

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149489.10 - phase: 0
(638 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
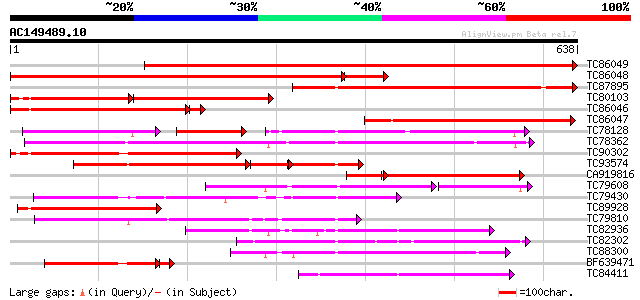
Score E
Sequences producing significant alignments: (bits) Value
TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative tra... 938 0.0
TC86048 weakly similar to PIR|H96561|H96561 probable peptide tra... 658 0.0
TC87895 similar to GP|20466248|gb|AAM20441.1 putative transport ... 478 e-135
TC80103 weakly similar to PIR|H96561|H96561 probable peptide tra... 264 e-131
TC86046 similar to PIR|H96561|H96561 probable peptide transporte... 374 e-107
TC86047 weakly similar to PIR|T48516|T48516 probable oligopeptid... 355 2e-98
TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T1... 194 2e-95
TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter... 343 9e-95
TC90302 weakly similar to PIR|H96561|H96561 probable peptide tra... 335 4e-92
TC93574 weakly similar to GP|21536862|gb|AAM61194.1 unknown {Ara... 207 2e-81
CA919816 similar to GP|20466248|gb putative transport protein {A... 232 2e-74
TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {... 153 1e-52
TC79430 similar to PIR|F86358|F86358 Similar to peptide transpor... 203 2e-52
TC89928 weakly similar to GP|20466248|gb|AAM20441.1 putative tra... 200 1e-51
TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter... 192 3e-49
TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptid... 192 4e-49
TC82302 similar to PIR|F84663|F84663 probable nitrate transporte... 182 3e-46
TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affini... 181 8e-46
BF639471 weakly similar to GP|20466248|gb| putative transport pr... 164 9e-44
TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersico... 165 4e-41
>TC86049 weakly similar to GP|20466248|gb|AAM20441.1 putative transport
protein {Arabidopsis thaliana}, partial (43%)
Length = 1836
Score = 938 bits (2425), Expect = 0.0
Identities = 471/487 (96%), Positives = 479/487 (97%)
Frame = +2
Query: 152 LILIAIGNGGITCSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALTGIVYIQ 211
LILIAIGNGGITCSL+FGA QVNRKDNP+ YRVLEIFFSWYYAF TIAVIIALTGIVYIQ
Sbjct: 2 LILIAIGNGGITCSLSFGAYQVNRKDNPDGYRVLEIFFSWYYAFTTIAVIIALTGIVYIQ 181
Query: 212 DHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKLSLPP 271
DHLGW++GFGVPA LMLISTVLFFLASPLYVKI Q+TSL TGFAQVSVAAYKNRKL LPP
Sbjct: 182 DHLGWKVGFGVPAILMLISTVLFFLASPLYVKIKQKTSLFTGFAQVSVAAYKNRKLPLPP 361
Query: 272 KTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK 331
KTSPEFYH +KDS+LV+PTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK
Sbjct: 362 KTSPEFYHQQKDSELVVPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELK 541
Query: 332 AIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWI 391
AIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWI
Sbjct: 542 AIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWI 721
Query: 392 IIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLND 451
IIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLND
Sbjct: 722 IIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLND 901
Query: 452 THGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLV 511
THGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLV
Sbjct: 902 THGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLV 1081
Query: 512 SSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD 571
SSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD
Sbjct: 1082SSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVD 1261
Query: 572 EVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQR 631
EVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQR
Sbjct: 1262EVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQR 1441
Query: 632 DLKEEDL 638
DLKEEDL
Sbjct: 1442DLKEEDL 1462
>TC86048 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (47%)
Length = 1364
Score = 658 bits (1698), Expect(2) = 0.0
Identities = 331/379 (87%), Positives = 351/379 (92%)
Frame = +3
Query: 1 MDKELQLCSVDAADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLM 60
