

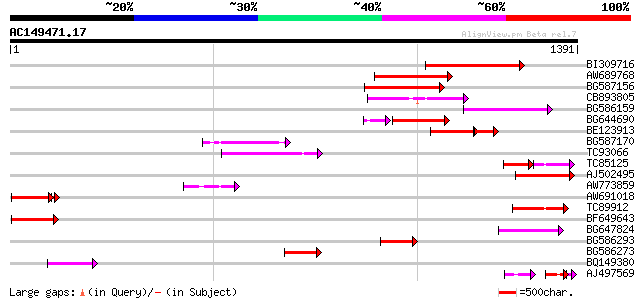
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149471.17 - phase: 0 /pseudo
(1391 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 282 6e-76
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 197 3e-50
BG587156 similar to PIR|G85055|G8 probable polyprotein [imported... 177 3e-44
CB893805 similar to GP|10177935|d copia-type polyprotein {Arabid... 166 6e-41
BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2... 161 1e-39
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 132 2e-38
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 103 6e-33
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 138 2e-32
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 125 1e-28
TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-relate... 75 3e-25
AJ502495 weakly similar to GP|18071369|g putative gag-pol polypr... 112 1e-24
AW773859 108 1e-23
AW691018 weakly similar to GP|2462935|em open reading frame 1 {B... 105 4e-23
TC89912 weakly similar to PIR|B84512|B84512 probable retroelemen... 106 5e-23
BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element pol... 99 1e-20
BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F... 97 4e-20
BG586293 weakly similar to PIR|E84473|E84 probable retroelement ... 92 2e-18
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 89 9e-18
BQ149380 89 2e-17
AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F... 59 1e-16
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 282 bits (722), Expect = 6e-76
Identities = 139/242 (57%), Positives = 177/242 (72%)
Frame = +2
Query: 1020 QVCKLQKSLYGLKQASRKWYEKLTSVLSHHHYIQASSDHSLFVKKTSSSFTILLVYVDDI 1079
+VC+LQKS+YGLKQASR+WY KL+ L Y+Q+SSD SLF K SSFT LLVYVDDI
Sbjct: 17 KVCELQKSIYGLKQASRQWYSKLSESLISFGYLQSSSDFSLFTKFKDSSFTTLLVYVDDI 196
Query: 1080 IIAGDSLTEFTYIKSVLDASFKIKDLGQLKYFLGIEVAHSKLGISLCQRKYCLDLLADSG 1139
++AG+ ++E ++K L FKIKDLG L+YFLG+EVA SK GI L QRKY L+LL DSG
Sbjct: 197 VLAGNDISEIQHVKCFLIDRFKIKDLGSLRYFLGLEVARSKQGILLNQRKYTLELLEDSG 376
Query: 1140 TIDSKPVSTPSDSSIKLHQDSSPSYADIPSYRRLVGRLLYLNTTRPDITFITQQLSQFLS 1199
+ K TP D S+KLH SP Y D YRRL+G+L+YL TTRPDI+F QQLSQF+S
Sbjct: 377 NLAVKSTLTPYDISLKLHNSDSPLYNDETQYRRLIGKLIYLTTTRPDISFAVQQLSQFVS 556
Query: 1200 QPTQAHHTAALRVLRYLKGCPGRGLFFPRNSSINLQGFSDADWAGCLDTRRSISGQCFFL 1259
+P Q H+ AA+RVL+YLK P +GLF+ S++ L F+D+DWA C TR+S++G FL
