

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
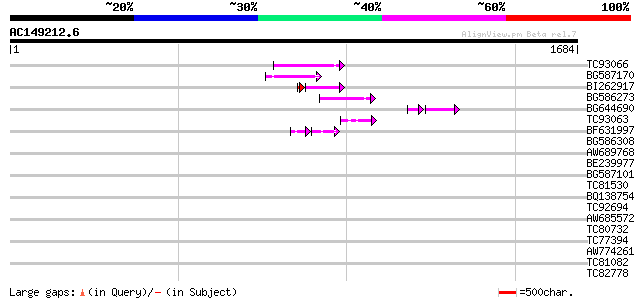
Query= AC149212.6 - phase: 0 /pseudo
(1684 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 123 5e-28
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 105 1e-22
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 100 3e-22
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 79 2e-14
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 47 7e-10
TC93063 similar to PIR|T08852|T08852 lustrin A - California red ... 51 4e-06
BF631997 weakly similar to GP|18542925|gb Putative pol polyprote... 35 1e-05
BG586308 weakly similar to PIR|F84528|F8 probable retroelement p... 42 0.003
AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsi... 41 0.003
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 40 0.006
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 38 0.038
TC81530 similar to GP|10177440|dbj|BAB10736. gb|AAC55944.1~gene_... 35 0.19
BQ138754 weakly similar to GP|14587221|db contains ESTs AU108104... 35 0.25
TC92694 35 0.25
AW685572 similar to GP|22830607|dbj PHD-finger family homeodomai... 35 0.25
TC80732 similar to GP|21553690|gb|AAM62783.1 unknown {Arabidopsi... 35 0.32
TC77394 similar to GP|4874305|gb|AAD31367.1| expressed protein {... 34 0.55
AW774261 weakly similar to EGAD|146423|156 vitellogenin {Anolis ... 33 0.94
TC81082 similar to GP|17385678|dbj|BAB78631. putative co-repress... 33 0.94
TC82778 similar to PIR|T04011|T04011 hypothetical protein T5L19.... 33 1.2
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 123 bits (309), Expect = 5e-28
Identities = 72/214 (33%), Positives = 109/214 (50%), Gaps = 2/214 (0%)
Frame = +1
Query: 783 LELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEK 842
L+ +H DL+G TS G +Y + I+DD+ R WV F++ K+ F + ++++
Sbjct: 97 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 276
Query: 843 ELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMI 902
+ K+ +D+ EF + F FC HGI + PR PQQNGV ER RTL E AR M+
Sbjct: 277 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 456
Query: 903 HENNL--AKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYIL 960
L + W EA +T+C++ NR + K +++ G + S FGC Y L
Sbjct: 457 SNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYAL 636
Query: 961 NTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSE 994
K +A IFL Y+ SK YR++ S+
Sbjct: 637 VNDG---KLAPRAGECIFLSYASESKGYRLWCSD 729
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 105 bits (262), Expect = 1e-22
Identities = 60/167 (35%), Positives = 95/167 (55%)
Frame = -3
Query: 760 ACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGQVNTTSLYGSKYGLVIVDDYSRWTWVK 819
AC GK K+ F V + +L++ DL+ + S KY + +D+ S++TW+
Sbjct: 497 ACILGKHCKNVFPRTSTVYENC-FDLIYTDLW-TAPSLSRDNHKYFVTFIDEKSKYTWLT 324
Query: 820 FIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPR 879
I SKD + F +F + + KI +RSD+GGE+ + F+S + HGILH+ S P
Sbjct: 323 LIPSKDRVIDAFKNFQAYVTNHYHAKIKILRSDNGGEYTSYAFKSHLDHHGILHQTSCPY 144
Query: 880 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRI 926