MDKELQLCSVDAADEEEMVSQQKP+RSKGGLVTMPFII NEALA+MAS GL PNMILYLM
Sbjct: 63 MDKELQLCSVDAADEEEMVSQQKPERSKGGLVTMPFIIANEALARMASTGLGPNMILYLM 242
Query: 61 GSYRLHLGISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTA 120
GSY LHL +TQILL+SSAA NFTP++ AFIADSY+GRFLGVGIGS ISFLGM++LWLTA
Sbjct: 243 GSYNLHLATATQILLISSAAGNFTPLVAAFIADSYIGRFLGVGIGSIISFLGMAMLWLTA 422
Query: 121 MIPAARPSACNHPSEGCESATPGQLAMLFSALILIAIGNGGITCSLAFGADQVNRKDNPN 180
M+PAA+P AC++P EGC SA PGQ+AML SALILI IGNGGI+CSLAFGADQV RK+N N
Sbjct: 423 MVPAAQPPACSNPPEGCISAKPGQMAMLLSALILIGIGNGGISCSLAFGADQVKRKENSN 602
Query: 181 NYRVLEIFFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPL 240
N R LEIFF+WYYA TIAVI AL GIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPL
Sbjct: 603 NNRALEIFFTWYYASTTIAVIAALFGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPL 782
Query: 241 YVKITQRTSLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKAC 300
YVKITQRTSLLTGFAQVSVAA+KNRKLSLPPKTSPEFYHHKKDS LV+PTDKLRFLNKAC
Sbjct: 783 YVKITQRTSLLTGFAQVSVAAFKNRKLSLPPKTSPEFYHHKKDSVLVVPTDKLRFLNKAC 962
Query: 301 VIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGSFGLLQAK 360
VIKDHEQDIASDGS INRWSLCTVDQVEELKAIIKVIPLWST ITMSINIGGSFGLLQA
Sbjct: 963 VIKDHEQDIASDGSIINRWSLCTVDQVEELKAIIKVIPLWSTGITMSINIGGSFGLLQAL 1142
Query: 361 SLDRHIISSSNFEVPAGSF 379
SLDRHIIS+SNFEVPAG F
Sbjct: 1143SLDRHIISNSNFEVPAGIF 1199
Score = 72.8 bits (177), Expect(2) = 0.0
Identities = 37/50 (74%), Positives = 40/50 (80%)
Frame = +1
Query: 377 GSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNF 426
GSF++ILI ILIWIIIYDRVLIPLASKIRGKP ISP RMG + F F
Sbjct: 1192 GSFTIILIFVILIWIIIYDRVLIPLASKIRGKPASISPMIRMGTWVVF*F 1341
>TC87895 similar to GP|20466248|gb|AAM20441.1 putative transport protein
{Arabidopsis thaliana}, partial (7%)
Length = 1265
Score = 478 bits (1229), Expect = e-135
Identities = 235/320 (73%), Positives = 271/320 (84%)
Frame = +1
Query: 319 WSLCTVDQVEELKAIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGS 378
W LCTVDQVEELKA+++VIPLWST I MS+NIGGSFGLLQAKSLDRHI +SNFEVPAGS
Sbjct: 1 WRLCTVDQVEELKALVRVIPLWSTGIMMSLNIGGSFGLLQAKSLDRHI--TSNFEVPAGS 174
Query: 379 FSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETV 438
FSVI++ +I IWI++YDRVLIPLASK+RGKPV ISPK RMGIGLF NFLHL+TAA FE++
Sbjct: 175 FSVIMVGSIFIWIVLYDRVLIPLASKLRGKPVRISPKIRMGIGLFLNFLHLVTAAAFESI 354
Query: 439 RRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAA 498
RRK+AI+ G+LNDTHGVLKMSA+WLAPQLCL GI+E FN IGQNEFYYKEFP++MSS++A
Sbjct: 355 RRKKAIQAGFLNDTHGVLKMSALWLAPQLCLGGISEAFNAIGQNEFYYKEFPRTMSSISA 534
Query: 499 SLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLY 558
SL GL M GNLVSS V + IE+ T GG EGW+SDNINKG +DKYY VI G++ALNLLY
Sbjct: 535 SLCGLGMAAGNLVSSFVFNTIENVTSRGGKEGWISDNINKGRYDKYYMVIAGVSALNLLY 714
Query: 559 YLVCSWAYGPTVDEVSNVSKENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNG 618
+LVCSWAYGPTVD+VS VS EN SK ++STEFK +NP FD ETSS KELTE KNG
Sbjct: 715 FLVCSWAYGPTVDQVSKVSDENDSKEKDSTEFKKVNPLFD-----ETSSNGKELTEIKNG 879
Query: 619 AQVEKVFKNSEQRDLKEEDL 638
Q EKVFK SE+ KEE+L
Sbjct: 880 GQAEKVFKISEENGSKEEEL 939
>TC80103 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (23%)
Length = 1236
Score = 264 bits (674), Expect(2) = e-131
Identities = 126/159 (79%), Positives = 146/159 (91%), Gaps = 2/159 (1%)
Frame = +3
Query: 140 ATPGQLAMLFSALILIAIGNGGITCSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIA 199
AT GQ+AML SA L++IGNGG++CS+AFGADQVNRKDNPNN+RVLEIFFSWYYAF TI+
Sbjct: 645 ATTGQMAMLLSAFGLMSIGNGGLSCSIAFGADQVNRKDNPNNHRVLEIFFSWYYAFTTIS 824
Query: 200 VIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSV 259
+I+ALT IVYIQ+HLGW+IGFGVPAALML+ST+ FFLASPLYVKIT+RT L+TGFAQV+
Sbjct: 825 IILALTVIVYIQEHLGWKIGFGVPAALMLLSTICFFLASPLYVKITKRTXLITGFAQVTT 1004
Query: 260 AAYKNRKLSLPPKTSPEFYHHKKDS--DLVIPTDKLRFL 296
AAYKNRKL LPPK SP+FYHHKKDS DLVIPTDKLR++
Sbjct: 1005AAYKNRKLQLPPKNSPQFYHHKKDSDLDLVIPTDKLRYI 1121
Score = 223 bits (567), Expect(2) = e-131
Identities = 112/139 (80%), Positives = 126/139 (90%)
Frame = +2