Sbjct: 557 KPQQVHYQAAIRVLQYLKTAPAKGLFYSATSNLKLSSFADSDWATCPTTRKSVTGYWVFL 736
Query: 1260 GN 1261
G+
Sbjct: 737 GS 742
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 197 bits (500), Expect = 3e-50
Identities = 94/191 (49%), Positives = 128/191 (66%)
Frame = +1
Query: 895 WKQAMQVELQALEKTGTWQLVDLPSNIKPIGCRWIYKVKYHADGSIERHKARLVAKGYNQ 954
W QAM+ E +AL TW LV LP + K IGC+W+Y+VK + DGS+ + KARLVAKG++Q
Sbjct: 97 WLQAMKTEYKALIDNKTWDLVPLPPHKKAIGCKWVYRVKENPDGSVNKFKARLVAKGFSQ 276
Query: 955 IEGLDYFDTYSPVAKLTTIRLVIALSSIHNWHLHQLDVNNAFLHGDLQEDVYMLIPPGIK 1014
G DY +T+SPV K TIRL++ ++ + W + Q+D+NNAFL+G LQE+VYM P G +
Sbjct: 277 TLGCDYTETFSPVIKPVTIRLILTIAITYKWEIQQIDINNAFLNGFLQEEVYMSQPQGFE 456
Query: 1015 SNKPNQVCKLQKSLYGLKQASRKWYEKLTSVLSHHHYIQASSDHSLFVKKTSSSFTILLV 1074
+ + VCKL KSLYGLKQA R WYE LTS + ++ D SL + + + L +
Sbjct: 457 AANKSLVCKLNKSLYGLKQAPRAWYEXLTSAQIQFGFTKSRCDPSLLIYNQNGACIYLXI 636
Query: 1075 YVDDIIIAGDS 1085
YVDDI+I G S
Sbjct: 637 YVDDILITGSS 669
>BG587156 similar to PIR|G85055|G8 probable polyprotein [imported] -
Arabidopsis thaliana, partial (17%)
Length = 618
Score = 177 bits (448), Expect = 3e-44
Identities = 87/199 (43%), Positives = 131/199 (65%), Gaps = 1/199 (0%)
Frame = -1
Query: 870 YSSFVMSLHTTTEPKSYAEASKHDCWKQAMQVELQALEKTGTWQLVDLPSNIKPIGCRWI 929
+ +F+++L+ P+SY EA + WK+++ E A+ K TW +LP K + RWI
Sbjct: 597 HCAFMVNLNENHIPRSYEEAMEDKEWKESVGAEAGAMIKNDTWYESELPKGKKAVSSRWI 418
Query: 930 YKVKYHADGSIERHKARLVAKGYNQIEGLDYFDTYSPVAKLTTIRLVIALSSIHNWHLHQ 989
+ +KY ADGSIER K RLVA+G+ G DY +T++PVAKL TIR+V++L+ W L Q
Sbjct: 417 FTIKYKADGSIERKKTRLVARGFTLTYGEDYIETFAPVAKLHTIRIVLSLAVNLGWGLWQ 238
Query: 990 LDVNNAFLHGDLQEDVYMLIPPGIKS-NKPNQVCKLQKSLYGLKQASRKWYEKLTSVLSH 1048
+DV NAFL G+L+++VYM PPG++ K V +L+K++YGLKQ+ R WY KL++ L+
Sbjct: 237 MDVKNAFLQGELEDEVYMYPPPGLEHLVKRGNVLRLKKAIYGLKQSPRAWYNKLSTTLNG 58
Query: 1049 HHYIQASSDHSLFVKKTSS 1067
+ ++ DH+LF T S
Sbjct: 57 RGFRKSELDHTLFTLTTPS 1
>CB893805 similar to GP|10177935|d copia-type polyprotein {Arabidopsis
thaliana}, partial (14%)
Length = 778
Score = 166 bits (420), Expect = 6e-41
Identities = 91/259 (35%), Positives = 152/259 (58%), Gaps = 9/259 (3%)
Frame = +3
Query: 877 LHTTTEPKSYAEASKHDCWKQAMQVELQALEKTGTWQLVDLPSNIKPIGCRWIYKVKYHA 936
L T++P ++ EA K + W+ +M E++A E+ TW+L DL S K IG +WI+K K +
Sbjct: 24 LTMTSDPTTFEEAVKSEKWRASMNNEMEATERNNTWELTDLRSGAKTIGLKWIFKTKLNE 203