TPQQNGV +RKN+ L E+AR+++ + N ++T+CY+ N I
Sbjct: 143 TPQQNGVAKRKNKHLMEVARSLMFQAN---------VSTACYLINWI 30
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 100 bits (250), Expect(2) = 3e-22
Identities = 49/113 (43%), Positives = 68/113 (59%)
Frame = +1
Query: 880 TPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYEL 939
TPQQNGV ER NRTL E R M+ +AK FWAEA+ T+CY+ NR + KT E+
Sbjct: 85 TPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTACYVINRSPSTVIDLKTPMEM 264
Query: 940 FKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYN 992
+KG + S H FGC Y++ K D K+++ IFLGY++ K Y +++
Sbjct: 265 WKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADNVKGYXLWD 423
Score = 24.3 bits (51), Expect(2) = 3e-22
Identities = 8/22 (36%), Positives = 16/22 (72%)
Frame = +3
Query: 855 GEFENETFESFCEKHGILHEFS 876
GE+ + F +FC++ GI+ +F+
Sbjct: 9 GEYVDGEFLAFCKQEGIVRQFT 74
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 78.6 bits (192), Expect = 2e-14
Identities = 47/172 (27%), Positives = 84/172 (48%), Gaps = 3/172 (1%)
Frame = -2
Query: 919 SCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIF 978
+CY+ NRI R + ++ +E+ +P+++Y FGC CY+L + K +A++++ +F
Sbjct: 704 ACYLINRIPTRVLKDQAPFEVLNQRKPSLTYMRVFGCLCYVLVPGELRNKLEARSRKAMF 525
Query: 979 LGYSERSKAYRVYNSETHCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLS 1038
+GYS K Y+ Y+ E V S VKF + + Q + T D
Sbjct: 524 IGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSDKAGVLRVILEG 345
Query: 1039 DSEKHSEVESSPEAEITPEAESNS--EAEPSPEIQNENASE-DIQDNSQQAI 1087
K ++ +S+ + PE SN A +P + +E SE + Q+N Q+ +
Sbjct: 344 LGIKMNQDQSTRSRQ--PEESSNEPRRAAQTPHLDHEGGSEPETQENGQEGV 195
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 47.0 bits (110), Expect(2) = 7e-10
Identities = 33/99 (33%), Positives = 48/99 (48%)
Frame = -3
Query: 1236 ETKPDLLLKVTVNKKALITLKHLLQLQDWKQSGYFYPMQLIMA*YCIKWMSKVVLLMVSL 1295
E P K T+ KK ++ L +WK + + C KWM +V LLM
Sbjct: 417 EINPSWWCKDTIKKKE*TMMRLFHLLPEWKLLEF**LLLHSWGSSCTKWM*RVHLLMEIS 238
Query: 1296 KKKFMSNNLLGLRILSILTMFINLRNHYMA*NKLPELGM 1334
K++ +S+NLL L++ M + HYM *+KL E GM
Sbjct: 237 KRRCLSSNLLDLKMQRYQIMCSD*IRHYMV*SKLQEHGM 121
Score = 36.2 bits (82), Expect(2) = 7e-10
Identities = 20/47 (42%), Positives = 26/47 (54%)
Frame = -1
Query: 1181 KKLSQMMVGY*LCKKN*ISFRGMMCGIWYPNLFRRTLLEQNGYSETS 1227
KK M G LCKKN IS + + G W+ +L + LE G+ ETS
Sbjct: 572 KKHCVMQTGSILCKKNSISLKEVRYGTWFLDLKAKQ*LELGGFLETS 432
>TC93063 similar to PIR|T08852|T08852 lustrin A - California red abalone,
partial (4%)
Length = 1058
Score = 50.8 bits (120), Expect = 4e-06
Identities = 35/106 (33%), Positives = 51/106 (48%)
Frame = -1
Query: 984 RSKAYRVYNSETHCVEESMHVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKH 1043
R+K RV + C E + ++ P K +SN +ED E+D +D E
Sbjct: 782 RTKCNRVKPTCDMCEEGDLQCQY----PLVKKQAPRKSNVSIGAAED--EADAEADDEPE 621
Query: 1044 SEVESSPEAEITPEAESNSEAEPSPEIQNENASEDIQDNSQQAIQP 1089
E E PEAE PEAE EAEP PE + E +E ++ ++ +P
Sbjct: 620 PEPEPEPEAEPEPEAEPEPEAEPEPEAEPEPEAEPEPEDEEEEQEP 483
Score = 31.2 bits (69), Expect = 3.6
Identities = 18/52 (34%), Positives = 24/52 (45%)