Query: 1 MDKELQLCSVDAADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLM 60
MDKELQLCSVD ++EMVSQQ PQR KGGL+TMPFII NEALA+MAS+GLLPNMILY M
Sbjct: 239 MDKELQLCSVD---DDEMVSQQ-PQRRKGGLITMPFIIANEALARMASLGLLPNMILYFM 406
Query: 61 GSYRLHLGISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTA 120
GSYRLHL +TQILLLSSAASNFTPV+GAFIADSYLGRFLGVG+GS +SFLGM +LWLTA
Sbjct: 407 GSYRLHLAKATQILLLSSAASNFTPVVGAFIADSYLGRFLGVGLGSFVSFLGMGMLWLTA 586
Query: 121 MIPAARPSACNHPSEGCES 139
MIPAARP CN+P++GC+S
Sbjct: 587 MIPAARPPPCNNPTKGCKS 643
>TC86046 similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (22%)
Length = 710
Score = 374 bits (960), Expect(2) = e-107
Identities = 190/203 (93%), Positives = 198/203 (96%)
Frame = +2
Query: 1 MDKELQLCSVDAADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLM 60
MDKELQLCSVDAA+EEEMVSQQ PQR KGGL+TMPFIIGNEALA+MAS+GLLPNMILYLM
Sbjct: 53 MDKELQLCSVDAANEEEMVSQQ-PQRRKGGLITMPFIIGNEALARMASLGLLPNMILYLM 229
Query: 61 GSYRLHLGISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTA 120
GSYRLHLGISTQILLLSSAASNFTPVIGAFIADS+LGRFLGVGIGSSISFLGMSLLWLTA
Sbjct: 230 GSYRLHLGISTQILLLSSAASNFTPVIGAFIADSFLGRFLGVGIGSSISFLGMSLLWLTA 409
Query: 121 MIPAARPSACNHPSEGCESATPGQLAMLFSALILIAIGNGGITCSLAFGADQVNRKDNPN 180
MIP+ARP ACNHPSEGCESATPGQLAMLFSALILIAIGNGGI+CSLAFGADQVNRKDNPN
Sbjct: 410 MIPSARPPACNHPSEGCESATPGQLAMLFSALILIAIGNGGISCSLAFGADQVNRKDNPN 589
Query: 181 NYRVLEIFFSWYYAFITIAVIIA 203
N RVLEIFFSWYYAF TIAVII+
Sbjct: 590 NRRVLEIFFSWYYAFTTIAVIIS 658
Score = 33.1 bits (74), Expect(2) = e-107
Identities = 14/18 (77%), Positives = 14/18 (77%)
Frame = +3
Query: 203 ALTGIVYIQDHLGWRIGF 220
ALTGIVY QDHLGW F
Sbjct: 657 ALTGIVYXQDHLGWXSWF 710
>TC86047 weakly similar to PIR|T48516|T48516 probable oligopeptide
transporter protein - Arabidopsis thaliana, partial
(11%)
Length = 983
Score = 355 bits (912), Expect = 2e-98
Identities = 180/238 (75%), Positives = 199/238 (82%), Gaps = 1/238 (0%)
Frame = +3
Query: 400 PLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMS 459
PLASKIRGKP ISP RM G F H + +IFE++RRK+AI+ G+LN+THGVLKMS
Sbjct: 3 PLASKIRGKPASISPMIRMDWGCFLISSHC-SGSIFESIRRKKAIQAGFLNNTHGVLKMS 179
Query: 460 AMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSII 519
AMWLAPQLCLAGIAE FNVIGQNEFYYKEFPK+MSSV+++LSGLAM GNLVSS V S I
Sbjct: 180 AMWLAPQLCLAGIAEAFNVIGQNEFYYKEFPKTMSSVSSALSGLAMAAGNLVSSFVFSTI 359
Query: 520 ESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVS-K 578
E+TT SGGNEGW+SDNINKGHFDKY WVI GIN LNLLYYLVCSWAYGPT D+VS VS +
Sbjct: 360 ENTTSSGGNEGWISDNINKGHFDKYLWVIAGINVLNLLYYLVCSWAYGPTADQVSKVSEE 539
Query: 579 ENGSKVEESTEFKHMNPHFDDKVSGETSSKEKELTEFKNGAQVEKVFKNSEQRDLKEE 636
ENGSK E STEFK++NP DDKVS ETSSKEKELTEFKNG QVEKV K SE+ KEE
Sbjct: 540 ENGSKEENSTEFKNVNPQIDDKVSDETSSKEKELTEFKNGGQVEKVSKVSEENGSKEE 713
>TC78128 weakly similar to GP|21928025|gb|AAM78041.1 At1g69870/T17F3_10
{Arabidopsis thaliana}, partial (77%)
Length = 2022
Score = 194 bits (494), Expect(3) = 2e-95
Identities = 109/303 (35%), Positives = 176/303 (57%), Gaps = 6/303 (1%)
Frame = +2
Query: 289 PTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAIT--M 346
PT+ R L+KA ++ E D+ DG+ +N+W+L ++ QVEE+K + + +P+W+ I
Sbjct: 1046 PTNS-RVLDKAALVM--EGDLNPDGTIVNQWNLVSIQQVEEVKCLARTLPIWAAGILGFT 1216
Query: 347 SINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIR 406
++ G+F + QA +DRH+ F++PAGS VI ++ I +W+ YDR+ +P KI
Sbjct: 1217 AMAQQGTFIVSQAMKMDRHL--GPKFQIPAGSLGVISLIVIGLWVPFYDRICVPSLRKIT 1390
Query: 407 GKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDT-HGVLKMSAMWLAP 465
I+ +R+GIG+ F+ + +I A + E VRR A N T G+ MS MWL P
Sbjct: 1391 KNEGGITLLQRIGIGMVFSIISMIVAGLVEKVRRDVANS----NPTPQGIAPMSVMWLFP 1558
Query: 466 QLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPS 525
QL L G E FN+IG EF+ ++FP M S+A +L + + N VSS+++ + S T +
Sbjct: 1559 QLVLMGFCEAFNIIGLIEFFNRQFPDHMRSIANALFSCSFALANYVSSILVITVHSVTKT 1738
Query: 526 GGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWAY---GPTVDEVSNVSKENGS 582
+ W+++NIN+G D +Y+++ G+ LNL+Y+L S Y G + + E GS
Sbjct: 1739 HNHPDWLTNNINEGRLDYFYYLLAGVGVLNLVYFLYVSQRYHYKGSVDIQEKPMDVELGS 1918