Query: 937 DGSIERHKARLVAKGYNQIEGLDYFDTYSPVAKLTTIRLVIALSSIHNWHLHQLDVNNAF 996
+G IE++KARLVAKGY+Q G+DY + ++PVA+ TIR+VIAL++ Q+ +
Sbjct: 204 NGEIEKYKARLVAKGYSQQYGVDYTEVFAPVARWDTIRMVIALAA-------QIKRDGVC 362
Query: 997 L------HGDLQEDV--YMLIPPGIKSNKPNQVCKLQKSLYGLKQASRKWYEKLTSVLSH 1048
+ H ++ + ++LI + + +++++LYGLKQA R WY ++ + +
Sbjct: 363 IS*M*KAHSCMEN*MRKFLLINHRVM*RRVIS-*RVKRALYGLKQAPRAWYSRIEAYFTK 539
Query: 1049 HHYIQASSDHSLFVK-KTSSSFTILLVYVDDIIIAGDSLTEFTYIKSVLDASFKIKDLGQ 1107
+ + +H+LFVK I+ +YVDD+I G+ F K + F + DLG+
Sbjct: 540 EGFEKCPYEHTLFVKLSEGGKILIISLYVDDLIFIGNDENMFEEFKKSMKKEFNMSDLGK 719
Query: 1108 LKYFLGIEVAHSKLGISLC 1126
+ YFLG+EV ++ GI +C
Sbjct: 720 MHYFLGVEVTQNEKGIYIC 776
>BG586159 weakly similar to PIR|T47841|T4 hypothetical protein T2O9.150 -
Arabidopsis thaliana, partial (11%)
Length = 732
Score = 161 bits (408), Expect = 1e-39
Identities = 82/217 (37%), Positives = 126/217 (57%)
Frame = +1
Query: 1114 IEVAHSKLGISLCQRKYCLDLLADSGTIDSKPVSTPSDSSIKLHQDSSPSYADIPSYRRL 1173
+EV ++ GI +CQRKY DLL G S P KL +D + D Y+++
Sbjct: 1 VEVIQNEEGIYICQRKYVTDLLERFGMEKSNLSRNPIAPRCKLIKDENGVKVDATKYKQI 180
Query: 1174 VGRLLYLNTTRPDITFITQQLSQFLSQPTQAHHTAALRVLRYLKGCPGRGLFFPRNSSIN 1233
VG L+YL TRPD+ ++ +S+F++ PT+ H A RVLRYL G G+ + RN S
Sbjct: 181 VGCLMYLAATRPDLMYVLSLISRFMNCPTELHMHAVKRVLRYLNGTINLGIMYKRNGSEK 360
Query: 1234 LQGFSDADWAGCLDTRRSISGQCFFLGNSLISWRTKKQITVSRSSSEAEYRALASATCEL 1293
L+ ++D+D+AG LD R+S SG F L + +SW +KKQ V+ S+++AE+ A A C+
Sbjct: 361 LEAYTDSDYAGDLDDRKSTSGYVFMLSSGAVSWSSKKQPVVTLSTTKAEFIAAAFCACQS 540
Query: 1294 QWILYLLQDIHISCPKLPVLYCDNQSALHIAANPVFH 1330
W+ +L+ + + +YCDN S + ++ NPV H
Sbjct: 541 VWMRRVLEKLGYTQSGSITMYCDNNSTIKLSKNPVLH 651
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 132 bits (333), Expect(2) = 2e-38
Identities = 64/141 (45%), Positives = 100/141 (70%), Gaps = 1/141 (0%)
Frame = -2
Query: 940 IERHKARLVAKGYNQIEGLDYFDTYSPVAKLTTIRLVIALSSIHNWHLHQLDVNNAFLHG 999
I R+K++LV +GYNQ EG+DY + +SPVA++ IR++IA ++ + L+Q+DV +AF++G
Sbjct: 424 ITRNKSKLVVQGYNQKEGIDYDEAFSPVARMEAIRILIAFAAFMGFKLYQMDVKSAFING 245
Query: 1000 DLQEDVYMLIPPGIK-SNKPNQVCKLQKSLYGLKQASRKWYEKLTSVLSHHHYIQASSDH 1058
DL+E+V++ PPG + + PN V +L K+LYGLKQA R WYE+L+ L + + + D+
Sbjct: 244 DLKEEVFVKQPPGFEDAEVPNHVFRLNKTLYGLKQAPRAWYERLSKFLLKNGFKRGKIDN 65
Query: 1059 SLFVKKTSSSFTILLVYVDDI 1079
+LF+ K I+ VYVDDI