Frame = -1
Query: 1027 DSEDASESDQLSDSEKHSEVESSPEAEITPEAESNSEAEPSPEIQNENASED 1078
+ E E++ ++E E E PEAE P EAEP PE + E D
Sbjct: 617 EPEPEPEAEPEPEAEPEPEAEPEPEAEPEP------EAEPEPEDEEEEQEPD 480
>BF631997 weakly similar to GP|18542925|gb Putative pol polyprotein {Oryza
sativa}, partial (6%)
Length = 650
Score = 34.7 bits (78), Expect(2) = 1e-05
Identities = 25/85 (29%), Positives = 39/85 (45%)
Frame = -1
Query: 896 EMARTMIHENNLAKHFWAEAINTSCYIQNRIYIRPMLEKTAYELFKGTRPNISYFHQFGC 955
E AR M L + F AEAI+T CY+ N + P+L T + N+ GC
Sbjct: 413 ERARCMFSNAGLNRSFQAEAISTKCYLVN---VLPLLL*TVRLHLRYGLINLLITQILGC 243
Query: 956 TCYILNTKDYLKKFDAKAQRGIFLG 980
Y + K + ++++G+F G
Sbjct: 242 PAYYHVNEG---KLEPRSKKGLFWG 177
Score = 34.3 bits (77), Expect(2) = 1e-05
Identities = 17/60 (28%), Positives = 33/60 (54%)
Frame = -2
Query: 834 FCTQIQSEKELKILKVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRT 893
F ++++ + + K+ ++G E + F C++ I +++ TPQ+NGV ER N T
Sbjct: 592 FWSRVKQGRR*NVFKL--NNGLEICSAEFNELCKEEHITRQYTVRNTPQKNGVAERMNIT 419
>BG586308 weakly similar to PIR|F84528|F8 probable retroelement pol
polyprotein [imported] - Arabidopsis thaliana, partial
(7%)
Length = 686
Score = 41.6 bits (96), Expect = 0.003
Identities = 21/70 (30%), Positives = 37/70 (52%)
Frame = -2
Query: 848 KVRSDHGGEFENETFESFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNL 907
++ +D+G F + F FCE+ I +SPR PQ NG E N+ + + + + +L
Sbjct: 670 EIVTDNGSHFISNKFREFCERWRIRLNTASPRYPQSNGQAEASNKIIIDGLKKRL---DL 500
Query: 908 AKHFWAEAIN 917
K WA+ ++
Sbjct: 499 KKGCWADELD 470
>AW689768 weakly similar to GP|10177485|d polyprotein {Arabidopsis thaliana},
partial (9%)
Length = 675
Score = 41.2 bits (95), Expect = 0.003
Identities = 42/129 (32%), Positives = 61/129 (46%)
Frame = +2
Query: 1206 GIWYPNLFRRTLLEQNGYSETS*MNKER*PETKPDLLLKVTVNKKALITLKHLLQLQDWK 1265
GI + L R LL +G+ E + KPD L+V+V +ITLK L +
Sbjct: 149 GILFLFLHTRKLLAASGFIELKKTQMDLLTNLKPD*WLRVSVKHLDVITLKLSLL**NLS 328
Query: 1266 QSGYFYPMQLIMA*YCIKWMSKVVLLMVSLKKKFMSNNLLGLRILSILTMFINLRNHYMA 1325
SG F P+ ++ K +S + MV K+K + NL L++L I + NH MA
Sbjct: 329 LSGSFSPLLSLINGKFNKLISTMPF*MVFFKRKCICLNLRDLKLL-INLWCAS*TNHSMA 505
Query: 1326 *NKLPELGM 1334
*+K GM
Sbjct: 506 *SKHHVHGM 532
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 40.4 bits (93), Expect = 0.006
Identities = 20/58 (34%), Positives = 30/58 (51%)
Frame = -2
Query: 953 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETHCVEESMHVKFDDRE 1010
FGCT ++ D KFD +A + +F+ YS K YR Y+ + S V F ++E
Sbjct: 450 FGCTSFVHIHSDGRSKFDHRALKCVFIRYSSTQKGYRCYHPPSRKYFVSRDVTFHEQE 277
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 37.7 bits (86), Expect = 0.038
Identities = 27/86 (31%), Positives = 40/86 (46%)
Frame = +2
Query: 798 SLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEF 857
S Y +KY LV VD S+ WV+ I S V + + V SD G F
Sbjct: 350 SSYNNKYILVAVDYVSK--WVEAIASPTNDATVVVKMFKSVIFPRFGVPRVVISDGGSHF 523
Query: 858 ENETFESFCEKHGILHEFSSPRTPQQ 883
N+ FE +K+G+ H+ ++ PQ+
Sbjct: 524 INKVFEKLLKKNGVRHKVATAYHPQK 601
>TC81530 similar to GP|10177440|dbj|BAB10736.