Query: 583 KVE 585
K E
Sbjct: 1919 KGE 1927
Score = 114 bits (284), Expect(3) = 2e-95
Identities = 60/159 (37%), Positives = 93/159 (57%), Gaps = 4/159 (2%)
Frame = +2
Query: 15 EEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQIL 74
E+E ++ ++ GG MPFI+GNE+ ++A+ GLL N ++YL + L ++ IL
Sbjct: 191 EKEPNYEKHGKKKPGGWKAMPFILGNESFERLAAFGLLANFMVYLTREFHLEQVHASNIL 370
Query: 75 LLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPS 134
+ SNF P++GAFI+D+Y GRF + S S LGM+ + LTA +P +P +C
Sbjct: 371 NIWGGISNFAPLLGAFISDTYTGRFKTIAFASFFSLLGMTAVTLTAWLPKLQPPSCTPQQ 550
Query: 135 EG---CESATPGQLAMLFSALILIAIGNGGI-TCSLAFG 169
+ C +A Q+ LF LI ++IG+ GI CS+ FG
Sbjct: 551 QALNQCVTANSSQVGFLFMGLIFLSIGSSGIRPCSIPFG 667
Score = 81.3 bits (199), Expect(3) = 2e-95
Identities = 34/79 (43%), Positives = 50/79 (63%)
Frame = +3
Query: 188 FFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQR 247
FF+WYY T+ ++ T IVYIQD + W+ GF +P ML S +LFF+ + +YV +
Sbjct: 720 FFNWYYTSFTVVLLFTQTVIVYIQDSISWKFGFAIPTLCMLXSIILFFIGTKIYVHVKPE 899
Query: 248 TSLLTGFAQVSVAAYKNRK 266
S+ + AQV VA++K RK
Sbjct: 900 GSIFSSIAQVLVASFKKRK 956
>TC78362 similar to GP|15391731|emb|CAA93316. nitrite transporter {Cucumis
sativus}, partial (88%)
Length = 1871
Score = 343 bits (881), Expect = 9e-95
Identities = 199/600 (33%), Positives = 333/600 (55%), Gaps = 26/600 (4%)
Frame = +3
Query: 17 EMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLL 76
+MV ++ ++ +GG+ T+PFI+ NE + AS G NMI YL + L ++ L
Sbjct: 51 KMVEKESKRQQRGGIRTLPFILANEVCDRFASAGFHANMITYLTQQLNMPLVSASNTLSN 230
Query: 77 SSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSE- 135
S S+ TP++GAFIADS+ GRF + + I LG+ + +A++P RP C P++
Sbjct: 231 FSGLSSLTPLLGAFIADSFAGRFWTIVFATLIYELGLITITTSAIVPHFRPPPC--PTQV 404
Query: 136 GCESATPGQLAMLFSALILIAIGNGGI-TCSLAFGADQVNRKDNPNNYRVLEIFFSWYYA 194
C+ A QL +L+ AL L ++G+GGI +C + F DQ + R +F +WY+
Sbjct: 405 NCQEAKSSQLWILYLALFLTSLGSGGIRSCVVPFSGDQFDMTKKGVESRKWNLF-NWYFF 581
Query: 195 FITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGF 254
+ A + ALT +VYIQDH+GW G G+P M I+ + F L S LY + S
Sbjct: 582 CMGFASLSALTIVVYIQDHMGWGWGLGIPTIAMFIAIIAFLLGSRLYKTLKPSGSPFLRL 761
Query: 255 AQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIP-----TDKLRFLNKACVIKDHEQDI 309
AQV VAA++ R +LP + + + DS + + TD+ ++L+KA ++ D E
Sbjct: 762 AQVIVAAFRKRNDALPNDPKLLYQNLELDSSISLEGRLSHTDQYKWLDKAAIVTDEEAKN 941
Query: 310 ASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAITM--SINIGGSFGLLQAKSLDRHII 367
+ N W+L TV +VEELK +++++P+W++ I + + + SF ++QA+++DRH+
Sbjct: 942 LN--KLPNLWNLATVHRVEELKCLVRMLPIWASGILLITASSSQHSFVIVQARTMDRHL- 1112
Query: 368 SSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFL 427
S FE+ S ++ ++ ++ +I+Y+R+ +P + P I+ +RMGIG N +
Sbjct: 1113-SHTFEISPASMAIFSVLTMMTGVILYERLFVPFIRRFTKNPAGITCLQRMGIGFVINII 1289
Query: 428 HLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYK 487
I +A+ E R+K A K L+ ++ +S WL PQ L G+AE+F +G EF Y
Sbjct: 1290ATIVSALLEIKRKKVASKYHLLDSPKAIIPISVFWLVPQYFLHGVAEVFMNVGHLEFLYD 1469
Query: 488 EFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSD-NINKGHFDKYYW 546
+ P+SM S A +L +A+ +G+ + +L+++++ T G W+ D N+N+G + YY+
Sbjct: 1470QSPESMRSSATALYCIAIAIGHFIGTLLVTLVHKYT--GKERNWLPDRNLNRGRLEYYYF 1643
Query: 547 VIVGINALNLLYYLVCSWAYG----------------PTVDEVSNVSKENGSKVEESTEF 590
++ GI +N +YY++C+W Y T E+S VS G KV+E EF
Sbjct: 1644LVCGIQVINFIYYVICAWIYNYKPLEEINENNQGDLEQTNGELSLVSLNEG-KVDEKREF 1820
>TC90302 weakly similar to PIR|H96561|H96561 probable peptide transporter
[imported] - Arabidopsis thaliana, partial (25%)
Length = 933
Score = 335 bits (858), Expect = 4e-92
Identities = 173/261 (66%), Positives = 206/261 (78%)
Frame = +2
Query: 1 MDKELQLCSVDAADEEEMVSQQKPQRSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLM 60
M+KE Q + D +EM SQ P+R KGGL+TMPFII NEAL AS+G+LPNMILYLM
Sbjct: 194 MNKEAQY----SGDADEMASQ--PRRRKGGLITMPFIIANEALGNTASLGILPNMILYLM 355
Query: 61 GSYRLHLGISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTA 120
G Y LHLG + QILLLS+AA F PV+GAF+ADSYLGRFL VG+GS++SFLGM+LLWLTA