Sbjct: 64 TLFLLKRE*ELLIIQVYVDDI 2
Score = 46.6 bits (109), Expect(2) = 2e-38
Identities = 23/65 (35%), Positives = 36/65 (55%)
Frame = -3
Query: 869 NYSSFVMSLHTTTEPKSYAEASKHDCWKQAMQVELQALEKTGTWQLVDLPSNIKPIGCRW 928
++S+F+ S+ EPK+ EA + W +MQ EL E++ W LV P IG RW
Sbjct: 615 SFSAFISSI----EPKNVKEALRDADWINSMQEELHQFERSKVWYLVPRPEGKTVIGTRW 448
Query: 929 IYKVK 933
+++ K
Sbjct: 447 VFRNK 433
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 103 bits (258), Expect(2) = 6e-33
Identities = 54/118 (45%), Positives = 76/118 (63%), Gaps = 1/118 (0%)
Frame = +1
Query: 1033 QASRKWYEKLTSVLSHHHYIQASSDHSLFVKKTSS-SFTILLVYVDDIIIAGDSLTEFTY 1091
Q+ R W+++ T V+ YIQ +DH++F+K +S+ IL+VYVDDI + GD
Sbjct: 1 QSPRDWFDRFT*VVKKFGYIQCQTDHAMFIKHSSTVKKAILIVYVDDIFLTGDHGK*IKR 180
Query: 1092 IKSVLDASFKIKDLGQLKYFLGIEVAHSKLGISLCQRKYCLDLLADSGTIDSKPVSTP 1149
+K++L F+IKDLG LKYFLG+EVA K G S+ QRKY LDLL ++ I K + P
Sbjct: 181 LKNLLAEEFEIKDLGNLKYFLGMEVARWKKGSSISQRKYVLDLLKETRMIGCKTIRDP 354
Score = 57.0 bits (136), Expect(2) = 6e-33
Identities = 26/58 (44%), Positives = 40/58 (68%)
Frame = +2
Query: 1141 IDSKPVSTPSDSSIKLHQDSSPSYADIPSYRRLVGRLLYLNTTRPDITFITQQLSQFL 1198
+D KP TP D+++KL + + D Y+RLVG+L+YL+ TRPDI+F+ +SQF+
Sbjct: 329 LDVKPSETPMDATVKLGTLDNGTLVDKGRYQRLVGKLIYLSHTRPDISFVVCTMSQFM 502
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 138 bits (347), Expect = 2e-32
Identities = 78/217 (35%), Positives = 121/217 (54%), Gaps = 3/217 (1%)
Frame = -3
Query: 474 SCNSSSSIPSNALWHFRLGHLSHQRLHSMSLLYPNIISSNNKDVCDLCHFAKHKHLPFNS 533
S SSSS+ +ALWH RLGH H R +++L+ P ++ N C+ C KH F
Sbjct: 620 SFTSSSSLNKDALWHARLGH-PHGR--ALNLMLPGVVFENKN--CEACILGKHCKNVFPR 456
Query: 534 SISHASTNFELLHLDIWGPLSIASVHGHRYFLTIVDDHSRFLWVILLKSKAEVSTHVINF 593
+ + F+L++ D+W S+ S H+YF+T +D+ S++ W+ L+ SK V NF
Sbjct: 455 TSTVYENCFDLIYTDLWTAPSL-SRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKNF 279
Query: 594 ITMIQTQFHITPKFIRTDNGPEFMLSTFYAS---HGIIHQKSCVETPQQNGRVERKHQHI 650
+ +H K +R+DNG E+ F + HGI+HQ SC TPQQNG +RK++H+
Sbjct: 278 QAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHL 99
Query: 651 LNVGRALLFQSKLPPSFWSYAILHAVFLINRVPTPIL 687
+ V R+L+FQ+ + A +LIN +PT +L
Sbjct: 98 MEVARSLMFQAN---------VSTACYLINWIPTKVL 15
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 125 bits (313), Expect = 1e-28
Identities = 85/255 (33%), Positives = 124/255 (48%), Gaps = 7/255 (2%)
Frame = +1
