144 QVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSA 203
QV VA+ K RK+ LP + +DS + +++ RFL KA ++ + + D GS
Sbjct: 9 QVIVASIKKRKMELPYNVGSLYEDTPEDSR-IEQSEQFRFLEKAAIVVEGDFDKDLYGSG 185
Query: 204 INRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG--SFGLLQAKSLDRHIISSSNFE 261
N W LC++ +VEE+K +++++P+W+T I +F + QA +++R++ NF+
Sbjct: 186 PNPWKLCSLTRVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQAATMERNV---GNFQ 356
Query: 262 VPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAA 321
+PAGS +V +VAIL+ + + DR+++PL K+ GKP S +R IGL + + + A+
Sbjct: 357 IPAGSLTVFFVVAILLTLAVNDRIIMPLWKKLNGKPGF-SNLQRNAIGLLLSTIGMAAAS 533
Query: 322 IFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSM 381
+ E V+R K N T L +S L PQ L G E F GQ +F+ + PK M
Sbjct: 534 LIE-VKRLSVAKGVKGNQT--TLPISVFLLVPQFFLVGSGEAFIYTGQLDFFITQSPKGM 704
Query: 382 SSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINA 441
+++ L + +G VSS ++S+++ T + +GW++D+INKG D +Y ++ ++
Sbjct: 705 KTMSTGLFLTTLSLGFFVSSFLVSVVKKVTGTRDGQGWLADHINKGRLDLFYALLTILSF 884
Query: 442 LNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEE 474
+N + +LVC++ Y P + S + S VEE
Sbjct: 885 INFVAFLVCAFWYKPKKPKPS-MQINGSSSVEE 980
>TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affinity nitrate
transporter {Nicotiana plumbaginifolia}, partial (56%)
Length = 1169
Score = 181 bits (458), Expect = 8e-46
Identities = 106/327 (32%), Positives = 182/327 (55%), Gaps = 12/327 (3%)
Frame = +1
Query: 137 SLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSEL------VVPTDK-LRFLNKACV 189
S T A V VAA++ R + LP +S F + E+ V+P K RFL+KA +
Sbjct: 61 SPLTQIAVVYVAAWRKRNMELPYDSSLLFNVDDIEDEMLRKKKQVLPHSKQFRFLDKAAI 240
Query: 190 IKDHEQDIASDGSAIN---RWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG--SFGL 244
+D +DG+ IN +W L T+ VEE+K +++++P+W+T I +F +
Sbjct: 241 -----KDPKTDGNEINVVRKWYLSTLTDVEEVKLVLRMLPIWATTIMFWTVYAQMTTFSV 405
Query: 245 LQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKK 304
QA +L+RHI +F++P S + I +IL+ I IYDRV++P+ KI P ++P +
Sbjct: 406 SQATTLNRHI--GKSFQIPPASLTAFFIGSILLTIPIYDRVIVPITRKIFKNPQGLTPLQ 579
Query: 305 RMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMF 364
R+G+GL F+ ++ AA+ E R + A ++ + + MS WL PQ G E F
Sbjct: 580 RIGVGLVFSIFAMVAAALTELKRMRMAHLHNLTHNPNSEIPMSVFWLIPQFFFVGSGEAF 759
Query: 365 NVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNI 424
IGQ +F+ +E PK M +++ L + +G SSL+++++ T ++ W++DN+
Sbjct: 760 TYIGQLDFFLRECPKGMKTMSTGLFLSTLSLGFFFSSLLVTLVHKVTSQ--HKPWLADNL 933
Query: 425 NKGHFDKYYWVIVGINALNLLYYLVCS 451
N+ +YW++ ++ LNL+ YL+C+
Sbjct: 934 NEAKLYNFYWLLALLSVLNLVIYLLCA 1014
>TC79430 similar to PIR|F86358|F86358 Similar to peptide transporter
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1303
Score = 166 bits (419), Expect = 3e-41
Identities = 109/335 (32%), Positives = 180/335 (53%), Gaps = 6/335 (1%)
Frame = +2
Query: 1 MSLLWLTAMIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGIS-CSLAFGAD 59
+SLL L+A +PS +G Q + F +L L+A GG C AFGAD
Sbjct: 386 LSLLTLSATLPS----------DG-----DAQAILFFFSLYLVAFAQGGHKPCVQAFGAD 520
Query: 60 QVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIST 119