Sbjct: 356 GPYNLHLGQANQILLLSAAAGKFMPVVGAFVADSYLGRFLSVGLGSAVSFLGMALLWLTA 535
Query: 121 MIPAARPSACNHPSEGCESATPGQLAMLFSALILIAIGNGGITCSLAFGADQVNRKDNPN 180
MI P+EG +S T ++AML SA ILI+I +GG++CS+AFGADQVN KDNPN
Sbjct: 536 MIK---------PTEGDQSPTSWEMAMLISAFILISIASGGVSCSMAFGADQVNIKDNPN 688
Query: 181 NYRVLEIFFSWYYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPL 240
N RVLE+FFSWYYAF +I+ IIALT IVYI DHLGW+IGFGVPAALM +ST+LFFLASPL
Sbjct: 689 NNRVLEMFFSWYYAFASISAIIALTVIVYIPDHLGWKIGFGVPAALMFLSTLLFFLASPL 868
Query: 241 YVKITQRTSLLTGFAQVSVAA 261
YVKI +R +L F QV VA+
Sbjct: 869 YVKIQKRNNLFASFGQVIVAS 931
>TC93574 weakly similar to GP|21536862|gb|AAM61194.1 unknown {Arabidopsis
thaliana}, partial (35%)
Length = 980
Score = 207 bits (528), Expect(3) = 2e-81
Identities = 106/200 (53%), Positives = 139/200 (69%), Gaps = 1/200 (0%)
Frame = +2
Query: 73 ILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNH 132
I+ L +A SNF P+ GA ++DS LGRF + G+ I LG+ +LWLTA+I ARP CN+
Sbjct: 8 IIFLWNAGSNFMPLFGALLSDSCLGRFRVIAWGTIIDLLGLIVLWLTAIIRHARPPECNN 187
Query: 133 PSEGCESATPGQLAMLFSALILIAIGNGGIT-CSLAFGADQVNRKDNPNNYRVLEIFFSW 191
E C SAT Q LFS+L L+A G GGI CSLAF ADQ+N NP N R+++ FF+W
Sbjct: 188 -GESCTSATGMQFLFLFSSLALMAFGAGGIRPCSLAFAADQINNPKNPKNERIMKSFFNW 364
Query: 192 YYAFITIAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLL 251
YY + ++V++++ IVYIQ GW IGFG+P LML S+V+FFL S +YVK+ SLL
Sbjct: 365 YYVSVGVSVMVSMVFIVYIQVKAGWVIGFGIPVGLMLFSSVVFFLGSFMYVKVKPNKSLL 544
Query: 252 TGFAQVSVAAYKNRKLSLPP 271
GFAQV VA++KNR L+LPP
Sbjct: 545 AGFAQVIVASWKNRHLTLPP 604
Score = 83.6 bits (205), Expect(3) = 2e-81
Identities = 42/87 (48%), Positives = 61/87 (69%), Gaps = 1/87 (1%)
Frame = +1
Query: 313 GSAINRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIG-GSFGLLQAKSLDRHIISSSN 371
G + SLCTV QVEELKA+I+V+PLWST IT+++ I SF ++QA +++R + N
Sbjct: 727 GCQLIHGSLCTVRQVEELKAVIQVLPLWSTGITIAVTISQQSFSVVQATTMNRMV---HN 897
Query: 372 FEVPAGSFSVILIVAILIWIIIYDRVL 398
FE+P SFS I+ + IW+ IYDR++
Sbjct: 898 FEIPPTSFSAFGILTLTIWVAIYDRII 978
Score = 52.0 bits (123), Expect(3) = 2e-81
Identities = 22/48 (45%), Positives = 35/48 (72%)
Frame = +3
Query: 272 KTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRW 319
+T+ ++ + S+LV PTDK RFLNKAC++K+ E+D+ S+G I+ W
Sbjct: 603 QTTAGLWYFQSGSNLVQPTDKARFLNKACIMKNREKDLDSNGMPIDPW 746
>CA919816 similar to GP|20466248|gb putative transport protein {Arabidopsis
thaliana}, partial (20%)
Length = 819
Score = 232 bits (592), Expect(2) = 2e-74
Identities = 106/161 (65%), Positives = 128/161 (78%)
Frame = -1
Query: 419 GIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNV 478
G + F+FLHL+TA+I ET+RR+ AI EGY ND H VL MSAMWL PQLCL GIAE FNV
Sbjct: 699 GTRIVFSFLHLVTASIVETIRRRRAINEGYGNDPHSVLNMSAMWLFPQLCLGGIAEAFNV 520
Query: 479 IGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINK 538
IGQNEFYY EFPK+MSS+A+SL GL M VG ++SS + SI+E T SGG +GW+SDNINK
Sbjct: 519 IGQNEFYYTEFPKTMSSIASSLFGLGMAVGYVISSFLFSIVERVTSSGGKDGWISDNINK 340
Query: 539 GHFDKYYWVIVGINALNLLYYLVCSWAYGPTVDEVSNVSKE 579
G FDKYYW+I ++ +N+LYY+VCSWAYGPT D S VS +
Sbjct: 339 GRFDKYYWLIATVSGVNILYYVVCSWAYGPTADVESKVSAD 217
Score = 65.9 bits (159), Expect(2) = 2e-74
Identities = 32/47 (68%), Positives = 37/47 (78%)
Frame = -2
Query: 380 SVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNF 426
SVI+I I IWI +YDRV+IPLA K+RGKPV IS K RMG+GLF F
Sbjct: 815 SVIMIFTIFIWIALYDRVVIPLARKLRGKPVRISAKTRMGLGLFSLF 675
>TC79608 similar to GP|20147231|gb|AAM10330.1 At1g68570/F24J5_7 {Arabidopsis
thaliana}, partial (59%)
Length = 1489
Score = 153 bits (387), Expect(2) = 1e-52
Identities = 91/267 (34%), Positives = 146/267 (54%), Gaps = 7/267 (2%)
Frame = +2
Query: 221 GVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHH 280
G+P MLIS + F PLY + S T +V VAA++ +K+ P ++ + +
Sbjct: 5 GIPTLAMLISIIAFIGGYPLYRNLNPEGSPFTRLVKVGVAAFRKKKIPKVPNSTLLYQND 184
Query: 281 KKDSDL-----VIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTVDQVEELKAIIK 335
+ D+ + ++ +D+L+FL+KA ++ + + D W L TV +VEELK+II+
Sbjct: 185 ELDASITLGGKLLHSDQLKFLDKAAIVTEEDNTKTPD-----LWRLSTVHRVEELKSIIR 349