Query: 520 LCHFAKHKHLPFNSSISHASTNFELLHLDIWGPLSIASVHGHRYFLTIVDDHSRFLWVIL 579
L F K + F+++ + +H D+WGP + S G RY +TI+DD R +WV
Sbjct: 31 LLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYF 210
Query: 580 LKSKAEVSTHVINFITMIQTQFHITPKFIRTDNGPEFMLS---TFYASHGIIHQKSCVET 636
L+ K E + +++TQ K + TDN EF S F +HGI K+
Sbjct: 211 LRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRN 390
Query: 637 PQQNGRVERKHQHILNVGRALLFQSKL--PPSFWSYAILHAVFLINRVPTPILHNQSPYF 694
PQQNG ER + +L R +L + L W A A L+NR P L + P
Sbjct: 391 PQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPED 570
Query: 695 VLHHQLPALNLFKVFGCLCYASTLQSHRTKLQPRARKSIFLGYKSGFKGFTLY--DIQSR 752
+ L + ++FGC YA + KL PRA + IFL Y S KG+ L+ D +S+
Sbjct: 571 IWSGNLVDYSNLRIFGCPAYALV---NDGKLAPRAGECIFLSYASESKGYRLWCSDPKSQ 741
Query: 753 EIFVSRHVTFHETFL 767
++ +SR VTF+E L
Sbjct: 742 KLILSRDVTFNEDAL 786
>TC85125 weakly similar to SP|P10978|POLX_TOBAC Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
(EC 3.4.23.-);, partial (7%)
Length = 705
Score = 74.7 bits (182), Expect(2) = 3e-25
Identities = 40/102 (39%), Positives = 59/102 (57%)
Frame = +3
Query: 1285 ALASATCELQWILYLLQDIHISCPKLPVLYCDNQSALHIAANPVFHERTKHLEIDCHIVR 1344
+L A E W+ L++++ ++ V YCD+QSALHIA NP FH RTKH+ I H VR
Sbjct: 228 SLPQACKEAIWMQRLMEELGHKQEQITV-YCDSQSALHIARNPAFHSRTKHIGIQYHFVR 404
Query: 1345 EKVQAGILKLLPVSSQDQVADFFTKALLPKPFNILLSKMGLI 1386
E V+ G + + + + D +AD TK++ F S GL+
Sbjct: 405 EVVEEGSVDMQKIHTNDNLADAMTKSINTDKFIWCRSSYGLL 530
Score = 60.1 bits (144), Expect(2) = 3e-25
Identities = 30/75 (40%), Positives = 47/75 (62%)
Frame = +1
Query: 1211 RVLRYLKGCPGRGLFFPRNSSINLQGFSDADWAGCLDTRRSISGQCFFLGNSLISWRTKK 1270
R++RY+KG G + F S + ++G+ D+D+AG D R+S +G F L +SW +K
Sbjct: 7 RIMRYIKGTSGVAVCFG-GSELTVRGYVDSDFAGDHDKRKSTTGYVFTLAGGAVSWLSKL 183
Query: 1271 QITVSRSSSEAEYRA 1285
Q V+ S++EAEY A
Sbjct: 184 QTVVALSTTEAEYMA 228
>AJ502495 weakly similar to GP|18071369|g putative gag-pol polyprotein {Oryza
sativa}, partial (9%)
Length = 542
Score = 112 bits (279), Expect = 1e-24
Identities = 56/147 (38%), Positives = 89/147 (60%)
Frame = +2
Query: 1240 ADWAGCLDTRRSISGQCFFLGNSLISWRTKKQITVSRSSSEAEYRALASATCELQWILYL 1299
+DWAG +TR+S SG F LG ISW +KKQ V+ S++EAEY A S + W+ +
Sbjct: 2 SDWAGDTETRKSTSGYAFHLGTGAISWSSKKQPVVAFSTAEAEYIASTSCATQTVWLRRI 181
Query: 1300 LQDIHISCPKLPVLYCDNQSALHIAANPVFHERTKHLEIDCHIVREKVQAGILKLLPVSS 1359