gb|AAC55944.1~gene_id:K9H21.2~similar to unknown protein
{Arabidopsis thaliana}, partial (24%)
Length = 1135
Score = 35.4 bits (80), Expect = 0.19
Identities = 26/87 (29%), Positives = 39/87 (43%), Gaps = 1/87 (1%)
Frame = +1
Query: 1013 SKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNSEAEPS-PEIQ 1071
S + SES + + DS+ +S S D+ K SE SS E + +PS PE
Sbjct: 31 SSSSSSSESGSSSSDSDSSSSSGSELDTAKASEPLSSKENVGPGLTYDQNRGDPSNPETG 210
Query: 1072 NENASEDIQDNSQQAIQPKFKHKSSHL 1098
N++ + IQ + +P SHL
Sbjct: 211 NDSTNLGIQADQSLQTKPDTIESESHL 291
>BQ138754 weakly similar to GP|14587221|db contains ESTs AU108104(C12321)
C26435(C12321)~similar to Arabidopsis thaliana chromosome
3 F24P17., partial (4%)
Length = 639
Score = 35.0 bits (79), Expect = 0.25
Identities = 27/98 (27%), Positives = 40/98 (40%), Gaps = 14/98 (14%)
Frame = -2
Query: 976 GIFLGYSERSKAYRVYNSETHCVEESMHVKFDDREPGSKTPEQSESNAGTIDSED----- 1030
G+ LGYSE A N E V +++ VK GSK S+ + ++ +ED
Sbjct: 581 GVHLGYSEFDPASLPMNLEEIAVTQNLXVKTGHSSCGSKNARTSKRSKPSMSAEDIVSIL 402
Query: 1031 ---------ASESDQLSDSEKHSEVESSPEAEITPEAE 1059
D L D + S+PEA P+A+
Sbjct: 401 KVKVETSNLTEVKDSLDDMDDCHASASTPEAFEIPDAQ 288
>TC92694
Length = 777
Score = 35.0 bits (79), Expect = 0.25
Identities = 22/73 (30%), Positives = 38/73 (51%), Gaps = 3/73 (4%)
Frame = +1
Query: 1020 ESNAGTIDSEDASESDQLSDS--EKHSEVESSPEAEITPEAESNSEAEPSPEIQNE-NAS 1076
+SNA I+S + E D +D+ E EVE P + E + E +PS ++ E +A
Sbjct: 157 QSNAMEIESSNTMEVDPSTDAKMEPLHEVEFQPSPNVKTETSAEVEIQPSLNLKTEPSAE 336
Query: 1077 EDIQDNSQQAIQP 1089
E++Q + ++P
Sbjct: 337 EEVQLSPNVKMEP 375
>AW685572 similar to GP|22830607|dbj PHD-finger family homeodomain protein
{Oryza sativa (japonica cultivar-group)}, partial (3%)
Length = 508
Score = 35.0 bits (79), Expect = 0.25
Identities = 23/83 (27%), Positives = 41/83 (48%)
Frame = +3
Query: 1007 DDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNSEAEP 1066
D PG+K E+S S+ T DSED + + + + S +V S+P ++ S+ +
Sbjct: 153 DAPNPGAKDIEESSSSDFTSDSEDLAATIKDNMSTGQDDVTSAPLDDVKNFKGSSKQNRK 332
Query: 1067 SPEIQNENASEDIQDNSQQAIQP 1089
P I +E +S D ++ + P
Sbjct: 333 KPSITDELSSLAEPDLGEEDLTP 401
>TC80732 similar to GP|21553690|gb|AAM62783.1 unknown {Arabidopsis thaliana},
partial (16%)
Length = 760
Score = 34.7 bits (78), Expect = 0.32
Identities = 18/52 (34%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Frame = +2
Query: 1036 QLSDSEKHSEVESSPEAEI---TPEAESNSEAEPSPEIQNENASEDIQDNSQ 1084
++S+ E +E + SPE + PE ESNS EP P+ + E + Q+ Q
Sbjct: 119 KMSEPESQNESQPSPEPQSQNGAPEPESNSNHEPDPQPETEPEQPESQEQVQ 274
Score = 30.8 bits (68), Expect = 4.7
Identities = 19/85 (22%), Positives = 37/85 (43%)
Frame = +2
Query: 1003 HVKFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNS 1062
H K + E +++ E + E S S+ D + +E E PE++ + E S
Sbjct: 113 HFKMSEPESQNESQPSPEPQSQNGAPEPESNSNHEPDPQPETEPEQ-PESQEQVQLEPES 289
Query: 1063 EAEPSPEIQNENASEDIQDNSQQAI 1087
+ P PE+ ++ ++ NS +
Sbjct: 290 GSRPDPEVNSDADLKETAINSNDVV 364
>TC77394 similar to GP|4874305|gb|AAD31367.1| expressed protein {Arabidopsis
thaliana}, partial (61%)
Length = 1396
Score = 33.9 bits (76), Expect = 0.55
Identities = 26/96 (27%), Positives = 40/96 (41%)
Frame = +3