Query: 336 VIPLWSTAITMSINIG--GSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIII 393
+ P+W++ I + G+F L QAK+++RH+ S FE+PAGS SV I+ +L +
Sbjct: 350 MGPIWASGILLITAYAQQGTFSLQQAKTMNRHLTKS--FEIPAGSMSVFTILTMLFTTAL 523
Query: 394 YDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTH 453
YDRVLI +A + G I+ RMGIG + A E R+K A++ G + +
Sbjct: 524 YDRVLIRVARRFTGLDRGITFLHRMGIGFVISLFATFVAGFVEMKRKKVAMEHGLIEHSS 703
Query: 454 GVLKMSAMWLAPQLCLAGIAEMFNVIG 480
++ +S WL PQ L G+AE F IG
Sbjct: 704 EIIPISVFWLVPQYSLHGMAEAFMSIG 784
Score = 72.0 bits (175), Expect(2) = 1e-52
Identities = 34/111 (30%), Positives = 65/111 (57%), Gaps = 5/111 (4%)
Frame = +1
Query: 483 EFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDN-INKGHF 541
EF+Y + P+SM+S A + ++ +GN +S+L+++++ T W+ DN +NKG
Sbjct: 793 EFFYDQAPESMTSTAMAFFWTSISLGNYISTLLVTLVHKFTKGPNGTNWLPDNNLNKGKL 972
Query: 542 DKYYWVIVGINALNLLYYLVCSWAYGPTVDEV----SNVSKENGSKVEEST 588
+ +YW+I + +NL+YYL+C+ Y +V N S++N ++ +T
Sbjct: 973 EYFYWLITLLQFINLIYYLICAKMYTYKQIQVHHKGENSSEDNNIELASTT 1125
>TC79430 similar to PIR|F86358|F86358 Similar to peptide transporter
[imported] - Arabidopsis thaliana, partial (55%)
Length = 1303
Score = 203 bits (516), Expect = 2e-52
Identities = 133/421 (31%), Positives = 221/421 (51%), Gaps = 6/421 (1%)
Frame = +2
Query: 27 SKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPV 86
S GG FII E + A G+ N+I YL G + + + + S ++ P+
Sbjct: 128 SSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAAENVNIWSGTASLLPL 307
Query: 87 IGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQLA 146
+GAF+ADS+LGR+ + I S + L +SLL L+A + PS+G Q
Sbjct: 308 LGAFLADSFLGRYRTIIIASLVYILALSLLTLSATL----------PSDG-----DAQAI 442
Query: 147 MLFSALILIAIGNGG-ITCSLAFGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIALT 205
+ F +L L+A GG C AFGADQ + ++P R FF+W+Y T +++ ++
Sbjct: 443 LFFFSLYLVAFAQGGHKPCVQAFGADQFD-INHPQERRSRSSFFNWWYFTFTAGLLVTVS 619
Query: 206 GIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLY---VKITQRTSLLTGFAQVSVAAY 262
+ Y+QD++GW +GFG+P M+I+ LF L + Y + Q+ + +V + A
Sbjct: 620 ILNYVQDNVGWVLGFGIPWIAMIIALSLFSLGTWTYRFSIPGNQQRGPFSRIGRVFITAL 799
Query: 263 KNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLC 322
+N + + + P H P+ + FLNKA IASDGS N +C
Sbjct: 800 RNFRTTEEEEPRPSLLHQ--------PSQQFSFLNKAL--------IASDGSKEN-GKVC 928
Query: 323 TVDQVEELKAIIKVIPLWSTAITMSI--NIGGSFGLLQAKSLDRHIISSSNFEVPAGSFS 380
+V +VEE KAI++++P+W+T++ +I + +F Q +LDR I+ F VP S
Sbjct: 929 SVSEVEEAKAILRLVPIWATSLIFAIVFSQSSTFFTKQGVTLDRKIL--PGFYVPPASLQ 1102
Query: 381 VILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRR 440
+ ++I+++I +YDR+++P+A GKP I+ +R+G G+ F+ + ++ AA E R
Sbjct: 1103SFISLSIVLFIPVYDRIIVPIARTFTGKPSGITMLQRIGAGILFSVISMVIAAFVEMKRS 1282
Query: 441 K 441
K
Sbjct: 1283K 1285
>TC89928 weakly similar to GP|20466248|gb|AAM20441.1 putative transport
protein {Arabidopsis thaliana}, partial (7%)
Length = 747
Score = 200 bits (509), Expect = 1e-51
Identities = 101/162 (62%), Positives = 129/162 (79%), Gaps = 1/162 (0%)
Frame = +3
Query: 10 VDAADEEEMVSQQKPQ-RSKGGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLG 68
+D+ ++ ++SQ PQ +KGG +TMPFII NE+LA++A++GLLPNMILYLMGSY LHL
Sbjct: 267 MDSQQKDTVISQ--PQLHNKGGFITMPFIIANESLARVATLGLLPNMILYLMGSYNLHLS 440
Query: 69 ISTQILLLSSAASNFTPVIGAFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPS 128
+ ILLLS A +NF P+IGAF+ADS+LGRFL VG+GS I+F+GM LLWLTAMIP ARP
Sbjct: 441 KANHILLLSVATTNFMPLIGAFVADSHLGRFLAVGLGSIITFIGMILLWLTAMIPQARPP 620
Query: 129 ACNHPSEGCESATPGQLAMLFSALILIAIGNGGITCSLAFGA 170
C +E C SAT Q+AML S+L L++IGNGG+ CS+AFGA
Sbjct: 621 PCIPATEKCISATTAQMAMLISSLALMSIGNGGLQCSIAFGA 746
>TC79810 similar to GP|11994403|dbj|BAB02362. nitrate transporter
{Arabidopsis thaliana}, partial (60%)
Length = 1236
Score = 192 bits (489), Expect = 3e-49
Identities = 120/374 (32%), Positives = 207/374 (55%), Gaps = 6/374 (1%)
Frame = +3
Query: 29 GGLVTMPFIIGNEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPVIG 88
GG + I+G E ++ +G+ N++ YL+G LH S I+ N ++G