L+ +H +YCDN+SA+ ++ NPVFH R+KH++I H +RE + + + +
Sbjct: 182 LEVMHHEQNTPTKIYCDNKSAIALSKNPVFHGRSKHIDIQFHKIRELIAEKEVVIEYCPT 361
Query: 1360 QDQVADFFTKALLPKPFNILLSKMGLI 1386
++++AD FTK L + F L +G++
Sbjct: 362 EEKIADIFTKPLKIESFYKLKKMLGMM 442
>AW773859
Length = 538
Score = 108 bits (271), Expect = 1e-23
Identities = 57/137 (41%), Positives = 77/137 (55%)
Frame = -3
Query: 426 IIQDTMSLKMIGLAKQLDGLYKYTPSSCSSNSVFSSVSHKSCNVVATISCNSSSSIPSNA 485
+IQ+ SLKMIG + +GLY T S ++ SS + +P A
Sbjct: 383 LIQEKRSLKMIGSGELFEGLYFLTTQDTPSATIASSQVQPQ---------PQPTFLPQEA 231
Query: 486 LWHFRLGHLSHQRLHSMSLLYPNIISSNNKDVCDLCHFAKHKHLPFNSSISHASTNFELL 545
LWHFRLGHLS+++L S+ +P I N VCD+CH+++HK LPF S + AS +EL
Sbjct: 230 LWHFRLGHLSNRKLLSLHSNFPFITIDQNS-VCDICHYSRHKKLPFQLSTNRASKCYELF 54
Query: 546 HLDIWGPLSIASVHGHR 562
H DIWGP S S+H R
Sbjct: 53 HFDIWGPFSTQSIHNQR 3
>AW691018 weakly similar to GP|2462935|em open reading frame 1 {Brassica
oleracea}, partial (12%)
Length = 639
Score = 105 bits (262), Expect(2) = 4e-23
Identities = 47/103 (45%), Positives = 69/103 (66%)
Frame = +2
Query: 4 SLGTKNKLGFINGVIPIPDADDLNRAAWERCNHLVQSWLINSVSDSIAQTIVFYDTAFEV 63
SL KNKLG I+G + P ADD W+RCN LV +W+++S+ IA++++F DTA V
Sbjct: 254 SLSAKNKLGLIDGTVKAPSADDPKLPLWKRCNDLVLTWILHSIEPDIARSVIFSDTAAAV 433
Query: 64 WHDLQERFSKVDRIRIANLRSTINNLKQGSKSVLDYFTEMKAL 106
W DL +RFS+ D RI +R I+ +QGS + DY+T++K+L
Sbjct: 434 WSDLHDRFSQGDESRIYQIRQEISECRQGSLLISDYYTKLKSL 562
Score = 22.3 bits (46), Expect(2) = 4e-23
Identities = 7/18 (38%), Positives = 11/18 (60%)
Frame = +3
Query: 104 KALWEELASHRPIPNCSC 121
+ W+EL S++ CSC
Sbjct: 555 RVFWDELGSYQEPITCSC 608
>TC89912 weakly similar to PIR|B84512|B84512 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(10%)
Length = 814
Score = 106 bits (265), Expect = 5e-23
Identities = 50/136 (36%), Positives = 91/136 (66%)
Frame = +1
Query: 1234 LQGFSDADWAGCLDTRRSISGQCFFLGNSLISWRTKKQITVSRSSSEAEYRALASATCEL 1293
L+G+ DAD+AG +DTR+S+SG F L + ISW+ +Q V+ S+++AEY A +
Sbjct: 88 LEGYVDADYAGNVDTRKSLSGFVFTLYGTTISWKANQQSVVTLSTTQAEYIAFVEGVKDA 267
Query: 1294 QWILYLLQDIHISCPKLPVLYCDNQSALHIAANPVFHERTKHLEIDCHIVREKVQAGILK 1353
W+ ++ ++ I+ + + +CD+QSA+H+A + V+HERTKH++I H +R+ +++ +
Sbjct: 268 IWLKGMIGELGITQEYVKI-HCDSQSAIHLANHQVYHERTKHIDIRLHFIRDMIESKEIV 444
Query: 1354 LLPVSSQDQVADFFTK 1369
+ ++S++ AD FTK
Sbjct: 445 VEKMASEENPADVFTK 492