Query: 1005 KFDDREPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNSEA 1064
+F D +P PE E E SE D L + ++ EVE + E PE
Sbjct: 594 QFSDPQPSEDKPEHFE--------ESESEEDILDEGDE--EVEEEQDHESEPE----YTV 731
Query: 1065 EPSPEIQNENASEDIQDNSQQAIQPKFKHKSSHLRS 1100
+P PE++ + +I PK K + +L+S
Sbjct: 732 KPEPEVKKNAEVPACTKGGRTSISPKRKGRRRNLKS 839
>AW774261 weakly similar to EGAD|146423|156 vitellogenin {Anolis pulchellus},
partial (40%)
Length = 495
Score = 33.1 bits (74), Expect = 0.94
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Frame = +1
Query: 1016 PEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEA-EITPEAESNSEAEPSPEIQNEN 1074
PE++ + ID E+ ++ ++ + ++ EVE E EI E E N E E + E
Sbjct: 259 PEKTVESDEKIDLEEENDPEEEMEEIEYEEVEEEEEVEEIEEEVEENEEDAEEEEEE*EK 438
Query: 1075 ASEDIQDN 1082
E+++++
Sbjct: 439 EEEEVEED 462
>TC81082 similar to GP|17385678|dbj|BAB78631. putative co-repressor protein
{Oryza sativa (japonica cultivar-group)}, partial (20%)
Length = 1461
Score = 33.1 bits (74), Expect = 0.94
Identities = 25/113 (22%), Positives = 50/113 (44%), Gaps = 5/113 (4%)
Frame = +3
Query: 986 KAYRVYNSETHCVEESMHVKFDDREPGSKTPEQS----ESNAGTIDSEDASESD-QLSDS 1040
+ Y+ E C E D + G ++P++S E+ +G +D + +D +
Sbjct: 270 RQYQNRRGEQVCGEARGENYADADDEGEESPQRSSEGSENASGNVDVSGSESADGEECSQ 449
Query: 1041 EKHSEVESSPEAEITPEAESNSEAEPSPEIQNENASEDIQDNSQQAIQPKFKH 1093
E+H + E +AE EAE ++A +++ + S + ++P KH
Sbjct: 450 EEHDDGEHDDKAESEGEAEGMADAH---DVEGDGTSLPFSERFLLNVKPLAKH 599
>TC82778 similar to PIR|T04011|T04011 hypothetical protein T5L19.200 -
Arabidopsis thaliana, partial (7%)
Length = 784
Score = 32.7 bits (73), Expect = 1.2
Identities = 20/91 (21%), Positives = 44/91 (47%), Gaps = 6/91 (6%)
Frame = +3
Query: 1005 KFDD--REPGSKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVESSPEAEITPEAESNS 1062
KF+D +P + +E++A + +D ++ +L D ++ ++ E + +
Sbjct: 120 KFEDLHSQPTESNTDGAETDAAAVAQDDVNKRPRLEDDNQNDLANTNGHQE-----KKVA 284
Query: 1063 EAEPSPEIQNENASEDIQ----DNSQQAIQP 1089
EAE + + SE++Q DNS++ +P
Sbjct: 285 EAETDTDTEENAPSEEVQDVSKDNSEETAEP 377
Score = 32.3 bits (72), Expect = 1.6
Identities = 15/70 (21%), Positives = 36/70 (51%), Gaps = 1/70 (1%)
Frame = +3
Query: 1013 SKTPEQSESNAGTIDSEDASESDQLSDSEKHSEVE-SSPEAEITPEAESNSEAEPSPEIQ 1071
+K P + N + + + + +++++E ++ E ++P E+ ++ NSE P
Sbjct: 207 NKRPRLEDDNQNDLANTNGHQEKKVAEAETDTDTEENAPSEEVQDVSKDNSEETAEPTDT 386
Query: 1072 NENASEDIQD 1081
NE ED+++
Sbjct: 387 NEILVEDVKE 416
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.359 0.159 0.563
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,914,893
Number of Sequences: 36976
Number of extensions: 708421
Number of successful extensions: 7736
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 2496
Number of HSP's successfully gapped in prelim test: 392
Number of HSP's that attempted gapping in prelim test: 5053
Number of HSP's gapped (non-prelim): 3238
length of query: 1684
length of database: 9,014,727
effective HSP length: 109
effective length of query: 1575
effective length of database: 4,984,343
effective search space: 7850340225
effective search space used: 7850340225
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 65 (29.6 bits)
Medicago: description of AC149212.6