Sbjct: 144 GGWLAAGLILGTELAERICVMGISMNLVTYLVGDLHLHSASSATIVTNFMGTLNLLGLLG 323
Query: 89 AFIADSYLGRFLGVGIGSSISFLGMSLLWLTAMIPAARPSACNH---PSEGCESATPGQL 145
F+AD+ LGR+L V I ++I+ +G+ +L L IP+ P C+ C A+ QL
Sbjct: 324 GFLADAKLGRYLTVAISATIATVGVCMLTLATTIPSMTPPPCSEVRRQHHQCIQASGKQL 503
Query: 146 AMLFSALILIAIGNGGITCSLA-FGADQVNRKDNPNNYRVLEIFFSWYYAFITIAVIIAL 204
++LF+AL IA+G GGI +++ FG+DQ + D P + + FF+ +Y FI+I + ++
Sbjct: 504 SLLFAALYTIALGGGGIKSNVSGFGSDQFDTND-PKEEKNMIFFFNRFYFFISIGSLFSV 680
Query: 205 TGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYVKITQRTSLLTGFAQVSVAAYKN 264
+VY+QD++G G+G+ A ML++ + +PLY + S LT +V A+K
Sbjct: 681 VVLVYVQDNIGRGWGYGISAGTMLVAVGILLCGTPLYRFKKPQGSPLTIIWRVLFLAWKK 860
Query: 265 RKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSAINRWSLCTV 324
R L +P + P + ++ V TDK R L+KA ++ ++ + DG+ N W + T+
Sbjct: 861 RTLPIP--SDPTLLNGYLEAK-VTYTDKFRSLDKAAIL---DETKSKDGNNENPWLVSTM 1022
Query: 325 DQVEELKAIIKVIPLWSTAITM--SINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVI 382
QVEE+K +IK++P+WST I + +F + QA ++R + + E+PAGS S
Sbjct: 1023TQVEEVKMVIKLLPIWSTCILFWTVYSQMNTFTIEQATFMNRKV---GSLEIPAGSLSAF 1193
Query: 383 LIVAILIWIIIYDR 396
L + IL++ + ++
Sbjct: 1194LFITILLFTSLNEK 1235
>TC82936 similar to PIR|G84829|G84829 probable PTR2 family peptide
transporter [imported] - Arabidopsis thaliana, partial
(53%)
Length = 1112
Score = 192 bits (487), Expect = 4e-49
Identities = 117/359 (32%), Positives = 198/359 (54%), Gaps = 11/359 (3%)
Frame = +3
Query: 198 IAVIIALTGIVYIQDHLGWRIGFGVPAALMLISTVLFFLASPLYV-KITQRTSLLTGFAQ 256
+ +IA G+VYIQ++LGW +G+G+P A +++S V+F++ +P+Y K+ S +
Sbjct: 75 LGALIATLGLVYIQENLGWGLGYGIPTAGLILSLVIFYIGTPIYRHKVRTSKSPAKDIIR 254
Query: 257 VSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIP-------TDKLRFLNKACVIKDHEQDI 309
V + A+K+RKL LP ++P H + VI T LRFL+KA + +D
Sbjct: 255 VFIVAFKSRKLQLP--SNPSELHEFQMEHCVIRGKRQVYHTPTLRFLDKAAIKED----- 413
Query: 310 ASDGSAINRWSLCTVDQVEELKAIIKVIPLWSTAI---TMSINIGGSFGLLQAKSLDRHI 366
+ GS+ R TV+QVE K I+ ++ +W + T+ I F + Q +LDR++
Sbjct: 414 PTTGSS--RRVPMTVNQVEGAKLILGMLLIWLVTLIPSTIWAQINTLF-VKQGTTLDRNL 584
Query: 367 ISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNF 426
+F++PA S + +++L+ + +YDR+ +P + G P I+ +R+GIG
Sbjct: 585 --GPDFKIPAASLGSFVTLSMLLSVPMYDRLFVPFMRQKTGHPRGITLLQRLGIGFSIQI 758
Query: 427 LHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYY 486
+ + A E VRR IKE ++ ++ MS WL PQ L GIA++FN IG EF+Y
Sbjct: 759 IAIAIAYAVE-VRRMHVIKENHIFGPKDIVPMSIFWLLPQYVLIGIADVFNAIGLLEFFY 935
Query: 487 KEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYY 545
+ P+ M S+ + +GVGN ++S ++++ + T G + W++DN+N H YY
Sbjct: 936 DQSPEDMQSLGTTFFTSGIGVGNFLNSFLVTMTDKITGRGDRKSWIADNLNDSHLXYYY 1112
>TC82302 similar to PIR|F84663|F84663 probable nitrate transporter
[imported] - Arabidopsis thaliana, partial (51%)
Length = 1133
Score = 182 bits (462), Expect = 3e-46
Identities = 107/333 (32%), Positives = 185/333 (55%), Gaps = 2/333 (0%)
Frame = +3
Query: 256 QVSVAAYKNRKLSLPPKTSPEFYHHKKDSDLVIPTDKLRFLNKACVIKDHEQDIASDGSA 315
QV VA+ K RK+ LP Y + + +++ RFL KA ++ + + D GS
Sbjct: 9 QVIVASIKKRKMELPYNVG-SLYEDTPEDSRIEQSEQFRFLEKAAIVVEGDFDKDLYGSG 185
Query: 316 INRWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG--SFGLLQAKSLDRHIISSSNFE 373
N W LC++ +VEE+K +++++P+W+T I +F + QA +++R++ NF+
Sbjct: 186 PNPWKLCSLTRVEEVKMMVRLLPIWATTIIFWTTYAQMITFSVEQAATMERNV---GNFQ 356
Query: 374 VPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAA 433
+PAGS +V +VAIL+ + + DR+++PL K+ GKP S +R IGL + + + A+
Sbjct: 357 IPAGSLTVFFVVAILLTLAVNDRIIMPLWKKLNGKPGF-SNLQRNAIGLLLSTIGMAAAS 533
Query: 434 IFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSM 493
+ E V+R K N T L +S L PQ L G E F GQ +F+ + PK M
Sbjct: 534 LIE-VKRLSVAKGVKGNQT--TLPISVFLLVPQFFLVGSGEAFIYTGQLDFFITQSPKGM 704
Query: 494 SSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINA 553
+++ L + +G VSS ++S+++ T + +GW++D+INKG D +Y ++ ++
Sbjct: 705 KTMSTGLFLTTLSLGFFVSSFLVSVVKKVTGTRDGQGWLADHINKGRLDLFYALLTILSF 884
Query: 554 LNLLYYLVCSWAYGPTVDEVSNVSKENGSKVEE 586
+N + +LVC++ Y P + S + S VEE