>BF649643 similar to PIR|H86486|H86 protein Ty1/copia-element polyprotein
[imported] - Arabidopsis thaliana, partial (2%)
Length = 608
Score = 98.6 bits (244), Expect = 1e-20
Identities = 46/118 (38%), Positives = 73/118 (60%), Gaps = 2/118 (1%)
Frame = +1
Query: 4 SLGTKNKLGFINGVIPIPDADD--LNRAAWERCNHLVQSWLINSVSDSIAQTIVFYDTAF 61
+L KNK+GFING I P D A W RCN ++ SWL +SV +A+ ++ TA
Sbjct: 220 ALTAKNKVGFINGSIKTPSEVDQPAEYALWNRCNSMILSWLTHSVEPDLAKGVIHAKTAC 399
Query: 62 EVWHDLQERFSKVDRIRIANLRSTINNLKQGSKSVLDYFTEMKALWEELASHRPIPNC 119
+VW D +++FS+ + I ++ ++ +L QG+ S YFT++K LW+EL ++R +P C
Sbjct: 400 QVWEDFKDQFSQKNIPAIYQIQKSLASLSQGTVSASTYFTKIKGLWDELETYRTLPTC 573
>BG647824 weakly similar to PIR|G96722|G96 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (5%)
Length = 721
Score = 97.1 bits (240), Expect = 4e-20
Identities = 62/161 (38%), Positives = 91/161 (56%), Gaps = 1/161 (0%)
Frame = -3
Query: 1200 QPTQAHHTAALRVLRYLKGCPGRGLFFPRNSSINLQGFSDADWAGCLDTRRSISGQCFFL 1259
Q T H AA+++L YLK P + +FFP S I ++ F D+D ++ Q
Sbjct: 605 QSTAQHPQAAIQIL-YLKISPSQ*IFFP--S*IQIKAFCDSD*ID--QAA*TLENQSVIF 441
Query: 1260 GNSLISWRTKKQITVSRSSSEAEYRALASATCELQWILYLLQDIHISCPKLPVLYCDNQS 1319
+S + + R + + YR++ S CE++W+ YLL D+ + K +LYCDNQS
Sbjct: 440 ASS*ATHSYAGNLKKKRYNFKI-YRSI*STICEIKWLTYLLNDLKFTFIKPAMLYCDNQS 264
Query: 1320 AL-HIAANPVFHERTKHLEIDCHIVREKVQAGILKLLPVSS 1359
A HIAAN F ERTKH+E+DCHIVR K+Q + +L + S
Sbjct: 263 AARHIAANSSFLERTKHIELDCHIVRVKLQLKLFHILHILS 141
>BG586293 weakly similar to PIR|E84473|E84 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 763
Score = 91.7 bits (226), Expect = 2e-18
Identities = 41/89 (46%), Positives = 67/89 (75%)
Frame = +2
Query: 911 TWQLVDLPSNIKPIGCRWIYKVKYHADGSIERHKARLVAKGYNQIEGLDYFDTYSPVAKL 970
T +LV P+ +KPIG RWIYK+K + DG++ ++KARLVAKGY + +G+D+ + ++PV ++
Sbjct: 56 TLKLVKKPTGVKPIGLRWIYKIKRNEDGTLIKYKARLVAKGYVKQQGIDFDEVFAPVVRI 235
Query: 971 TTIRLVIALSSIHNWHLHQLDVNNAFLHG 999
TI L++AL++ + +H +DV AFL+G
Sbjct: 236 ETI*LLLALAATNGC*IHHIDVKIAFLNG 322
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 89.4 bits (220), Expect = 9e-18
Identities = 41/90 (45%), Positives = 61/90 (67%)
Frame = -2
Query: 675 AVFLINRVPTPILHNQSPYFVLHHQLPALNLFKVFGCLCYASTLQSHRTKLQPRARKSIF 734
A +LINR+PT +L +Q+P+ VL+ + P+L +VFGCLCY R KL+ R+RK++F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 735 LGYKSGFKGFTLYDIQSREIFVSRHVTFHE 764