Q + ++P RR FF+W+Y T +++ ++ + Y+QD++GW +GFG+P I M+I+
Sbjct: 521 QFDI-NHPQERRSRSSFFNWWYFTFTAGLLVTVSILNYVQDNVGWVLGFGIPWIAMIIAL 697
Query: 120 VLFFLASPLY---VKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVV 176
LF L + Y + Q+ F+ +V + A +N + + P HQ
Sbjct: 698 SLFSLGTWTYRFSIPGNQQRGPFSRIGRVFITALRNFRTTEEEEPRPSLLHQ-------- 853
Query: 177 PTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSI 236
P+ + FLNKA IASDGS N +C+V +VEE KAI++++P+W+T++ +I
Sbjct: 854 PSQQFSFLNKAL--------IASDGSKEN-GKVCSVSEVEEAKAILRLVPIWATSLIFAI 1006
Query: 237 --NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIR 294
+ +F Q +LDR I+ F VP S + ++I+++I +YDR+++P+A
Sbjct: 1007VFSQSSTFFTKQGVTLDRKIL--PGFYVPPASLQSFISLSIVLFIPVYDRIIVPIARTFT 1180
Query: 295 GKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRK 329
GKP I+ +R+G G+ F+ + ++ AA E R K
Sbjct: 1181GKPSGITMLQRIGAGILFSVISMVIAAFVEMKRSK 1285
>TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersicon
esculentum}, partial (38%)
Length = 804
Score = 165 bits (418), Expect = 3e-41
Identities = 84/243 (34%), Positives = 143/243 (58%)
Frame = +1
Query: 214 QVEELKAIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIV 273
QVEELK +I++ P+W+T I S ++ L + S +F++ S S +
Sbjct: 1 QVEELKILIRMFPIWATGIIFS-SVYAQMSTLFVEQGTMMNTSIGSFKLSPASLSTFEVA 177
Query: 274 AILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIK 333
++++W+ +YD++L+P+ K GK IS +R+GIG F + L ++ AA E ++R + +
Sbjct: 178 SVVMWVPVYDKILVPIVKKFTGKKRGISVFQRIGIGPFISGLCMLAAAAVE-IKRLQLAR 354
Query: 334 EGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAM 393
E L D + +S +W PQ + G AE+F +GQ EF+Y+E P +M ++ +L L
Sbjct: 355 ELDLVDKPVGVPLSVLWQIPQYLILGAAEIFTFVGQLEFFYEESPDAMRTICGALPLLNF 534
Query: 394 GVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWA 453
+GN +SS +L+I+ T GG GW+ DN+NKGH D Y+ ++ G++ LN+L ++V +
Sbjct: 535 SLGNYLSSFILTIVTYFTTKGGRLGWIPDNLNKGHLDYYFLLLSGLSLLNMLVFIVAAKI 714
Query: 454 YGP 456
Y P
Sbjct: 715 YKP 723
>BE998753 similar to PIR|T45958|T45 oligopeptide transporter-like protein -
Arabidopsis thaliana, partial (44%)
Length = 809
Score = 156 bits (394), Expect = 2e-38
Identities = 88/268 (32%), Positives = 151/268 (55%), Gaps = 9/268 (3%)
Frame = +3
Query: 146 SVAAYKNRKLPLPPKTSPEFYHQQKDSELVVP-------TDKLRFLNKACVIKDHEQDIA 198
SV RK + T ++ D+E + T++LRF +KA V E D
Sbjct: 27 SVIVASIRKYRVDAPTDKSLLYEIADTESAIKGSRKLDHTNELRFFDKAAV--QGESDNL 200
Query: 199 SDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG--GSFGLLQAKSLDRHIIS 256
+ +IN W LCTV QVEELK+I++++P+W+T I + G + +LQ ++++ H+
Sbjct: 201 KE--SINPWRLCTVTQVEELKSILRLLPVWATGIIFATVYGQMSTLFVLQGQTMNTHV-G 371
Query: 257 SSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLH 316
+S+F++P S S+ ++++ W+ +YDR+++P+A K G ++ +RMG+GLF +
Sbjct: 372 NSSFKIPPASLSIFDTISVIFWVPVYDRIIVPIARKFTGHKNGLTQLQRMGVGLFISIFS 551
Query: 317 LITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKE 376
++ AA E VR + + Y + M+ W PQ L G AE+F IGQ EF+Y++
Sbjct: 552 MVAAAFLELVRLRTVRRNNYYELEE--IPMTIFWQVPQYFLIGCAEVFTFIGQLEFFYEQ 725
Query: 377 FPKSMSSVAASLSGLAMGVGNLVSSLVL 404
P +M S+ ++LS L + G +SSL++
Sbjct: 726 APDAMRSLCSALSLLTVAFGQYLSSLLV 809
>TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter
{Arabidopsis thaliana}, partial (60%)
Length = 1236
Score = 149 bits (377), Expect = 2e-36
Identities = 95/288 (32%), Positives = 162/288 (55%), Gaps = 6/288 (2%)
Frame = +3
Query: 3 LLWLTAMIPSARPPACNH---PSEGCESATPGQLAMLFSALILIAIGNGGISCSLA-FGA 58
+L L IPS PP C+ C A+ QL++LF+AL IA+G GGI +++ FG+
Sbjct: 402 MLTLATTIPSMTPPPCSEVRRQHHQCIQASGKQLSLLFAALYTIALGGGGIKSNVSGFGS 581
Query: 59 DQVNRKDNPNNRRVLEIFFSWYYAFTTIAVIIALTGIVYIQDHLGWKVGFGVPAILMLIS 118
DQ + D P + + FF+ +Y F +I + ++ +VY+QD++G G+G+ A ML++
Sbjct: 582 DQFDTND-PKEEKNMIFFFNRFYFFISIGSLFSVVVLVYVQDNIGRGWGYGISAGTMLVA 758
Query: 119 TVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPPKTSPEFYHQQKDSELVVPT 178
+ +PLY K + S T +V A+K R LP+P + P + +++ V T
Sbjct: 759 VGILLCGTPLYRFKKPQGSPLTIIWRVLFLAWKKRTLPIP--SDPTLLNGYLEAK-VTYT 929
Query: 179 DKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SI 236
DK R L+KA ++ ++ + DG+ N W + T+ QVEE+K +IK++P+WST I
Sbjct: 930 DKFRSLDKAAIL---DETKSKDGNNENPWLVSTMTQVEEVKMVIKLLPIWSTCILFWTVY 1100
Query: 237 NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDR 284
+ +F + QA ++R + + E+PAGS S L + IL++ + ++
Sbjct: 1101SQMNTFTIEQATFMNRKV---GSLEIPAGSLSAFLFITILLFTSLNEK 1235
>TC93408 similar to GP|11933407|dbj|BAB19758. putative nitrate transporter
NRT1-3 {Glycine max}, partial (44%)
Length = 909
Score = 149 bits (377), Expect = 2e-36
Identities = 97/308 (31%), Positives = 164/308 (52%), Gaps = 10/308 (3%)
Frame = +1
Query: 91 ALTGIVYIQDHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAY 150
A T +VYIQD++GW +G+ +P + + IS ++F +P Y S FT A+V VA+
Sbjct: 1 ANTVLVYIQDNVGWTLGYALPTLGLAISIMIFLGGTPFYRHKLPAGSTFTRMARVIVASL 180
Query: 151 KNRKLPLPPKTS-------PEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSA 203
+ K+P+P T E+ + ++ + T LRFL+KA V GS
Sbjct: 181 RKWKVPVPDDTKKLYELDMEEYAKKGSNTYRIDSTPTLRFLDKASV---------KTGST 333
Query: 204 INRWSLCTVDQVEELKAIIKVIPLWSTAI---TMSINIGGSFGLLQAKSLDRHIISSSNF 260
+ W LCTV VEE K ++++IP+ TM + F + Q +LDRHI +F
Sbjct: 334 -SPWMLCTVTHVEETKQMLRMIPILVATFVPSTMMAQVNTLF-VKQGTTLDRHI---GSF 498
Query: 261 EVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITA 320
++P S + + +++L+ +++YDR + + ++ P I+ +RMGIGL + + ++ A
Sbjct: 499 KIPPASLAAFVTLSLLVCVVLYDRFFVRIMQRLTKNPRGITLLQRMGIGLVLHTIIMVVA 678
Query: 321 AIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKS 380
++ E R + A KE L ++ G + +S L PQ L G A+ F + + EF+Y + P S
Sbjct: 679 SVTENYRLRVA-KEHGLVESGGQVPLSIFILLPQFILMGTADAFLEVAKIEFFYDQAPXS 855
Query: 381 MSSVAASL 388
M S+ S+
Sbjct: 856 MKSIGTSI 879
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,301,556
Number of Sequences: 36976
Number of extensions: 251128
Number of successful extensions: 1436
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 1353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1367
length of query: 526
length of database: 9,014,727
effective HSP length: 101
effective length of query: 425
effective length of database: 5,280,151
effective search space: 2244064175
effective search space used: 2244064175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149489.11