Sbjct: 885 INFVAFLVCAFWYKPKKPKPS-MQINGSSSVEE 980
>TC88300 similar to GP|9581817|emb|CAC00544.1 putative low-affinity nitrate
transporter {Nicotiana plumbaginifolia}, partial (56%)
Length = 1169
Score = 181 bits (459), Expect = 8e-46
Identities = 106/327 (32%), Positives = 183/327 (55%), Gaps = 12/327 (3%)
Frame = +1
Query: 249 SLLTGFAQVSVAAYKNRKLSLPPKTSPEFYHHKKDSDL------VIPTDK-LRFLNKACV 301
S LT A V VAA++ R + LP +S F + ++ V+P K RFL+KA +
Sbjct: 61 SPLTQIAVVYVAAWRKRNMELPYDSSLLFNVDDIEDEMLRKKKQVLPHSKQFRFLDKAAI 240
Query: 302 IKDHEQDIASDGSAIN---RWSLCTVDQVEELKAIIKVIPLWSTAITMSINIGG--SFGL 356
+D +DG+ IN +W L T+ VEE+K +++++P+W+T I +F +
Sbjct: 241 -----KDPKTDGNEINVVRKWYLSTLTDVEEVKLVLRMLPIWATTIMFWTVYAQMTTFSV 405
Query: 357 LQAKSLDRHIISSSNFEVPAGSFSVILIVAILIWIIIYDRVLIPLASKIRGKPVIISPKK 416
QA +L+RHI +F++P S + I +IL+ I IYDRV++P+ KI P ++P +
Sbjct: 406 SQATTLNRHI--GKSFQIPPASLTAFFIGSILLTIPIYDRVIVPITRKIFKNPQGLTPLQ 579
Query: 417 RMGIGLFFNFLHLITAAIFETVRRKEAIKEGYLNDTHGVLKMSAMWLAPQLCLAGIAEMF 476
R+G+GL F+ ++ AA+ E R + A ++ + + MS WL PQ G E F
Sbjct: 580 RIGVGLVFSIFAMVAAALTELKRMRMAHLHNLTHNPNSEIPMSVFWLIPQFFFVGSGEAF 759
Query: 477 NVIGQNEFYYKEFPKSMSSVAASLSGLAMGVGNLVSSLVLSIIESTTPSGGNEGWVSDNI 536
IGQ +F+ +E PK M +++ L + +G SSL+++++ T ++ W++DN+
Sbjct: 760 TYIGQLDFFLRECPKGMKTMSTGLFLSTLSLGFFFSSLLVTLVHKVTSQ--HKPWLADNL 933
Query: 537 NKGHFDKYYWVIVGINALNLLYYLVCS 563
N+ +YW++ ++ LNL+ YL+C+
Sbjct: 934 NEAKLYNFYWLLALLSVLNLVIYLLCA 1014
>BF639471 weakly similar to GP|20466248|gb| putative transport protein
{Arabidopsis thaliana}, partial (8%)
Length = 522
Score = 164 bits (416), Expect(2) = 9e-44
Identities = 83/130 (63%), Positives = 102/130 (77%)
Frame = +1
Query: 40 NEALAKMASVGLLPNMILYLMGSYRLHLGISTQILLLSSAASNFTPVIGAFIADSYLGRF 99
NEAL AS+G+LPNMILYLMG Y LHLG + QILLLS+AA F PV+GAF+ADSYLGRF
Sbjct: 109 NEALGNTASLGILPNMILYLMGPYNLHLGQANQILLLSAAAGKFMPVVGAFVADSYLGRF 288
Query: 100 LGVGIGSSISFLGMSLLWLTAMIPAARPSACNHPSEGCESATPGQLAMLFSALILIAIGN 159
L VG+GS++SFLGM+LLWLTAMI P+EG +S T ++AML SA ILI+I +
Sbjct: 289 LSVGLGSAVSFLGMALLWLTAMI---------KPTEGDQSPTSWEMAMLISAFILISIAS 441
Query: 160 GGITCSLAFG 169
GG++CS+AFG
Sbjct: 442 GGVSCSMAFG 471
Score = 31.2 bits (69), Expect(2) = 9e-44
Identities = 14/17 (82%), Positives = 14/17 (82%)
Frame = +2
Query: 169 GADQVNRKDNPNNYRVL 185
GADQVN DNPNN RVL
Sbjct: 470 GADQVNITDNPNNNRVL 520
>TC84411 similar to GP|4102839|gb|AAD01600.1| LeOPT1 {Lycopersicon
esculentum}, partial (38%)
Length = 804
Score = 165 bits (418), Expect = 4e-41
Identities = 84/243 (34%), Positives = 143/243 (58%)
Frame = +1
Query: 326 QVEELKAIIKVIPLWSTAITMSINIGGSFGLLQAKSLDRHIISSSNFEVPAGSFSVILIV 385
QVEELK +I++ P+W+T I S ++ L + S +F++ S S +
Sbjct: 1 QVEELKILIRMFPIWATGIIFS-SVYAQMSTLFVEQGTMMNTSIGSFKLSPASLSTFEVA 177
Query: 386 AILIWIIIYDRVLIPLASKIRGKPVIISPKKRMGIGLFFNFLHLITAAIFETVRRKEAIK 445
++++W+ +YD++L+P+ K GK IS +R+GIG F + L ++ AA E ++R + +
Sbjct: 178 SVVMWVPVYDKILVPIVKKFTGKKRGISVFQRIGIGPFISGLCMLAAAAVE-IKRLQLAR 354
Query: 446 EGYLNDTHGVLKMSAMWLAPQLCLAGIAEMFNVIGQNEFYYKEFPKSMSSVAASLSGLAM 505
E L D + +S +W PQ + G AE+F +GQ EF+Y+E P +M ++ +L L
Sbjct: 355 ELDLVDKPVGVPLSVLWQIPQYLILGAAEIFTFVGQLEFFYEESPDAMRTICGALPLLNF 534
Query: 506 GVGNLVSSLVLSIIESTTPSGGNEGWVSDNINKGHFDKYYWVIVGINALNLLYYLVCSWA 565
+GN +SS +L+I+ T GG GW+ DN+NKGH D Y+ ++ G++ LN+L ++V +
Sbjct: 535 SLGNYLSSFILTIVTYFTTKGGRLGWIPDNLNKGHLDYYFLLLSGLSLLNMLVFIVAAKI 714
Query: 566 YGP 568
Y P
Sbjct: 715 YKP 723
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.321 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,329,648
Number of Sequences: 36976
Number of extensions: 297523
Number of successful extensions: 1598
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 1500
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1518
length of query: 638
length of database: 9,014,727
effective HSP length: 102
effective length of query: 536
effective length of database: 5,243,175
effective search space: 2810341800
effective search space used: 2810341800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149489.10