+GY + KG+ YD ++R + VSR V F E
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIE 435
>BQ149380
Length = 419
Score = 88.6 bits (218), Expect = 2e-17
Identities = 47/125 (37%), Positives = 74/125 (58%), Gaps = 3/125 (2%)
Frame = +3
Query: 94 KSVLDYFTEMKALWEELASHRPIPNCSCIHPCRCEASKVAKIHRNEDQIMQFLTGLNDQF 153
+S L+ +++ LW+EL +RP P C+C C C A + K ED+I++FL GLND +
Sbjct: 45 ESYLNSSPDLRGLWDELDQYRPKPQCTCPIQCTCFAMRNNKALTAEDRIIKFLMGLNDDY 224
Query: 154 SIVRTQVLLLDPLPSLNKVYSLVVQEESN---NASLSSLSVSDDSSIQINASDVRKFQGR 210
V ++VLL+ PLP +N V+S+V+Q+E N S + D + +NA D ++ GR
Sbjct: 225 QGVTSKVLLMYPLPQINMVFSMVMQQERKMQ*NVVSSPTPIDDTTCGLVNAMDGQRQFGR 404
Query: 211 GKNPS 215
G+ S
Sbjct: 405 GRGXS 419
>AJ497569 weakly similar to PIR|T04833|T04 hypothetical protein F21P8.50 -
Arabidopsis thaliana, partial (4%)
Length = 723
Score = 58.9 bits (141), Expect(3) = 1e-16
Identities = 33/58 (56%), Positives = 41/58 (69%)
Frame = +2
Query: 1314 YCDNQSALHIAANPVFHERTKHLEIDCHIVREKVQAGILKLLPVSSQDQVADFFTKAL 1371
YCDN SALHIAAN VFHERT H E D +IV+ + +L+L+P +S+DQ A TK L
Sbjct: 290 YCDNISALHIAANMVFHERT*HRETDPYIVQ---GSRMLQLMPSASKDQPAYSLTKPL 454
Score = 44.7 bits (104), Expect(3) = 1e-16
Identities = 35/78 (44%), Positives = 45/78 (56%)
Frame = +3
Query: 1213 LRYLKGCPGRGLFFPRNSSINLQGFSDADWAGCLDTRRSISGQCFFLGNSLISWRTKKQI 1272
L YLK PG+ +F S+ + S C + R + CF L +SLISW++KKQ
Sbjct: 15 LHYLK-TPGKCIFVSNASNPHFNRGS------CPYSIR*TTEFCF-LSSSLISWKSKKQC 170
Query: 1273 TVSRSSSEAEYRALASAT 1290
VSRS SEA RALA+AT
Sbjct: 171 VVSRSFSEA**RALANAT 224
Score = 21.9 bits (45), Expect(3) = 1e-16
Identities = 9/24 (37%), Positives = 14/24 (57%)
Frame = +1
Query: 1366 FFTKALLPKPFNILLSKMGLINIY 1389
F K P + LLSK+ ++NI+
Sbjct: 436 FINKTSAPGILHSLLSKLNMVNIH 507
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.133 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,995,285
Number of Sequences: 36976
Number of extensions: 982715
Number of successful extensions: 15436
Number of sequences better than 10.0: 375
Number of HSP's better than 10.0 without gapping: 8552
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11862
length of query: 1391
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1283
effective length of database: 5,021,319
effective search space: 6442352277
effective search space used: 6442352277
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC149471.17


